YY2 Promotes Osteoblast Differentiation by Upregulating Osterix Transcriptional Activity
Abstract
1. Introduction
2. Results
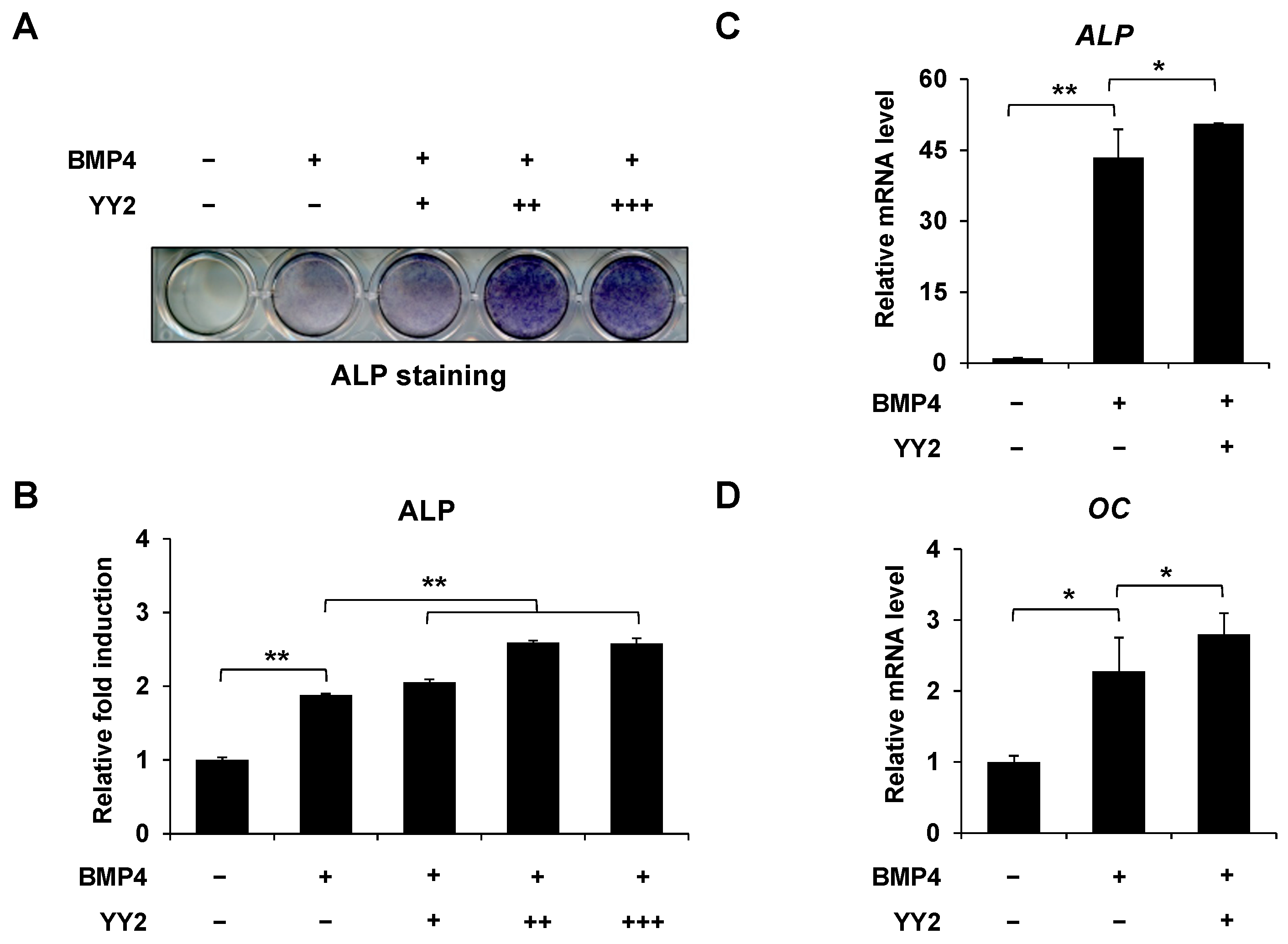
2.1. YY2 Promoted BMP4-Induced Osteoblast Differentiation
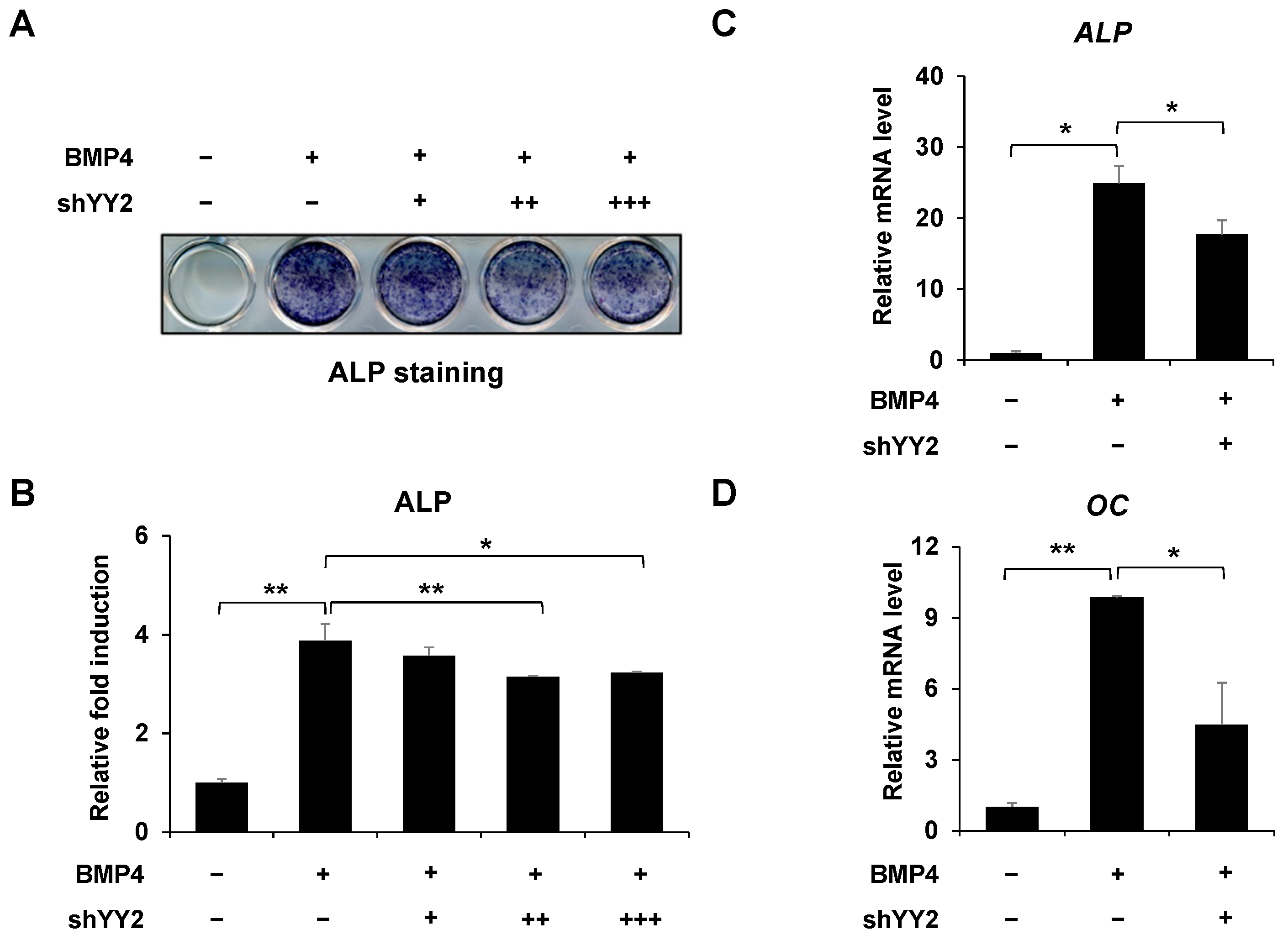
2.2. Knockdown of YY2 Attenuated Osteoblast Differentiation
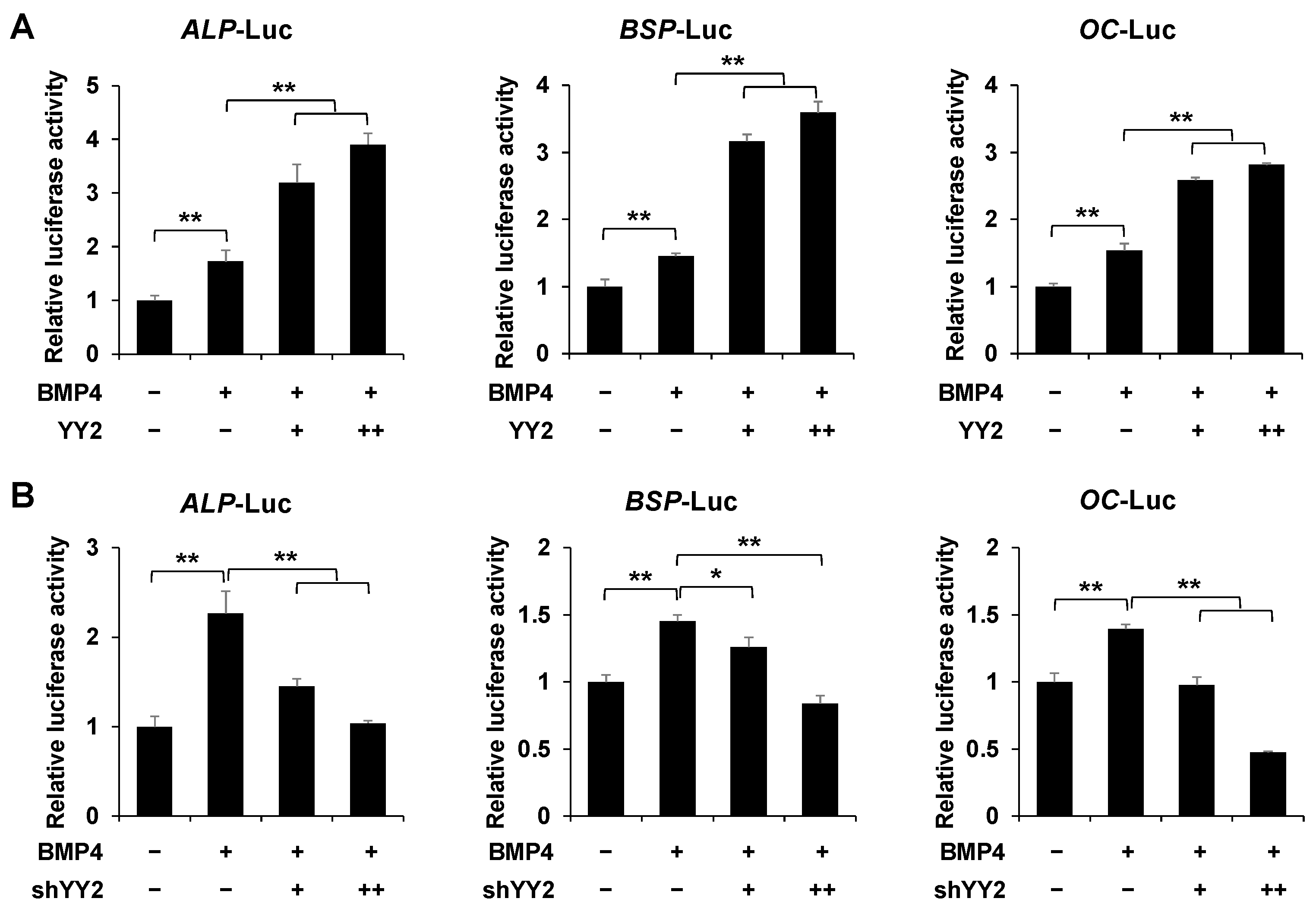
2.3. YY2 Postively Regulates the Promoter Activity of Osteoblast-Specific Marker Genes
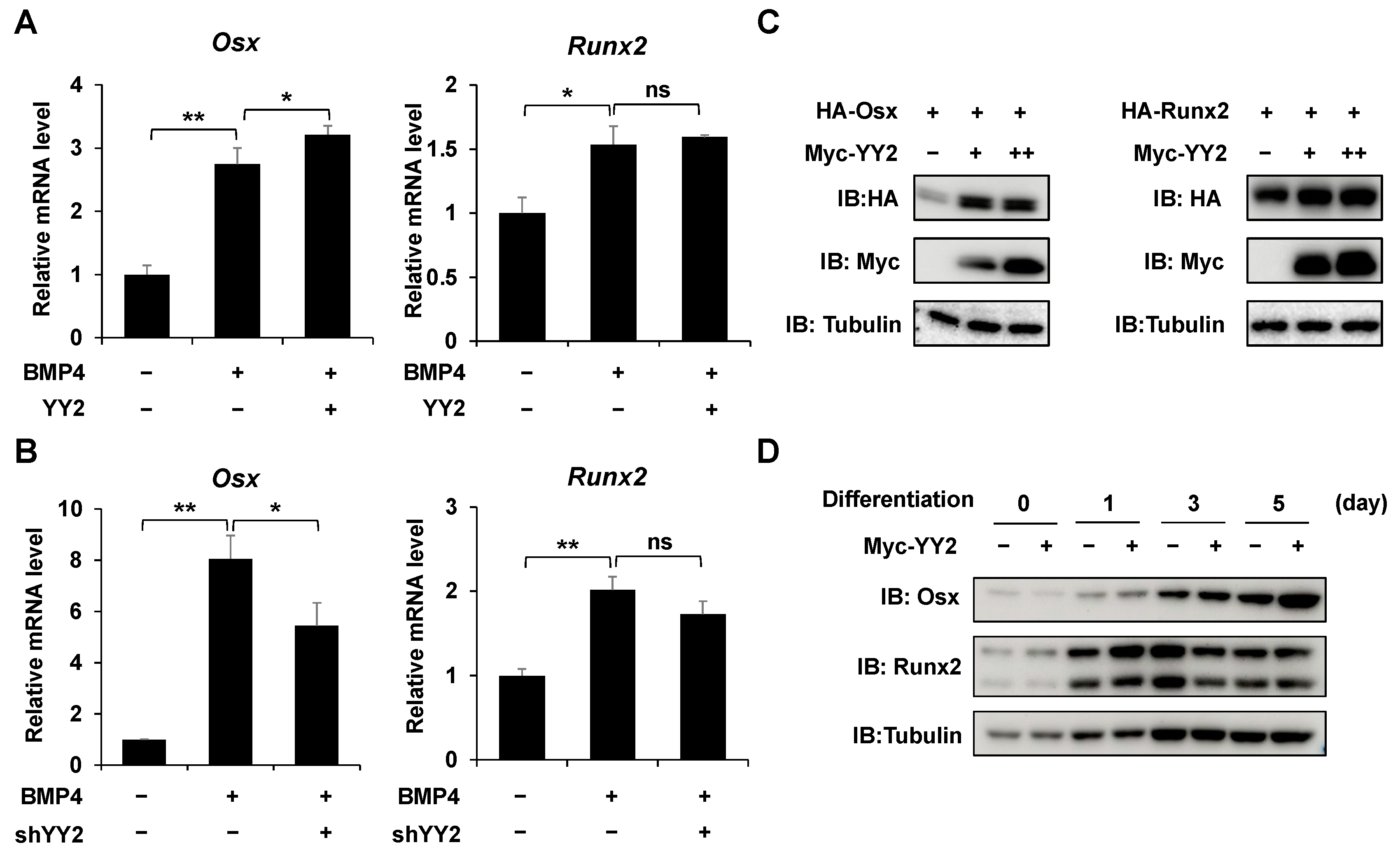
2.4. YY2 Upregulats the Expression of Osx
2.5. YY2 Activates the Osx Promoter Activity
2.6. YY2 Regulates Osx Promoter by Binding YY1 Consensus Motif
3. Discussion
4. Experimental Procedures
4.1. Cell Cultures and Transfection
4.2. Alkaline Phosphatase (ALP) Staining
4.3. Luciferase Assay
4.4. RNA Isolation and RT-qPCR
4.5. Immunoblotting
4.6. Construction of Human Osx Promoters with Deletions and Point Mutations
4.7. Chromatin Immunoprecipitation (ChIP)
4.8. Knockdown Experiment
4.9. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Takayanagi, H. Osteoimmunology: Shared mechanisms and crosstalk between the immune and bone systems. Nat. Rev. Immunol. 2007, 7, 292–304. [Google Scholar] [CrossRef] [PubMed]
- Manolagas, S.C.; Jilka, R.L.; Epstein, F.H. Bone Marrow, Cytokines, and Bone Remodeling—Emerging Insights into the Pathophysiology of Osteoporosis. N. Engl. J. Med. 1995, 332, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Bianco, P.; Robey, P.G. Stem cells in tissue engineering. Nature 2001, 414, 118–121. [Google Scholar] [CrossRef] [PubMed]
- Heino, T.; Hentunen, T.A. Differentiation of Osteoblasts and Osteocytes from Mesenchymal Stem Cells. Curr. Stem Cell Res. Ther. 2008, 3, 131–145. [Google Scholar] [CrossRef] [PubMed]
- Komori, T. Regulation of osteoblast differentiation by transcription factors. J. Cell. Biochem. 2006, 99, 1233–1239. [Google Scholar] [CrossRef]
- Lin, G.L.; Hankenson, K.D. Integration of BMP, Wnt, and notch signaling pathways in osteoblast differentiation. J. Cell. Biochem. 2011, 112, 3491–3501. [Google Scholar] [CrossRef]
- Fu, H.; Doll, B.; McNelis, T.; Hollinger, J.O. Osteoblast differentiation in vitro and in vivo promoted by Osterix. J. Biomed. Mater. Res. Part A Off. J. Soc. Biomater. Jpn. Soc. Biomater. Aust. Soc. Biomater. Korean Soc. Biomater. 2007, 83, 770–778. [Google Scholar]
- Nakashima, K.; Zhou, X.; Kunkel, G.; Zhang, Z.; Deng, J.M.; Behringer, R.R.; de Crombrugghe, B. The Novel Zinc Finger-Containing Transcription Factor Osterix Is Required for Osteoblast Differentiation and Bone Formation. Cell 2002, 108, 17–29. [Google Scholar] [CrossRef]
- Tu, Q.; Valverde, P.; Chen, J. Osterix enhances proliferation and osteogenic potential of bone marrow stromal cells. Biochem. Biophys. Res. Commun. 2006, 341, 1257–1265. [Google Scholar] [CrossRef]
- Zhang, C. Transcriptional regulation of bone formation by the osteoblast-specific transcription factor Osx. J. Orthop. Surg. Res. 2010, 5, 37. [Google Scholar] [CrossRef]
- Nakashima, K.; de Crombrugghe, B. Transcriptional mechanisms in osteoblast differentiation and bone formation. Trends Genet. 2003, 19, 458–466. [Google Scholar] [CrossRef]
- Nguyen, N.; Zhang, X.; Olashaw, N.; Seto, E. Molecular Cloning and Functional Characterization of the Transcription Factor YY2. J. Biol. Chem. 2004, 279, 25927–25934. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, Y.; Meliala, I.T.S.; Kasim, V.; Wu, S. Biological roles of Yin Yang 2: Its implications in physiological and pathological events. J. Cell. Mol. Med. 2020, 24, 12886–12899. [Google Scholar] [CrossRef] [PubMed]
- Kasim, V.; Xie, Y.D.; Wang, H.M.; Huang, C.; Yan, X.S.; Nian, W.Q.; Zheng, X.-D.; Miyagishi, M.; Wu, S.R. Transcription factor Yin Yang 2 is a novel regulator of the p53/p21 axis. Oncotarget 2017, 8, 54694–54707. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Cho, Y.-C.; Jeong, H.M.; Kim, K.H.; Choi, H.J.; Lee, K.Y.; Kang, B.Y. Yin-Yang 1 and Yin-Yang 2 exert opposing effects on the promoter activity of interleukin 4. Arch. Pharmacal Res. 2016, 39, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.M.; Choi, Y.H.; Lee, S.H.; Lee, K.Y. YY1 represses the transcriptional activity of Runx2 in C2C12 cells. Mol. Cell. Endocrinol. 2014, 383, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Ehara, A.; Ogata, K.; Imazato, S.; Ebisu, S.; Nakano, T.; Umakoshi, Y. Effects of alpha-TCP and TetCP on MC3T3-E1 proliferation, differentiation and mineralization. Biomaterials 2003, 24, 831–836. [Google Scholar] [CrossRef]
- Li, G.; Peng, H.; Corsi, K.; Usas, A.; Olshanski, A.; Huard, J. Differential Effect of BMP4 on NIH/3T3 and C2C12 Cells: Implications for Endochondral Bone Formation. J. Bone Miner. Res. 2005, 9, 1611–1623. [Google Scholar] [CrossRef]
- Pérez-Palacios, R.; Climent, M.; Santiago-Arcos, J.; Macías-Redondo, S.; Klar, M.; Muniesa, P.; Schoorlemmer, J. YY2 in Mouse Preimplantation Embryos and in Embryonic Stem Cells. Cells 2021, 10, 1123. [Google Scholar] [CrossRef]
- Gao, Y.; Jheon, A.; Nourkeyhani, H.; Kobayashi, H.; Ganss, B. Molecular cloning, structure, expression, and chromosomal localization of the human Osterix (SP7) gene. Gene 2004, 341, 101–110. [Google Scholar] [CrossRef]
- Kim, J.; Faulk, C. Retroposition and evolution of the DNA-binding motifs of YY1, YY2 and REX1. Nucleic Acids Res. 2007, 35, 3442–3452. [Google Scholar] [CrossRef] [PubMed]
- Karsenty, G.; Kronenberg, H.M.; Settembre, C. Genetic Control of Bone Formation. Annu. Rev. Cell Dev. Biol. 2009, 25, 629–648. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, T.; Kida, K.; Yamaguchi, A.; Hata, K.; Ichida, F.; Meguro, H.; Aburatani, H.; Nishimura, R.; Yoneda, T. BMP2 Regulates Osterix through Msx2 and Runx2 during Osteoblast Differentiation. J. Biol. Chem. 2008, 283, 29119–29125. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.H.; Kwon, T.G.; Park, H.S.; Wozney, J.M.; Ryoo, H.M. BMP-2-induced Osterix expression is mediated by Dlx5 but is independent of Runx2. Biochem. Biophys. Res. Commun. 2003, 309, 689–694. [Google Scholar] [CrossRef] [PubMed]
- Celil, A.B.; Campbell, P.G. BMP-2 and insulin-like growth factor-I mediate Osterix (Osx) expression in human mesen-chymal stem cells via the MAPK and protein kinase D signaling pathways. J. Biol. Chem. 2005, 280, 31353–31359. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Gilbert, L.; He, X.; Rubin, J.; Nanes, M.S. Transcriptional Regulation of the Osterix (Osx, Sp7) Promoter by Tumor Necrosis Factor Identifies Disparate Effects of Mitogen-activated Protein Kinase and NFκB Pathways. J. Biol. Chem. 2006, 281, 6297–6306. [Google Scholar] [CrossRef] [PubMed]
- Barbuto, R.; Mitchell, J. Regulation of the osterix (Osx, Sp7) promoter by osterix and its inhibition by parathyroid hormone. J. Mol. Endocrinol. 2013, 51, 99–108. [Google Scholar] [CrossRef]
- Sinha, K.M.; Zhou, X. Genetic and molecular control of osterix in skeletal formation. J. Cell. Biochem. 2013, 114, 975–984. [Google Scholar] [CrossRef]
- Figiel, M.; Łakomska, J.; Miłek, P.; Dziedzicka-Wasylewska, M.; Górecki, A. The transcription factor YY 2 has less momentous properties of an intrinsically disordered protein than its paralog YY 1. FEBS Lett. 2019, 593, 1787–1798. [Google Scholar] [CrossRef]
- Luo, C.; Lu, X.; Stubbs, L.; Kim, J. Rapid evolution of a recently retroposed transcription factor YY2 in mammalian genomes. Genomics 2006, 87, 348–355. [Google Scholar] [CrossRef][Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piao, M.; Lee, S.H.; Kim, M.J.; Kim, H.S.; Lee, K.Y. YY2 Promotes Osteoblast Differentiation by Upregulating Osterix Transcriptional Activity. Int. J. Mol. Sci. 2022, 23, 4303. https://doi.org/10.3390/ijms23084303
Piao M, Lee SH, Kim MJ, Kim HS, Lee KY. YY2 Promotes Osteoblast Differentiation by Upregulating Osterix Transcriptional Activity. International Journal of Molecular Sciences. 2022; 23(8):4303. https://doi.org/10.3390/ijms23084303
Chicago/Turabian StylePiao, Meiyu, Sung Ho Lee, Myeong Ji Kim, Hyung Sik Kim, and Kwang Youl Lee. 2022. "YY2 Promotes Osteoblast Differentiation by Upregulating Osterix Transcriptional Activity" International Journal of Molecular Sciences 23, no. 8: 4303. https://doi.org/10.3390/ijms23084303
APA StylePiao, M., Lee, S. H., Kim, M. J., Kim, H. S., & Lee, K. Y. (2022). YY2 Promotes Osteoblast Differentiation by Upregulating Osterix Transcriptional Activity. International Journal of Molecular Sciences, 23(8), 4303. https://doi.org/10.3390/ijms23084303






