Visualizing Cell Death in Live Retina: Using Calpain Activity Detection as a Biomarker for Retinal Degeneration
Abstract
:1. Introduction
2. Results
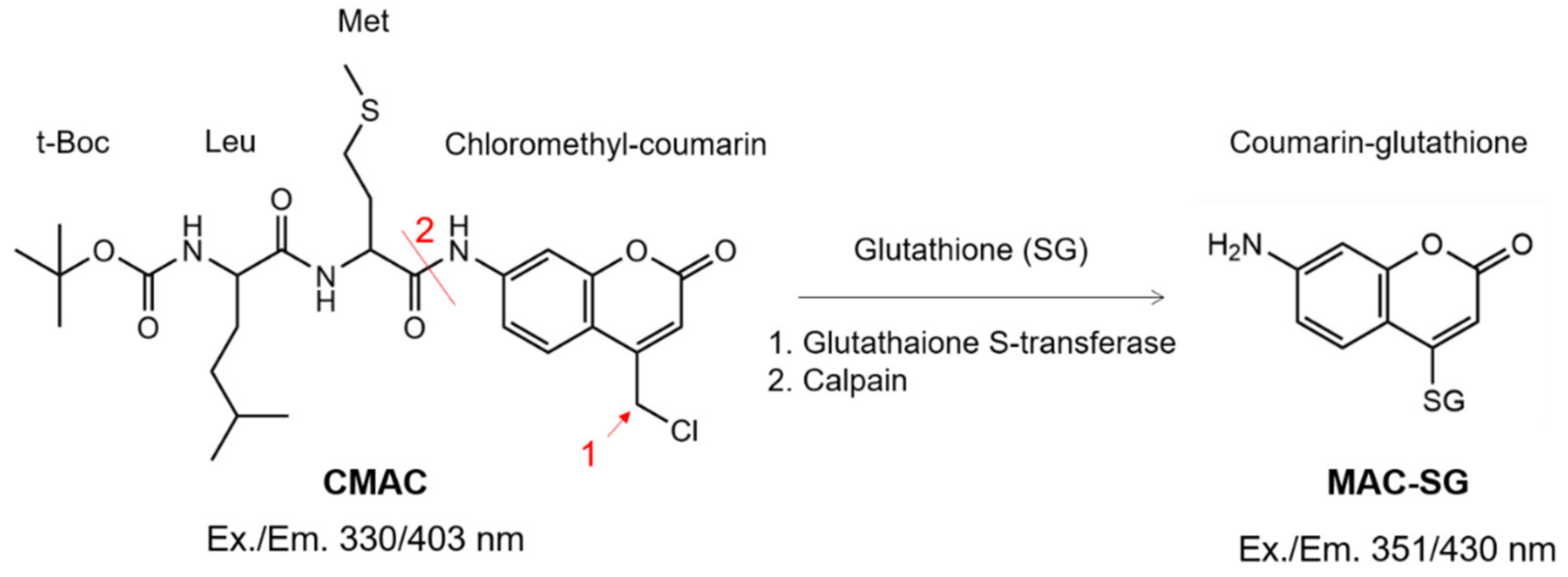
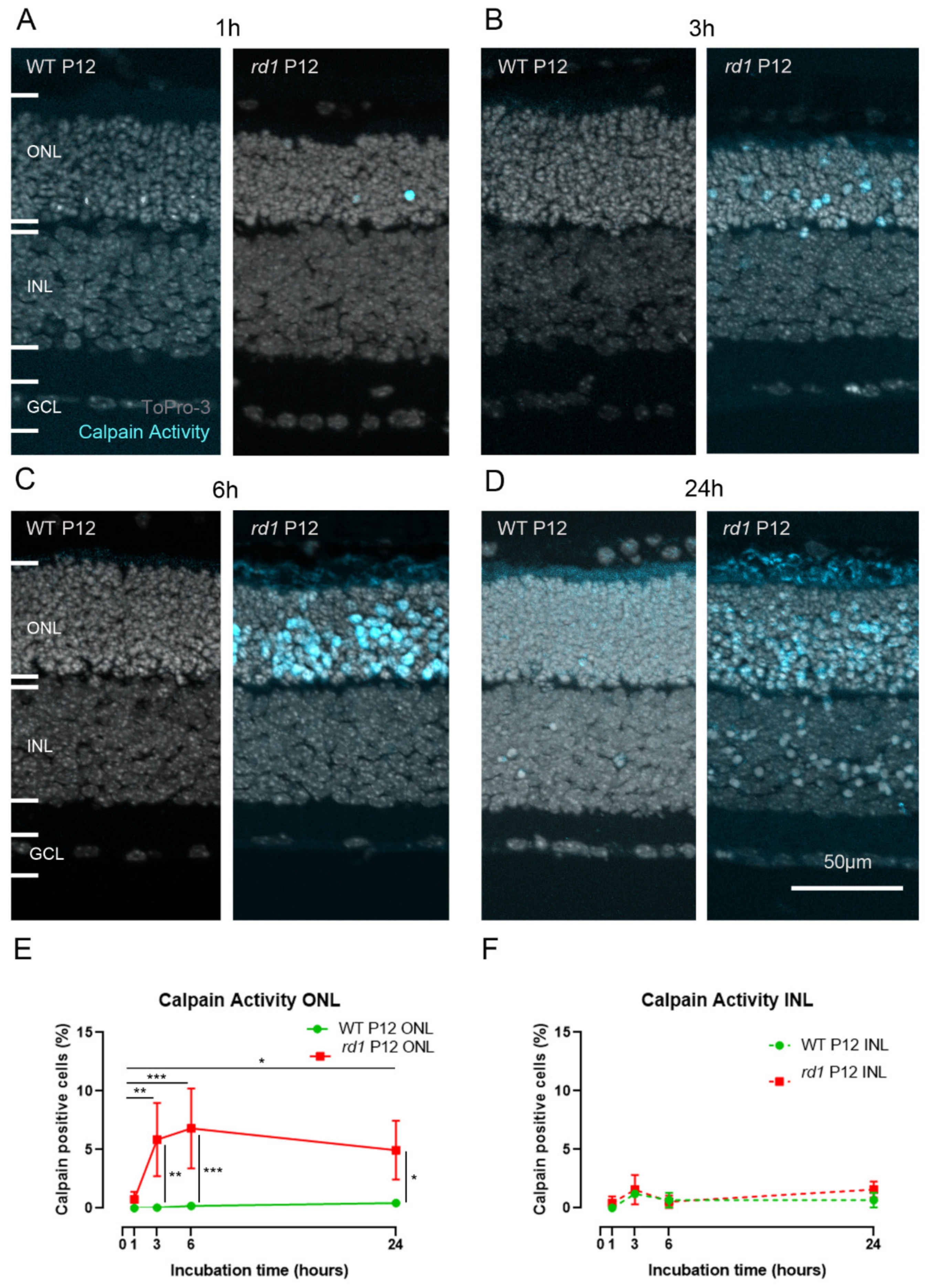
2.1. Calpain Activity Detection in Live Retina
2.2. Calpain Activity Assay Exhibits Low Toxicity in Live Retina
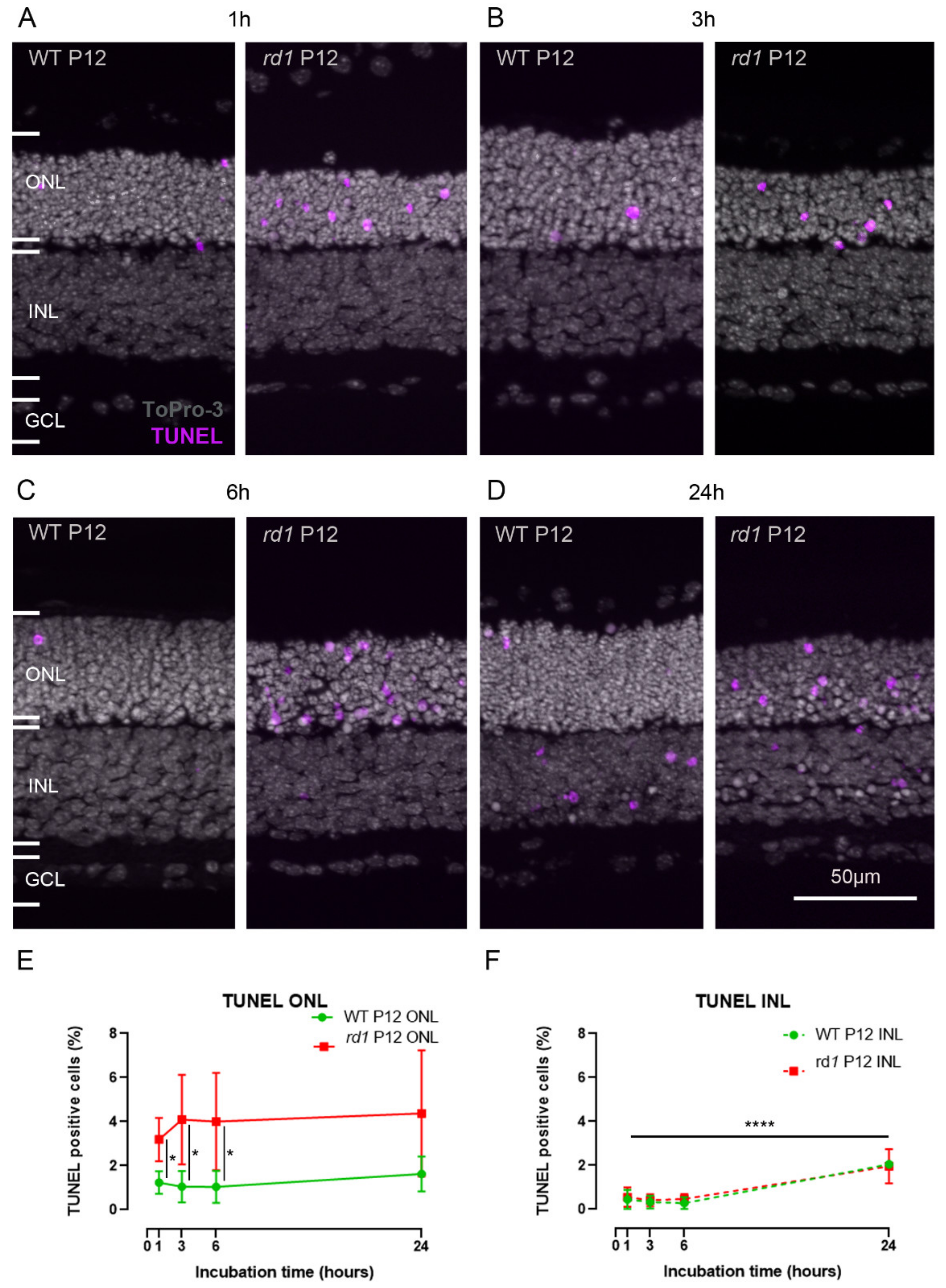
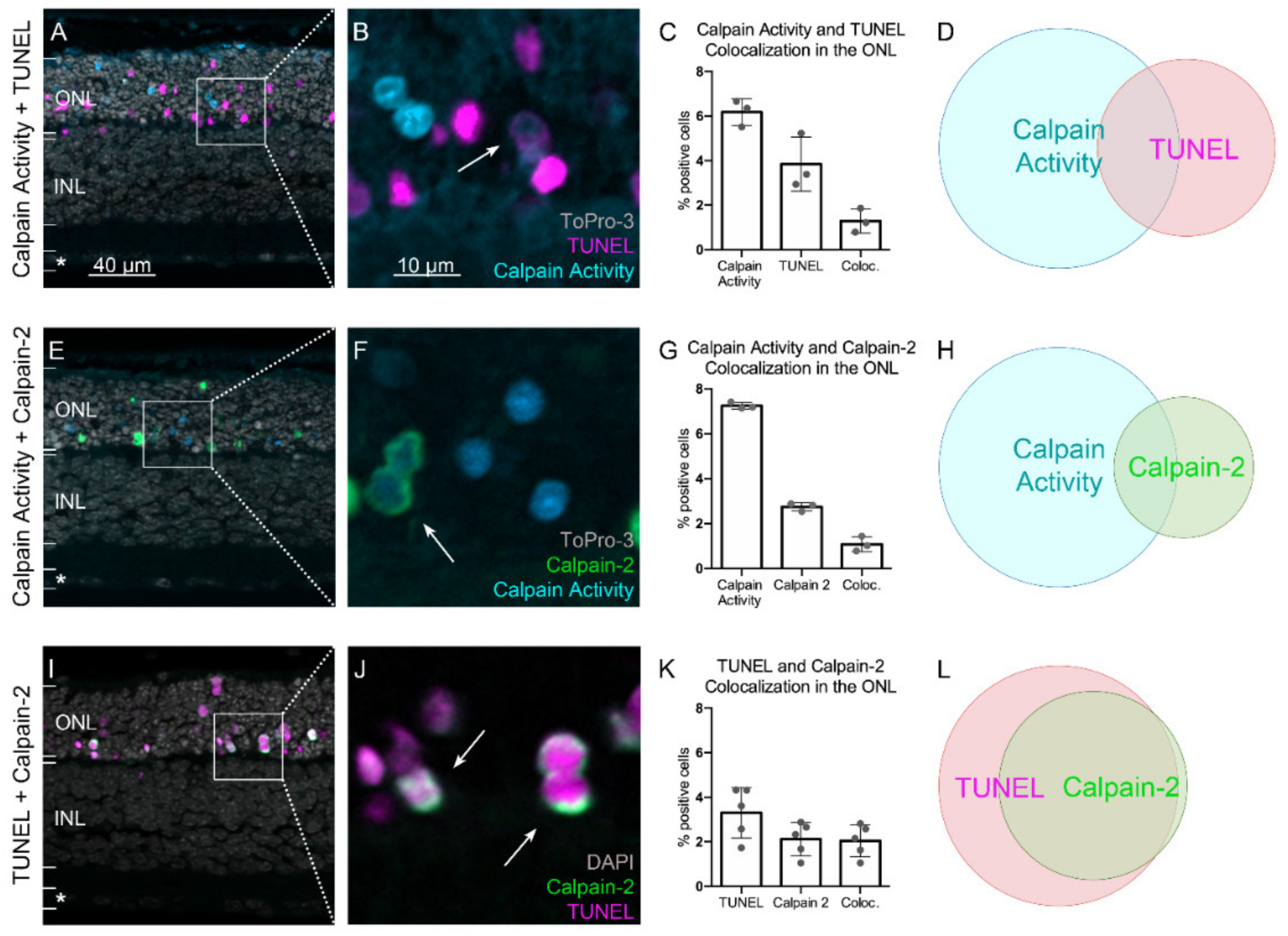
2.3. Calpain Activity and TUNEL Assay Partly Colocalize
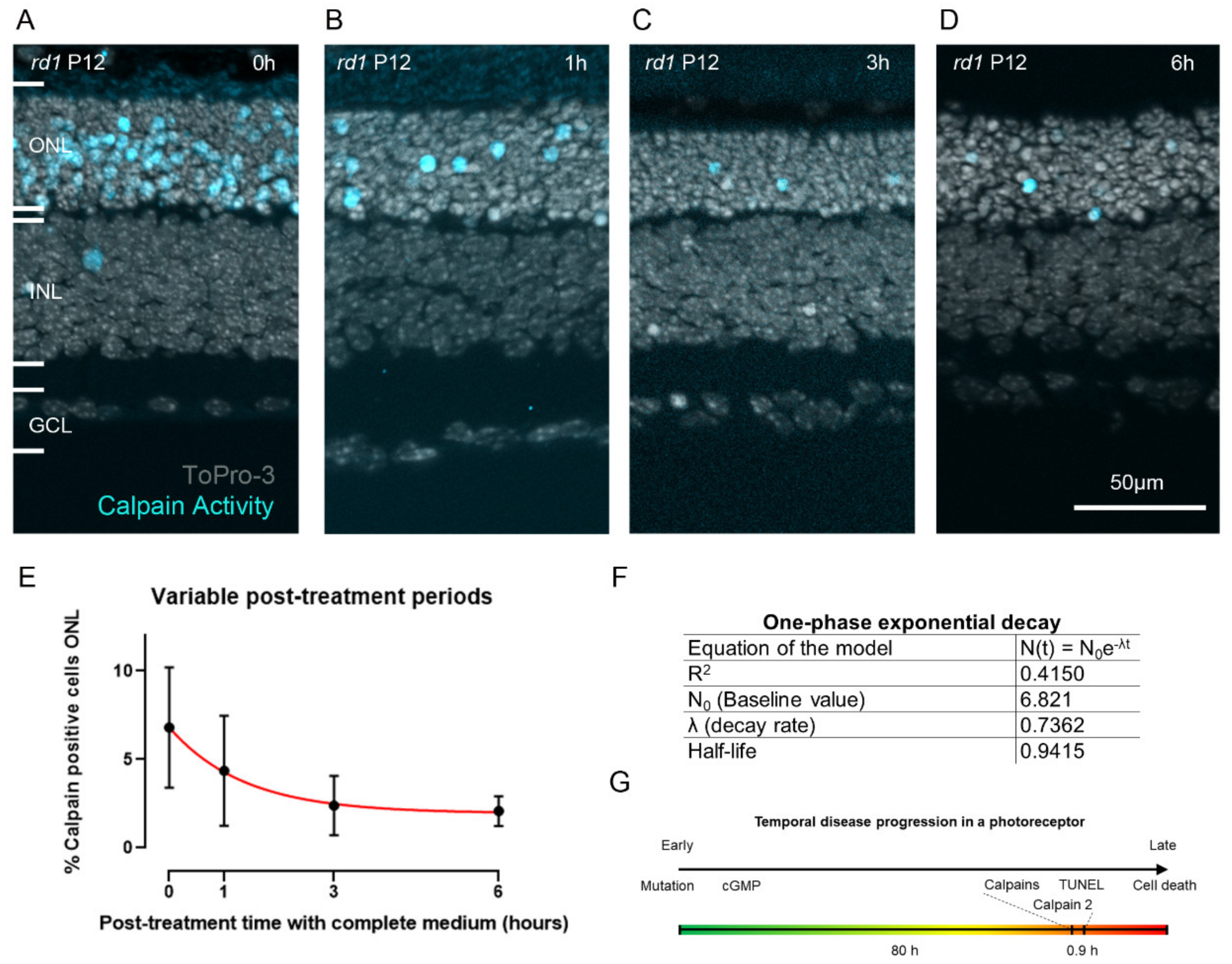
2.4. Estimating the Duration of Calpain Activity in Individual Dying Photoreceptors
3. Discussion
3.1. Detection of Calpain Activity
3.2. Calpain Activity Assay Exhibits Low Toxicity in Live Retinal Explant Cultures
3.3. Calpain-2 Is Activated in the Final Stages of Photoreceptor Cell Death
3.4. Calpain Activity: A Short Event in the Photoreceptor Death Process?
4. Materials and Methods
4.1. Animals
4.2. Organotypic Retinal Explants
4.3. TUNEL Assay
4.4. Immunohistochemistry
4.5. Detection of Calpain Activity in Retinal Explants
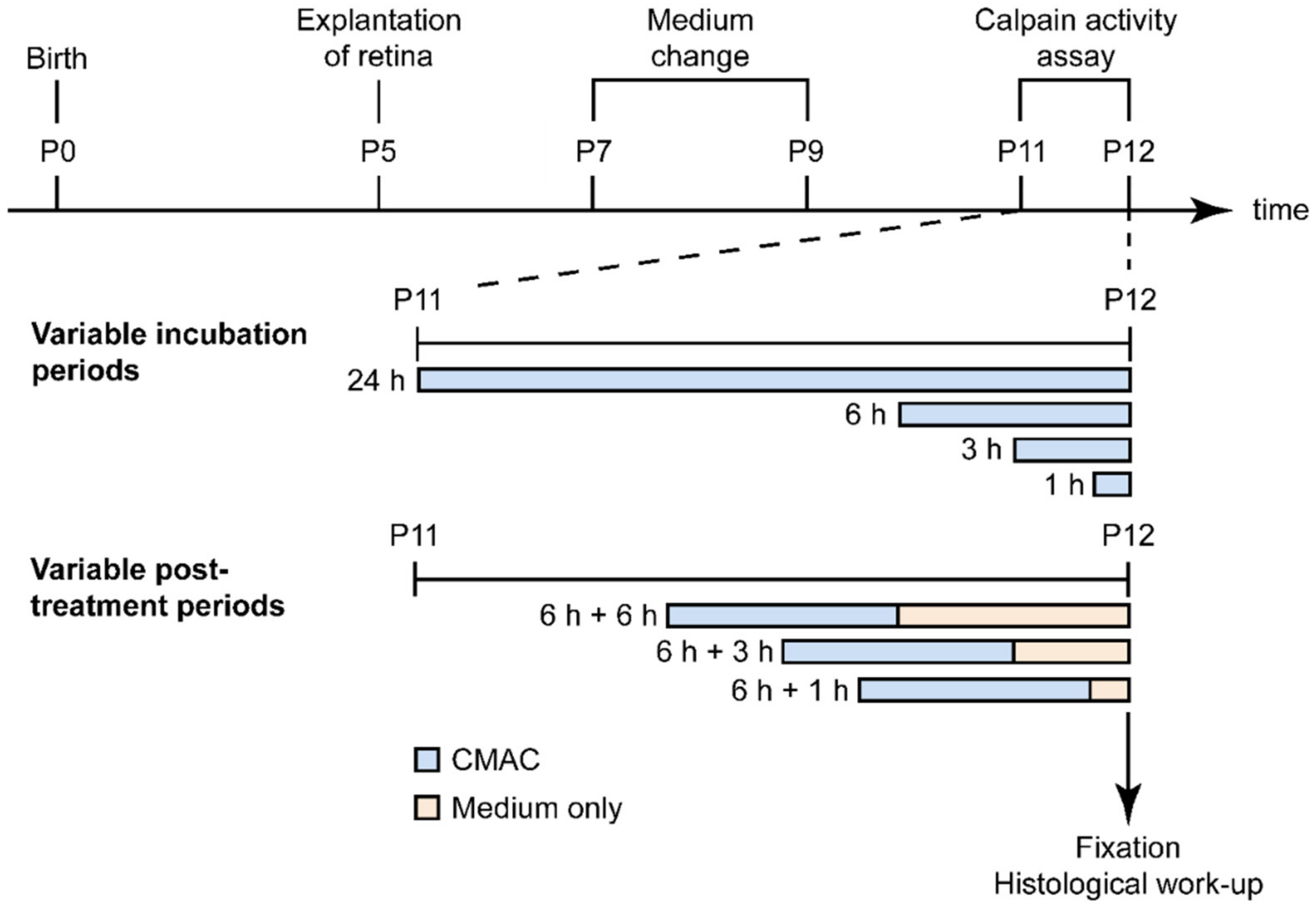
4.6. Microscopy, Cell Counting, and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bertelsen, M.; Jensen, H.; Bregnhøj, J.F.; Rosenberg, T. Prevalence of generalized retinal dystrophy in Denmark. Ophthalmic Epidemiol. 2014, 21, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Berger, W.; Kloeckener-Gruissem, B.; Neidhardt, J. The molecular basis of human retinal and vitreoretinal diseases. Prog. Retin. Eye Res. 2010, 29, 335–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Himawan, E.; Ekstrom, P.; Buzgo, M.; Gaillard, P.; Stefansson, E.; Marigo, V.; Loftsson, T.; Paquet-Durand, F. Drug delivery to retinal photoreceptors. Drug Discov. Today 2019, 24, 1637–1643. [Google Scholar] [CrossRef] [PubMed]
- Hansson, O. Biomarkers for neurodegenerative diseases. Nat. Med. 2021, 27, 954–963. [Google Scholar] [CrossRef]
- Farber, D.B.; Lolley, R.N. Cyclic guanosine monophosphate: Elevation in degenerating photoreceptor cells of the C3H mouse retina. Science 1974, 186, 449–451. [Google Scholar] [CrossRef]
- Power, M.; Das, S.; Schutze, K.; Marigo, V.; Ekstrom, P.; Paquet-Durand, F. Cellular mechanisms of hereditary photoreceptor degeneration-Focus on cGMP. Prog. Retin. Eye Res. 2020, 74, 100772. [Google Scholar] [CrossRef]
- Das, S.; Chen, Y.; Yan, J.; Christensen, G.; Belhadj, S.; Tolone, A.; Paquet-Durand, F. The role of cGMP-signalling and calcium-signalling in photoreceptor cell death: Perspectives for therapy development. Pflug. Arch. 2021, 473, 1411–1421. [Google Scholar] [CrossRef]
- Goll, D.E.; Thompson, V.F.; Li, H.; Wei, W.; Cong, J. The calpain system. Physiol. Rev. 2003, 83, 731–801. [Google Scholar] [CrossRef]
- Khorchid, A.; Ikura, M. How calpain is activated by calcium. Nat. Struct. Biol. 2002, 9, 239–241. [Google Scholar] [CrossRef]
- Baudry, M. Calpain-1 and Calpain-2 in the Brain: Dr. Jekill and Mr Hyde? Curr. Neuropharmacol. 2019, 17, 823–829. [Google Scholar] [CrossRef]
- Baudry, M.; Bi, X. Calpain-1 and Calpain-2: The Yin and Yang of Synaptic Plasticity and Neurodegeneration. Trends Neurosci. 2016, 39, 235–245. [Google Scholar] [CrossRef] [Green Version]
- Arango-Gonzalez, B.; Trifunovic, D.; Sahaboglu, A.; Kranz, K.; Michalakis, S.; Farinelli, P.; Koch, S.; Koch, F.; Cottet, S.; Janssen-Bienhold, U.; et al. Identification of a common non-apoptotic cell death mechanism in hereditary retinal degeneration. PLoS ONE 2014, 9, e112142. [Google Scholar] [CrossRef]
- Paquet-Durand, F.; Azadi, S.; Hauck, S.M.; Ueffing, M.; van Veen, T.; Ekström, P. Calpain is activated in degenerating photoreceptors in the rd1 mouse. J. Neurochem. 2006, 96, 802–814. [Google Scholar] [CrossRef]
- Carido, M.; Zhu, Y.; Postel, K.; Benkner, B.; Cimalla, P.; Karl, M.O.; Kurth, T.; Paquet-Durand, F.; Koch, E.; Munch, T.A.; et al. Characterization of a mouse model with complete RPE loss and its use for RPE cell transplantation. Investig. Ophthalmol. Vis. Sci. 2014, 55, 5431–5444. [Google Scholar] [CrossRef] [Green Version]
- Tao, Y.; Chen, T.; Fang, W.; Peng, G.; Wang, L.; Qin, L.; Liu, B.; Fei Huang, Y. The temporal topography of the N-Methyl- N-nitrosourea induced photoreceptor degeneration in mouse retina. Sci. Rep. 2015, 5, 18612. [Google Scholar] [CrossRef]
- Paquet-Durand, F.; Johnson, L.; Ekström, P. Calpain activity in retinal degeneration. J. Neurosci. Res. 2007, 85, 693–702. [Google Scholar] [CrossRef]
- Power, M.J.; Rogerson, L.E.; Schubert, T.; Berens, P.; Euler, T.; Paquet-Durand, F. Systematic spatiotemporal mapping reveals divergent cell death pathways in three mouse models of hereditary retinal degeneration. J. Comp. Neurol. 2020, 528, 1113–1139. [Google Scholar] [CrossRef] [Green Version]
- Azuma, M.; Sakamoto-Mizutani, K.; Nakajima, T.; Kanaami-Daibo, S.; Tamada, Y.; Shearer, T.R. Involvement of calpain isoforms in retinal degeneration in WBN/Kob rats. Comp. Med. 2004, 54, 533–542. [Google Scholar]
- Ekstrom, P.A.; Ueffing, M.; Zrenner, E.; Paquet-Durand, F. Novel in situ activity assays for the quantitative molecular analysis of neurodegenerative processes in the retina. Curr. Med. Chem. 2014, 21, 3478–3493. [Google Scholar] [CrossRef]
- Habig, W.H.; Pabst, M.J.; Jakoby, W.B. Glutathione S-transferases. The first enzymatic step in mercapturic acid formation. J. Biol. Chem. 1974, 249, 7130–7139. [Google Scholar] [CrossRef]
- Rosser, B.G.; Powers, S.P.; Gores, G.J. Calpain activity increases in hepatocytes following addition of ATP. Demonstration by a novel fluorescent approach. J. Biol. Chem. 1993, 268, 23593–23600. [Google Scholar] [CrossRef]
- Pitkanen, L.; Ranta, V.P.; Moilanen, H.; Urtti, A. Permeability of retinal pigment epithelium: Effects of permeant molecular weight and lipophilicity. Investig. Ophthalmol. Vis. Sci. 2005, 46, 641–646. [Google Scholar] [CrossRef] [Green Version]
- Comitato, A.; Schiroli, D.; Montanari, M.; Marigo, V. Calpain Activation Is the Major Cause of Cell Death in Photoreceptors Expressing a Rhodopsin Misfolding Mutation. Mol. Neurobiol. 2020, 57, 589–599. [Google Scholar] [CrossRef]
- Nakamura, P.A.; Shimchuk, A.A.; Tang, S.; Wang, Z.; DeGolier, K.; Ding, S.; Reh, T.A. Small molecule Photoregulin3 prevents retinal degeneration in the Rho(P23H) mouse model of retinitis pigmentosa. eLife 2017, 6, e30577. [Google Scholar] [CrossRef] [Green Version]
- Gavrieli, Y.; Sherman, Y.; Ben-Sasson, S.A. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J. Cell Biol. 1992, 119, 493–501. [Google Scholar] [CrossRef]
- Sahaboglu, A.; Barth, M.; Secer, E.; Amo, E.M.; Urtti, A.; Arsenijevic, Y.; Zrenner, E.; Paquet-Durand, F. Olaparib significantly delays photoreceptor loss in a model for hereditary retinal degeneration. Sci. Rep. 2016, 6, 39537. [Google Scholar] [CrossRef] [Green Version]
- Sahaboglu, A.; Paquet-Durand, O.; Dietter, J.; Dengler, K.; Bernhard-Kurz, S.; Ekström, P.A.; Hitzmann, B.; Ueffing, M.; Paquet-Durand, F. Retinitis pigmentosa: Rapid neurodegeneration is governed by slow cell death mechanisms. Cell Death Dis. 2013, 4, e488. [Google Scholar] [CrossRef]
- Suzuki, K.; Hata, S.; Kawabata, Y.; Sorimachi, H. Structure, activation, and biology of calpain. Diabetes 2004, 53 (Suppl. S1), S12–S18. [Google Scholar] [CrossRef] [Green Version]
- Clarke, G.; Collins, R.A.; Leavitt, B.R.; Andrews, D.F.; Hayden, M.R.; Lumsden, C.J.; McInnes, R.R. A one-hit model of cell death in inherited neuronal degenerations. Nature 2000, 406, 195–199. [Google Scholar] [CrossRef]
- Fischer, J.; Otto, T.; Delori, F.; Pace, L.; Staurenghi, G. Scanning Laser Ophthalmoscopy (SLO). In High Resolution Imaging in Microscopy and Ophthalmology: New Frontiers in Biomedical Optics; Bille, J.F., Ed.; Springer: Cham, Switzerland, 2019; pp. 35–57. [Google Scholar] [CrossRef] [Green Version]
- Podoleanu, A.G. Optical coherence tomography. J. Microsc. 2012, 247, 209–219. [Google Scholar] [CrossRef] [Green Version]
- Tian, X.; Inoue, K.; Zhang, Y.; Wang, Y.; Sperati, C.J.; Pedigo, C.E.; Zhao, T.; Yan, M.; Groener, M.; Moledina, D.G.; et al. Inhibiting calpain 1 and 2 in cyclin G associated kinase-knockout mice mitigates podocyte injury. JCI Insight 2020, 5, e142740. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Mun, S.; Park, A.; Kim, D.; Heun Cha, B.; Kang, H.G. Bicalutamide enhances fodrin-mediated apoptosis through calpain in LNCaP. Exp. Biol. Med. 2018, 243, 843–851. [Google Scholar] [CrossRef] [PubMed]
- Saatman, K.E.; Abai, B.; Grosvenor, A.; Vorwerk, C.K.; Smith, D.H.; Meaney, D.F. Traumatic axonal injury results in biphasic calpain activation and retrograde transport impairment in mice. J. Cereb. Blood Flow Metab. 2003, 23, 34–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartoli, M.; Bourg, N.; Stockholm, D.; Raynaud, F.; Delevacque, A.; Han, Y.; Borel, P.; Seddik, K.; Armande, N.; Richard, I. A mouse model for monitoring calpain activity under physiological and pathological conditions. J. Biol. Chem. 2006, 281, 39672–39680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stockholm, D.; Bartoli, M.; Sillon, G.; Bourg, N.; Davoust, J.; Richard, I. Imaging calpain protease activity by multiphoton FRET in living mice. J. Mol. Biol. 2005, 346, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Sohocki, M.M.; Daiger, S.P.; Bowne, S.J.; Rodriquez, J.A.; Northrup, H.; Heckenlively, J.R.; Birch, D.G.; Mintz-Hittner, H.; Ruiz, R.S.; Lewis, R.A.; et al. Prevalence of mutations causing retinitis pigmentosa and other inherited retinopathies. Hum. Mutat. 2001, 17, 42–51. [Google Scholar] [CrossRef] [Green Version]
- Marzia, M.; Chiusaroli, R.; Neff, L.; Kim, N.Y.; Chishti, A.H.; Baron, R.; Horne, W.C. Calpain is required for normal osteoclast function and is down-regulated by calcitonin. J. Biol. Chem. 2006, 281, 9745–9754. [Google Scholar] [CrossRef] [Green Version]
- Su, L.T.; Chen, H.C.; González-Pagán, O.; Overton, J.D.; Xie, J.; Yue, L.; Runnels, L.W. TRPM7 activates m-calpain by stress-dependent stimulation of p38 MAPK and c-Jun N-terminal kinase. J. Mol. Biol. 2010, 396, 858–869. [Google Scholar] [CrossRef] [Green Version]
- Tauskela, J.S.; Hewitt, K.; Kang, L.P.; Comas, T.; Gendron, T.; Hakim, A.; Hogan, M.; Durkin, J.; Morley, P. Evaluation of glutathione-sensitive fluorescent dyes in cortical culture. Glia 2000, 30, 329–341. [Google Scholar] [CrossRef]
- Hata, S.; Nishi, K.; Kawamoto, T.; Lee, H.J.; Kawahara, H.; Maeda, T.; Shintani, Y.; Sorimachi, H.; Suzuki, K. Both the conserved and the unique gene structure of stomach-specific calpains reveal processes of calpain gene evolution. J. Mol. Evol. 2001, 53, 191–203. [Google Scholar] [CrossRef]
- Moldoveanu, T.; Hosfield, C.M.; Lim, D.; Jia, Z.; Davies, P.L. Calpain silencing by a reversible intrinsic mechanism. Nat. Struct. Biol. 2003, 10, 371–378. [Google Scholar] [CrossRef]
- Dutt, P.; Croall, D.E.; Arthur, J.S.; Veyra, T.D.; Williams, K.; Elce, J.S.; Greer, P.A. m-Calpain is required for preimplantation embryonic development in mice. BMC Dev. Biol. 2006, 6, 3. [Google Scholar] [CrossRef] [Green Version]
- Takano, J.; Mihira, N.; Fujioka, R.; Hosoki, E.; Chishti, A.H.; Saido, T.C. Vital role of the calpain-calpastatin system for placental-integrity-dependent embryonic survival. Mol. Cell. Biol. 2011, 31, 4097–4106. [Google Scholar] [CrossRef] [Green Version]
- Franco, S.; Perrin, B.; Huttenlocher, A. Isoform specific function of calpain 2 in regulating membrane protrusion. Exp. Cell Res. 2004, 299, 179–187. [Google Scholar] [CrossRef]
- Santos, D.M.; Xavier, J.M.; Morgado, A.L.; Solá, S.; Rodrigues, C.M. Distinct regulatory functions of calpain 1 and 2 during neural stem cell self-renewal and differentiation. PLoS ONE 2012, 7, e33468. [Google Scholar] [CrossRef]
- Shinkai-Ouchi, F.; Shindo, M.; Doi, N.; Hata, S.; Ono, Y. Calpain-2 participates in the process of calpain-1 inactivation. Biosci. Rep. 2020, 40, BSR20200552. [Google Scholar] [CrossRef]
- Yoshimura, N.; Kikuchi, T.; Sasaki, T.; Kitahara, A.; Hatanaka, M.; Murachi, T. Two distinct Ca2+ proteases (calpain I and calpain II) purified concurrently by the same method from rat kidney. J. Biol. Chem. 1983, 258, 8883–8889. [Google Scholar] [CrossRef]
- Paquet-Durand, F.; Sanges, D.; McCall, J.; Silva, J.; van Veen, T.; Marigo, V.; Ekström, P. Photoreceptor rescue and toxicity induced by different calpain inhibitors. J. Neurochem. 2010, 115, 930–940. [Google Scholar] [CrossRef]
- Das, S.; Popp, V.; Power, M.; Groeneveld, K.; Melle, C.; Rogerson, L.; Achury, M.; Schwede, F.; Strasser, T.; Euler, T.; et al. Redefining the role of Ca2+-permeable channels in photoreceptor degeneration using diltiazem. bioRxiv 2021. [Google Scholar] [CrossRef]
- Sanyal, S.; Bal, A.K. Comparative light and electron microscopic study of retinal histogenesis in normal and rd mutant mice. Z. Anat. Entwicklungsgesch. 1973, 142, 219–238. [Google Scholar] [CrossRef]
- Sakami, S.; Maeda, T.; Bereta, G.; Okano, K.; Golczak, M.; Sumaroka, A.; Roman, A.J.; Cideciyan, A.V.; Jacobson, S.G.; Palczewski, K. Probing mechanisms of photoreceptor degeneration in a new mouse model of the common form of autosomal dominant retinitis pigmentosa due to P23H opsin mutations. J. Biol. Chem. 2011, 286, 10551–10567. [Google Scholar] [CrossRef] [Green Version]
- Belhadj, S.; Tolone, A.; Christensen, G.; Das, S.; Chen, Y.; Paquet-Durand, F. Long-Term, Serum-Free Cultivation of Organotypic Mouse Retina Explants with Intact Retinal Pigment Epithelium. J. Vis. Exp. 2020, 165, e61868. [Google Scholar] [CrossRef]
- Akyol, E.; Hagag, A.M.; Sivaprasad, S.; Lotery, A.J. Adaptive optics: Principles and applications in ophthalmology. Eye 2021, 35, 244–264. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Belhadj, S.; Hermann, N.S.; Zhu, Y.; Christensen, G.; Strasser, T.; Paquet-Durand, F. Visualizing Cell Death in Live Retina: Using Calpain Activity Detection as a Biomarker for Retinal Degeneration. Int. J. Mol. Sci. 2022, 23, 3892. https://doi.org/10.3390/ijms23073892
Belhadj S, Hermann NS, Zhu Y, Christensen G, Strasser T, Paquet-Durand F. Visualizing Cell Death in Live Retina: Using Calpain Activity Detection as a Biomarker for Retinal Degeneration. International Journal of Molecular Sciences. 2022; 23(7):3892. https://doi.org/10.3390/ijms23073892
Chicago/Turabian StyleBelhadj, Soumaya, Nina Sofia Hermann, Yu Zhu, Gustav Christensen, Torsten Strasser, and François Paquet-Durand. 2022. "Visualizing Cell Death in Live Retina: Using Calpain Activity Detection as a Biomarker for Retinal Degeneration" International Journal of Molecular Sciences 23, no. 7: 3892. https://doi.org/10.3390/ijms23073892
APA StyleBelhadj, S., Hermann, N. S., Zhu, Y., Christensen, G., Strasser, T., & Paquet-Durand, F. (2022). Visualizing Cell Death in Live Retina: Using Calpain Activity Detection as a Biomarker for Retinal Degeneration. International Journal of Molecular Sciences, 23(7), 3892. https://doi.org/10.3390/ijms23073892





