Plastid Transformation of Micro-Tom Tomato with a Hemipteran Double-Stranded RNA Results in RNA Interference in Multiple Insect Species
Abstract
1. Introduction
2. Results
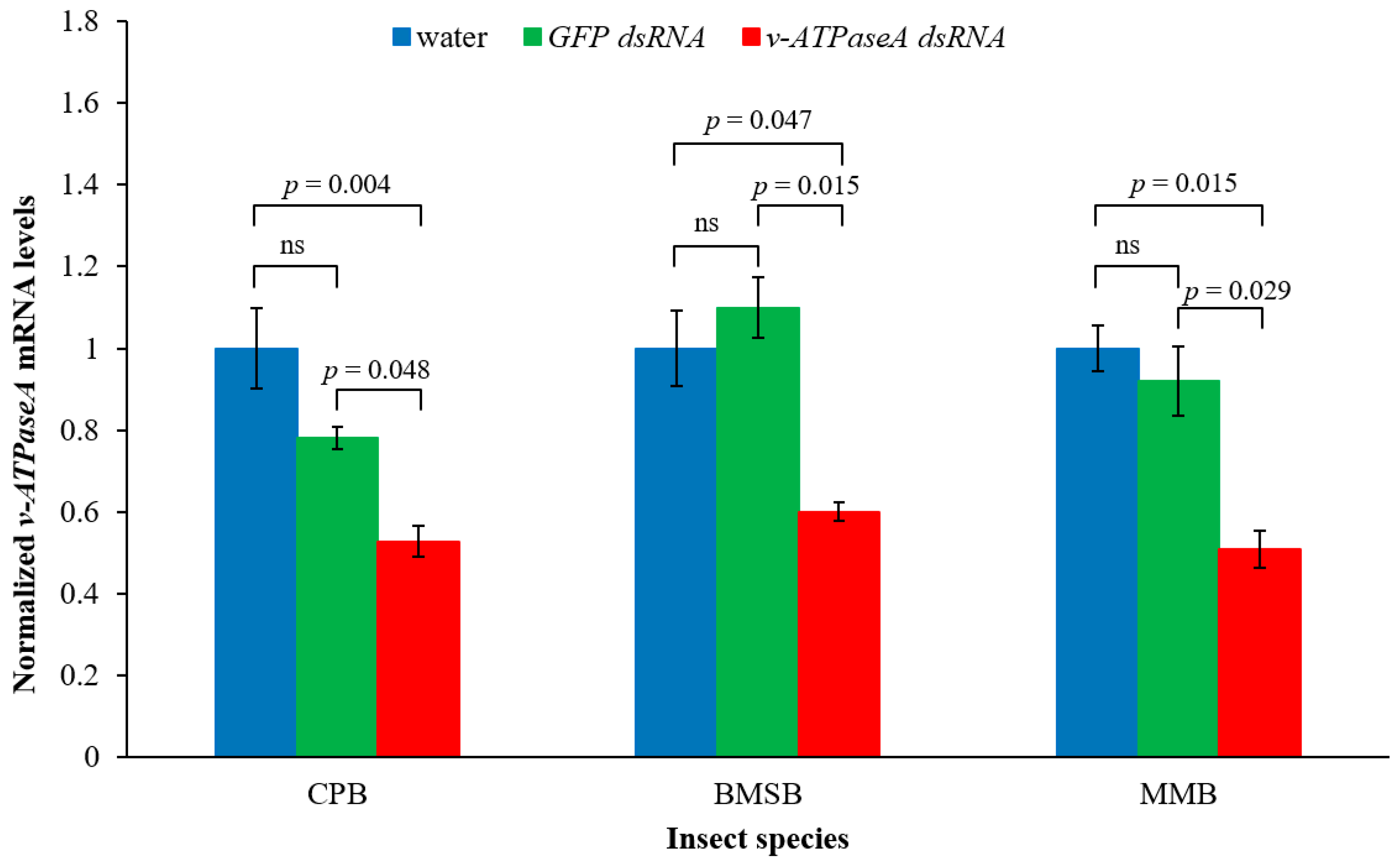
2.1. Cotton Mealybug-Specific dsRNA Down-Regulates v-ATPaseA Gene in Three Insect Species
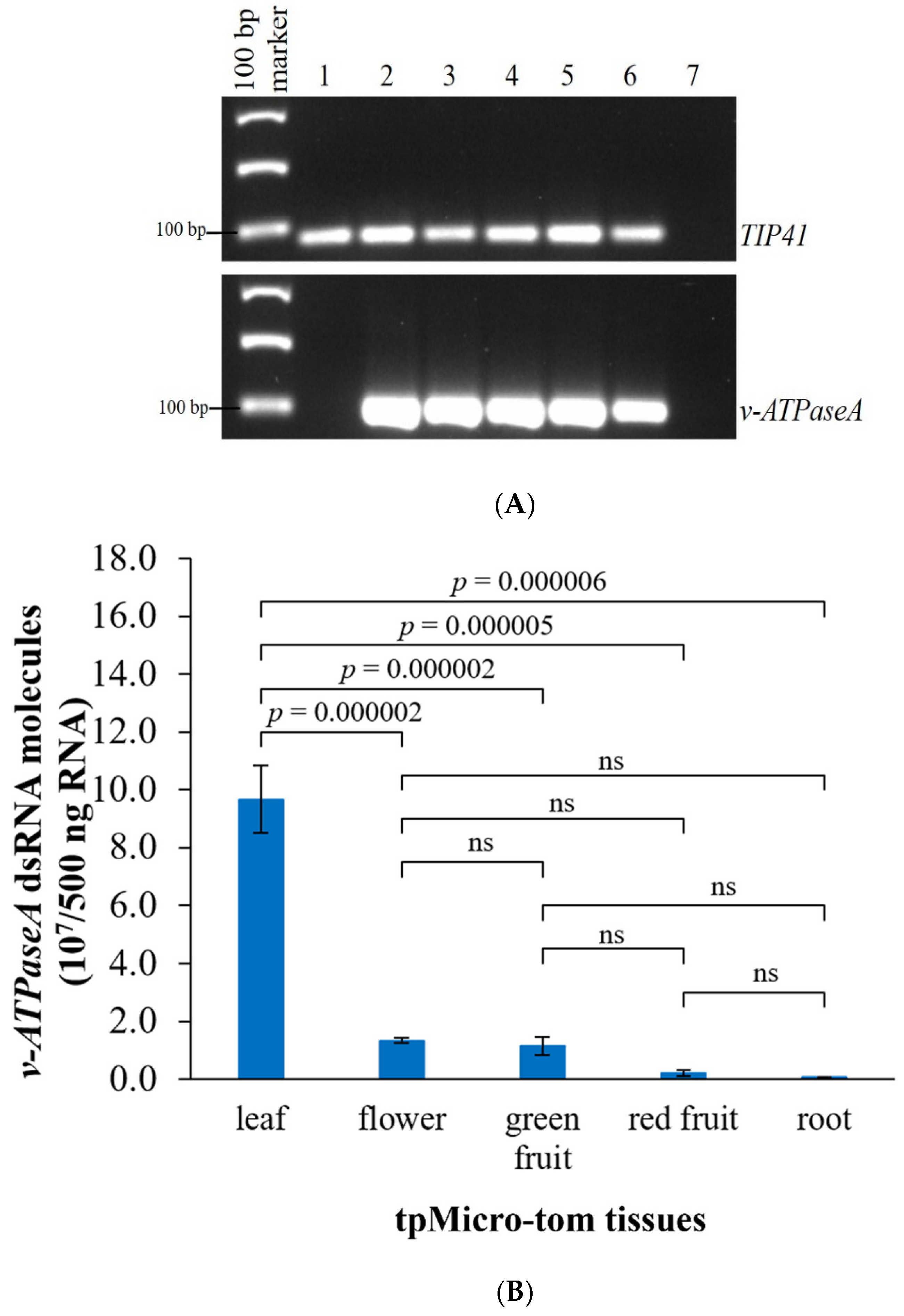
2.2. Development of tpMicro-Tom to Express dsRNA
2.3. dsRNA Concentrations Are Higher in the Leaves Compared with Other Tissues
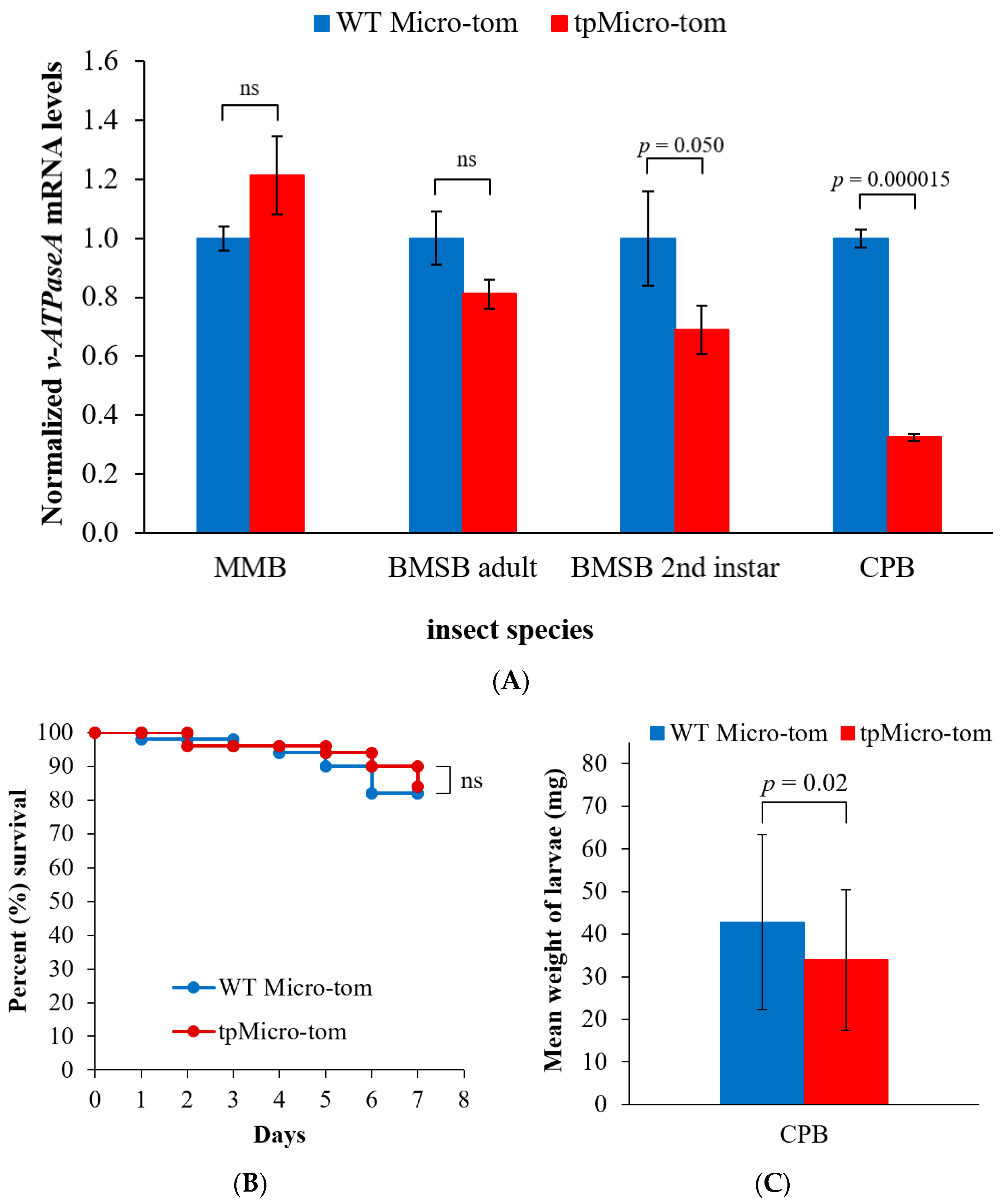
2.4. Insect Feeding Strategy Affects v-ATPaseA mRNA Levels Resulting from Exposure to Plastid-Produced dsRNA
3. Discussion
4. Materials and Methods
4.1. Bioassays Using In Vitro-Synthesized dsRNA
4.2. Micro-Tom Plant Growth
4.3. Micro-Tom Plastid Transformation
4.4. DNA and RNA Extraction
4.5. Southern Blot to Confirm Homoplastomy of tpMicro-Tom
4.6. cDNA Synthesis and End-Point PCR to Confirm dsRNA Production in Micro-Tom
4.7. Droplet Digital PCR (ddPCR) to Quantify dsRNA in Micro-Tom Tissues
4.8. Insect Bioassays Using WT and Transplastomic Micro-Tom
4.9. cDNA Synthesis from Insect RNA
4.10. Quantitative PCR to Measure Gene Down-Regulation in Insects
4.11. Comparing Sequence Identities of v-ATPaseA Genes in Insects
4.12. Statistical Analysis of the Data
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bronkhorst, A.W.; van Rij, R.P. The long and short of antiviral defense: Small RNA-based immunity in insects. Curr. Opin. Virol. 2014, 7, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Rosa, C.; Kuo, Y.W.; Wuriyanghan, H.; Falk, B.W. RNA Interference Mechanisms and Applications in Plant Pathology. Annu. Rev. Phytopathol. 2018, 56, 581–610. [Google Scholar] [CrossRef] [PubMed]
- Mamta, B.; Rajam, M.V. RNAi technology: A new platform for crop pest control. Physiol. Mol. Biol. Plants 2017, 23, 487–501. [Google Scholar] [CrossRef] [PubMed]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Kennerdell, J.R.; Carthew, R.W. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled2 act in the wingless pathway. Cell 1998, 95, 1017–1026. [Google Scholar] [CrossRef]
- Mao, Y.B.; Cai, W.J.; Wang, J.W.; Hong, G.J.; Tao, X.Y.; Wang, L.J.; Huang, Y.P.; Chen, X.Y. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 2007, 25, 1307–1313. [Google Scholar] [CrossRef] [PubMed]
- Timmons, L.; Court, D.L.; Fire, A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 2001, 263, 103–112. [Google Scholar] [CrossRef]
- Huvenne, H.; Smagghe, G. Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: A review. J. Insect Physiol. 2010, 56, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Saleh, M.-C.; van Rij, R.P.; Hekele, A.; Gillis, A.; Foley, E.; O’Farrell, P.H.; Andino, R. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat. Cell Biol. 2006, 8, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Winston, W.M.; Sutherlin, M.; Wright, A.J.; Feinberg, E.H.; Hunter, C.P. Caenorhabditis elegans SID-2 is required for environmental RNA interference. Proc. Natl. Acad. Sci. USA 2007, 104, 10565–10570. [Google Scholar] [CrossRef]
- Joga, M.R.; Zotti, M.J.; Smagghe, G.; Christiaens, O. RNAi efficiency, systemic properties, and novel delivery methods for pest insect control: What we know so far. Front. Physiol. 2016, 7, 553. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Khan, S.A.; Heckel, D.G.; Bock, R. Next-generation insect-resistant plants: RNAi-mediated crop protection. Trends Biotechnol. 2017, 35, 871–882. [Google Scholar] [CrossRef] [PubMed]
- Baum, J.A.; Bogaert, T.; Clinton, W.; Heck, G.R.; Feldmann, P.; Ilagan, O.; Johnson, S.; Plaetinck, G.; Munyikwa, T.; Pleau, M.; et al. Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 2007, 25, 1322–1326. [Google Scholar] [CrossRef] [PubMed]
- Terenius, O.; Papanicolaou, A.; Garbutt, J.S.; Eleftherianos, I.; Huvenne, H.; Kanginakudru, S.; Albrechtsen, M.; An, C.; Aymeric, J.L.; Barthel, A.; et al. RNA interference in Lepidoptera: An overview of successful and unsuccessful studies and implications for experimental design. J. Insect Physiol. 2011, 57, 231–245. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, J.; Dao, V.A.; Majumdar, U.; Schmitt-Engel, C.; Schwirz, J.; Schultheis, D.; Ströhlein, N.; Troelenberg, N.; Grossmann, D.; Richter, T.; et al. Large scale RNAi screen in Tribolium reveals novel target genes for pest control and the proteasome as prime target. BMC Genom. 2015, 16, 674. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Khan, S.A.; Hasse, C.; Ruf, S.; Heckel, D.G.; Bock, R. Full crop protection from an insect pest by expression of long double-stranded RNAs in plastids. Science 2015, 347, 991–994. [Google Scholar] [CrossRef] [PubMed]
- Jackson, A.L.; Bartz, S.R.; Schelter, J.; Kobayashi, S.V.; Burchard, J.; Mao, M.; Li, B.; Cavet, G.; Linsley, P.S. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003, 21, 635–637. [Google Scholar] [CrossRef] [PubMed]
- Burke, W.G.; Kaplanoglu, E.; Kolotilin, I.; Menassa, R.; Donly, C. RNA Interference in the tobacco hornworm, Manduca sexta, using plastid-encoded long double-stranded RNA. Front. Plant Sci. 2019, 10, 313. [Google Scholar] [CrossRef]
- Li, H.; Khajuria, C.; Rangasamy, M.; Gandra, P.; Fitter, M.; Geng, C.; Woosely, A.; Hasler, J.; Schulenberg, G.; Worden, S.; et al. Long dsRNA but not siRNA initiates RNAi in western corn rootworm larvae and adults. J. Appl. Entomol. 2015, 139, 432–445. [Google Scholar] [CrossRef]
- He, W.; Xu, W.; Xu, L.; Fu, K.; Guo, W.; Bock, R.; Zhang, J. Length-dependent accumulation of double-stranded RNAs in plastids affects RNA interference efficiency in the Colorado potato beetle. J. Exp. Bot. 2020, 71, 2670–2677. [Google Scholar] [CrossRef] [PubMed]
- Garbutt, J.S.; Belles, X.; Richards, E.H.; Reynolds, S.E. Persistence of double-stranded RNA in insect hemolymph as a potential determiner of RNA interference success: Evidence from Manduca sexta and Blattella germanica. J. Insect Physiol. 2013, 59, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Shukla, J.N.; Kalsi, M.; Sethi, A.; Narva, K.E.; Fishilevich, E.; Singh, S.; Mogilicherla, K.; Palli, S.R. Reduced stability and intracellular transport of dsRNA contribute to poor RNAi response in lepidopteran insects. RNA Biol. 2016, 13, 656–669. [Google Scholar] [CrossRef] [PubMed]
- Singh, I.K.; Singh, S.; Mogilicherla, K.; Shukla, J.N.; Palli, S.R. Comparative analysis of double-stranded RNA degradation and processing in insects. Sci. Rep. 2017, 7, 17059. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.-S.; Gurusamy, D.; Palli, S.R. Accumulation of dsRNA in endosomes contributes to inefficient RNA interference in the fall armyworm, Spodoptera frugiperda. Insect Biochem. Mol. Biol. 2017, 90, 53–60. [Google Scholar] [CrossRef]
- Palli, S.R. RNA interference in Colorado potato beetle: Steps toward development of dsRNA as a commercial insecticide. Curr. Opin. Insect Sci. 2014, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Tomoyasu, Y.; Miller, S.C.; Tomita, S.; Schoppmeier, M.; Grossmann, D.; Bucher, G. Exploring systemic RNA interference in insects: A genome-wide survey for RNAi genes in Tribolium. Genome Biol. 2008, 9, R10. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Xu, J.; Palli, R.; Ferguson, J.; Palli, S.R. Ingested RNA interference for managing the populations of the Colorado potato beetle, Leptinotarsa decemlineata. Pest Manag. Sci. 2011, 67, 175–182. [Google Scholar] [CrossRef]
- Yoon, J.S.; Mogilicherla, K.; Gurusamy, D.; Chen, X.; Chereddy, S.; Palli, S.R. Double-stranded RNA binding protein, Staufen, is required for the initiation of RNAi in coleopteran insects. Proc. Natl. Acad. Sci. USA 2018, 115, 8334–8339. [Google Scholar] [CrossRef] [PubMed]
- Baum, J.A.; Roberts, J.K. Progress towards RNAi-Mediated Insect Pest Management. In Advances in Insect Physiology; Dhadialla, T.S., Gill, S.S., Eds.; Elsevier BV: Amsterdam, The Netherlands, 2014; Volume 47, pp. 249–295. [Google Scholar]
- Bolognesi, R.; Ramaseshadri, P.; Anderson, J.; Bachman, P.; Clinton, W.; Flannagan, R.; Ilagan, O.; Lawrence, C.; Levine, S.; Moar, W.; et al. Characterizing the mechanism of action of double-stranded RNA activity against western corn rootworm (Diabrotica virgifera virgifera LeConte). PLoS ONE 2012, 7, e47534. [Google Scholar] [CrossRef] [PubMed]
- Bock, R. Engineering plastid genomes: Methods, tools, and applications in basic research and biotechnology. Annu. Rev. Plant Biol. 2015, 66, 211–241. [Google Scholar] [CrossRef]
- Bally, J.; McIntyre, G.J.; Doran, R.L.; Lee, K.; Perez, A.; Jung, H.; Naim, F.; Larrinua, I.M.; Narva, K.E.; Waterhouse, P.M. In-Plant protection against Helicoverpa armigera by production of long hpRNA in chloroplasts. Front. Plant Sci. 2016, 7, 1453. [Google Scholar] [CrossRef]
- Douglas, A.E. Phloem-sap feeding by animals: Problems and solutions. J. Exp. Bot. 2006, 57, 747–754. [Google Scholar] [CrossRef]
- Stafford, C.A.; Walker, G.P.; Ullman, D.E. Hitching a ride: Vector feeding and virus transmission. Commun. Integr. Biol. 2012, 5, 43–49. [Google Scholar] [CrossRef]
- Dong, Y.; Yang, Y.; Wang, Z.; Wu, M.; Fu, J.; Guo, J.; Chang, L.; Zhang, J. Inaccessibility to double-stranded RNAs in plastids restricts RNA interference in Bemisia tabaci (whitefly). Pest Manag. Sci. 2020, 76, 3168–3176. [Google Scholar] [CrossRef]
- Ruf, S.; Hermann, M.; Berger, I.J.; Carrer, H.; Bock, R. Stable genetic transformation of tomato plastids and expression of a foreign protein in fruit. Nat. Biotechnol. 2001, 19, 870–875. [Google Scholar] [CrossRef]
- Kaplanoglu, E.; Kolotilin, I.; Menassa, R.; Donly, C. Transplastomic Tomato Plants Expressing Insect-Specific Double-Stranded RNAs: A Protocol Based on Biolistic Transformation. Methods Mol. Biol. 2022, 2360, 235–252. [Google Scholar]
- Forgac, M. Vacuolar ATPases: Rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 2007, 8, 917–929. [Google Scholar] [CrossRef]
- Upadhyay, S.K.; Chandrashekar, K.; Thakur, N.; Verma, P.C.; Borgio, J.F.; Singh, P.K.; Tuli, R. RNA interference for the control of whiteflies (Bemisia tabaci) by oral route. J. Biosci. 2011, 36, 153–161. [Google Scholar] [CrossRef]
- Guo, W.; Guo, M.; Yang, C.; Liu, Z.; Chen, S.; Lu, J.; Qiu, B.; Zhang, Y.; Zhou, X.; Pan, H. RNA interference-mediated silencing of vATPase subunits A and E affect survival and development of the 28-spotted ladybeetle, Henosepilachna vigintioctopunctata. Insect Sci. 2021, 28, 1664–1676. [Google Scholar] [CrossRef]
- Lange, W.H.; Bronson, L. Insect pests of tomatoes. Annu. Rev. Entomol. 1981, 26, 345–371. [Google Scholar] [CrossRef]
- Exposito-Rodriguez, M.; Borges, A.A.; Borges-Perez, A.; Perez, J.A. Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol. 2008, 8, 131. [Google Scholar] [CrossRef]
- Guo, W.C.; Fu, K.Y.; Yang, S.; Li, X.X.; Li, G.Q. Instar-dependent systemic RNA interference response in Leptinotarsa decemlineata larvae. Pestic. Biochem. Physiol. 2015, 123, 64–73. [Google Scholar] [CrossRef]
- Jin, S.; Singh, N.D.; Li, L.; Zhang, X.; Daniell, H. Engineered chloroplast dsRNA silences cytochrome p450 monooxygenase, V-ATPase and chitin synthase genes in the insect gut and disrupts Helicoverpa zea larval development and pupation. Plant Biotechnol. J. 2015, 13, 435–446. [Google Scholar] [CrossRef]
- Bachman, P.M.; Bolognesi, R.; Moar, W.J.; Mueller, G.M.; Paradise, M.S.; Ramaseshadri, P.; Tan, J.; Uffman, J.P.; Warren, J.; Wiggins, B.E.; et al. Characterization of the spectrum of insecticidal activity of a double-stranded RNA with targeted activity against Western Corn Rootworm (Diabrotica virgifera virgifera LeConte). Transgenic Res. 2013, 22, 1207–1222. [Google Scholar] [CrossRef]
- Haller, S.; Widmer, F.; Siegfried, B.D.; Zhuo, X.; Romeis, J. Responses of two ladybird beetle species (Coleoptera: Coccinellidae) to dietary RNAi. Pest Manag. Sci. 2019, 75, 2652–2662. [Google Scholar] [CrossRef]
- Poreddy, S.; Li, J.; Baldwin, I.T. Plant-mediated RNAi silences midgut-expressed genes in congeneric lepidopteran insects in nature. BMC Plant Biol. 2017, 17, 199. [Google Scholar] [CrossRef]
- Powell, M.; Pyati, P.; Cao, M.; Bell, H.; Gatehouse, J.A.; Fitches, E. Insecticidal effects of dsRNA targeting the Diap1 gene in dipteran pests. Sci. Rep. 2017, 7, 15147. [Google Scholar] [CrossRef]
- Whyard, S.; Singh, A.D.; Wong, S. Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochem. Mol. Biol. 2009, 39, 824–832. [Google Scholar] [CrossRef]
- Pan, H.; Yang, X.; Bidne, K.; Hellmich, R.L.; Siegfried, B.D.; Zhou, X. Dietary Risk Assessment of v-ATPase A dsRNAs on Monarch Butterfly Larvae. Front. Plant Sci. 2017, 8, 242. [Google Scholar] [CrossRef]
- Daniell, H.; Ruiz, O.N.; Dhingra, A. Chloroplast genetic engineering to improve agronomic traits. In Transgenic Plants: Methods and Protocols; Peña, L., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 111–137. [Google Scholar]
- Soloway, S.B. Naturally occurring insecticides. Environ. Health Perspect. 1976, 14, 109–117. [Google Scholar] [CrossRef]
- Wurbs, D.; Ruf, S.; Bock, R. Contained metabolic engineering in tomatoes by expression of carotenoid biosynthesis genes from the plastid genome. Plant J. 2007, 49, 276–288. [Google Scholar] [CrossRef]
- Lu, Y.; Rijzaani, H.; Karcher, D.; Ruf, S.; Bock, R. Efficient metabolic pathway engineering in transgenic tobacco and tomato plastids with synthetic multigene operons. Proc. Natl. Acad. Sci. USA 2013, 110, E623–E632. [Google Scholar] [CrossRef]
- Guo, M.; Zhang, Y.L.; Meng, Z.J.; Jiang, J. Optimization of factors affecting Agrobacterium-mediated transformation of Micro-Tom tomatoes. Genet. Mol. Res. 2012, 11, 661–671. [Google Scholar] [CrossRef]
- Sun, H.J.; Uchii, S.; Watanabe, S.; Ezura, H. A highly efficient transformation protocol for Micro-Tom, a model cultivar for tomato functional genomics. Plant Cell Physiol. 2006, 47, 426–431. [Google Scholar] [CrossRef]
- Kahlau, S.; Bock, R. Plastid transcriptomics and translatomics of tomato fruit development and chloroplast-to-chromoplast differentiation: Chromoplast gene expression largely serves the production of a single protein. Plant Cell 2008, 20, 856–874. [Google Scholar] [CrossRef]
- Valkov, V.T.; Scotti, N.; Kahlau, S.; Maclean, D.; Grillo, S.; Gray, J.C.; Bock, R.; Cardi, T. Genome-wide analysis of plastid gene expression in potato leaf chloroplasts and tuber amyloplasts: Transcriptional and posttranscriptional control. Plant Physiol. 2009, 150, 2030–2044. [Google Scholar] [CrossRef]
- Caroca, R.; Howell, K.A.; Hasse, C.; Ruf, S.; Bock, R. Design of chimeric expression elements that confer high-level gene activity in chromoplasts. Plant J. 2013, 73, 368–379. [Google Scholar] [CrossRef]
- Sükösd, Z.; Knudsen, B.; Anderson, J.W.; Novák, Á.; Kjems, J.; Pedersen, C.N. Characterising RNA secondary structure space using information entropy. BMC Bioinform. 2013, 14, S22. [Google Scholar] [CrossRef]
- Feng, L.; Niu, D.K. Relationship between mRNA stability and length: An old question with a new twist. Biochem. Genet. 2007, 45, 131–137. [Google Scholar] [CrossRef]
- Bally, J.; Fishilevich, E.; Bowling, A.J.; Pence, H.E.; Narva, K.E.; Waterhouse, P.M. Improved insect-proofing: Expressing double-stranded RNA in chloroplasts. Pest Manag. Sci. 2018, 74, 1751–1758. [Google Scholar] [CrossRef]
- Thakur, N.; Upadhyay, S.K.; Verma, P.C.; Chandrashekar, K.; Tuli, R.; Singh, P.K. Enhanced whitefly resistance in transgenic tobacco plants expressing double stranded RNA of v-ATPase A gene. PLoS ONE 2014, 9, e87235. [Google Scholar] [CrossRef]
- Miles, P.W. The Saliva of Hemiptera. In Advances in Insect Physiology; Academic Press Inc.: London, UK, 1972; Volume 9, pp. 183–255. [Google Scholar]
- Mogilicherla, K.; Howell, J.L.; Palli, S.R. Improving RNAi in the Brown Marmorated Stink Bug: Identification of target genes and reference genes for RT-qPCR. Sci. Rep. 2018, 8, 3720. [Google Scholar] [CrossRef]
- Walker, G.P. Sieve element occlusion: Interactions with phloem sap-feeding insects. A review. J. Plant Physiol. 2022, 269, 153582. [Google Scholar] [CrossRef]
- Behnke, H.-D. Sieve-element characters. Nord. J. Bot. 1981, 1, 381–400. [Google Scholar] [CrossRef]
- Taylor, T.N.; Taylor, E.L.; Krings, M. Introduction to Vascular Plant Morphology and Anatomy. In Paleobotany, Biology and Evolution of Fossil Plants; Taylor, T.N., Taylor, E.L., Krings, M., Eds.; Academic Press Inc.: London, UK, 2009; Volume 2, pp. 201–222. [Google Scholar]
- Ren, B.; Cao, J.; He, Y.; Yang, S.; Zhang, J. Assessment on effects of transplastomic potato plants expressing Colorado potato beetle beta-Actin double-stranded RNAs for three non-target pests. Pestic. Biochem. Physiol. 2021, 178, 104909. [Google Scholar] [CrossRef]
- Sun, Y.; Sparks, C.; Jones, H.; Riley, M.; Francis, F.; Du, W.; Xia, L. Silencing an essential gene involved in infestation and digestion in grain aphid through plant-mediated RNA interference generates aphid-resistant wheat plants. Plant Biotechnol. J. 2019, 17, 852–854. [Google Scholar] [CrossRef]
- Ibrahim, A.B.; Monteiro, T.R.; Cabral, G.B.; Aragao, F.J.L. RNAi-mediated resistance to whitefly (Bemisia tabaci) in genetically engineered lettuce (Lactuca sativa). Transgenic Res. 2017, 26, 613–624. [Google Scholar] [CrossRef]
- Khan, A.M.; Ashfaq, M.; Kiss, Z.; Khan, A.A.; Mansoor, S.; Falk, B.W. Use of recombinant tobacco mosaic virus to achieve RNA interference in plants against the citrus mealybug, Planococcus citri (Hemiptera: Pseudococcidae). PLoS ONE 2013, 8, e73657. [Google Scholar] [CrossRef]
- Mermigka, G.; Verret, F.; Kalantidis, K. RNA silencing movement in plants. J. Integr. Plant Biol. 2016, 58, 328–342. [Google Scholar] [CrossRef]
- Pant, B.D.; Buhtz, A.; Kehr, J.; Scheible, W.R. MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 2008, 53, 731–738. [Google Scholar] [CrossRef]
- Melnyk, C.W.; Molnar, A.; Baulcombe, D.C. Intercellular and systemic movement of RNA silencing signals. EMBO J. 2011, 30, 3553–3563. [Google Scholar] [CrossRef]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Report. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Deepa, K.; Sheeja, T.E.; Santhi, R.; Sasikumar, B.; Cyriac, A.; Deepesh, P.V.; Prasath, D. A simple and efficient protocol for isolation of high quality functional RNA from different tissues of turmeric (Curcuma longa L.). Physiol. Mol. Biol. Plants 2014, 20, 263–271. [Google Scholar] [CrossRef]
- Southern, E. Southern blotting. Nat. Protoc. 2006, 1, 518–525. [Google Scholar] [CrossRef]
- Taylor, S.; Wakem, M.; Dijkman, G.; Alsarraj, M.; Nguyen, M. A practical approach to RT-qPCR-Publishing data that conform to the MIQE guidelines. Methods 2010, 50, S1–S5. [Google Scholar] [CrossRef]
- Taylor, S.C.; Carbonneau, J.; Shelton, D.N.; Boivin, G. Optimization of Droplet Digital PCR from RNA and DNA extracts with direct comparison to RT-qPCR: Clinical implications for quantification of Oseltamivir-resistant subpopulations. J. Virol. Methods 2015, 224, 58–66. [Google Scholar] [CrossRef]
- Bio-Rad. Droplet Digital PCR Applications Guide (Bulletin 6407, Version B); Bio-Rad Laboratories Inc.: Hercules, CA, USA, 2018; Available online: http://www.bio-rad.com/webroot/web/pdf/lsr/literature/Bulletin_6407.pdf (accessed on 2 February 2022).
- Kaplanoglu, E.; Chapman, P.; Scott, I.M.; Donly, C. Overexpression of a cytochrome P450 and a UDP-glycosyltransferase is associated with imidacloprid resistance in the Colorado potato beetle, Leptinotarsa decemlineata. Sci. Rep. 2017, 7, 1762. [Google Scholar] [CrossRef]
- Arya, S.K.; Jain, G.; Upadhyay, S.K.; Sarita; Singh, H.; Dixit, S.; Verma, P.C. Reference genes validation in Phenacoccus solenopsis under various biotic and abiotic stress conditions. Sci. Rep. 2017, 7, 13520. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]

| Insect Species | Order | Accession Number/Availability | % Identity |
|---|---|---|---|
| P. madeirensis | Hemiptera | MN364707.1 | 86 |
| H. halys | Hemiptera | XM_014417043 | 81 |
| L. decemlineata | Coleoptera | XM_023156517 | 76 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaplanoglu, E.; Kolotilin, I.; Menassa, R.; Donly, C. Plastid Transformation of Micro-Tom Tomato with a Hemipteran Double-Stranded RNA Results in RNA Interference in Multiple Insect Species. Int. J. Mol. Sci. 2022, 23, 3918. https://doi.org/10.3390/ijms23073918
Kaplanoglu E, Kolotilin I, Menassa R, Donly C. Plastid Transformation of Micro-Tom Tomato with a Hemipteran Double-Stranded RNA Results in RNA Interference in Multiple Insect Species. International Journal of Molecular Sciences. 2022; 23(7):3918. https://doi.org/10.3390/ijms23073918
Chicago/Turabian StyleKaplanoglu, Emine, Igor Kolotilin, Rima Menassa, and Cam Donly. 2022. "Plastid Transformation of Micro-Tom Tomato with a Hemipteran Double-Stranded RNA Results in RNA Interference in Multiple Insect Species" International Journal of Molecular Sciences 23, no. 7: 3918. https://doi.org/10.3390/ijms23073918
APA StyleKaplanoglu, E., Kolotilin, I., Menassa, R., & Donly, C. (2022). Plastid Transformation of Micro-Tom Tomato with a Hemipteran Double-Stranded RNA Results in RNA Interference in Multiple Insect Species. International Journal of Molecular Sciences, 23(7), 3918. https://doi.org/10.3390/ijms23073918





