Fragment-Based Discovery of AF9 YEATS Domain Inhibitors
Abstract
:1. Introduction
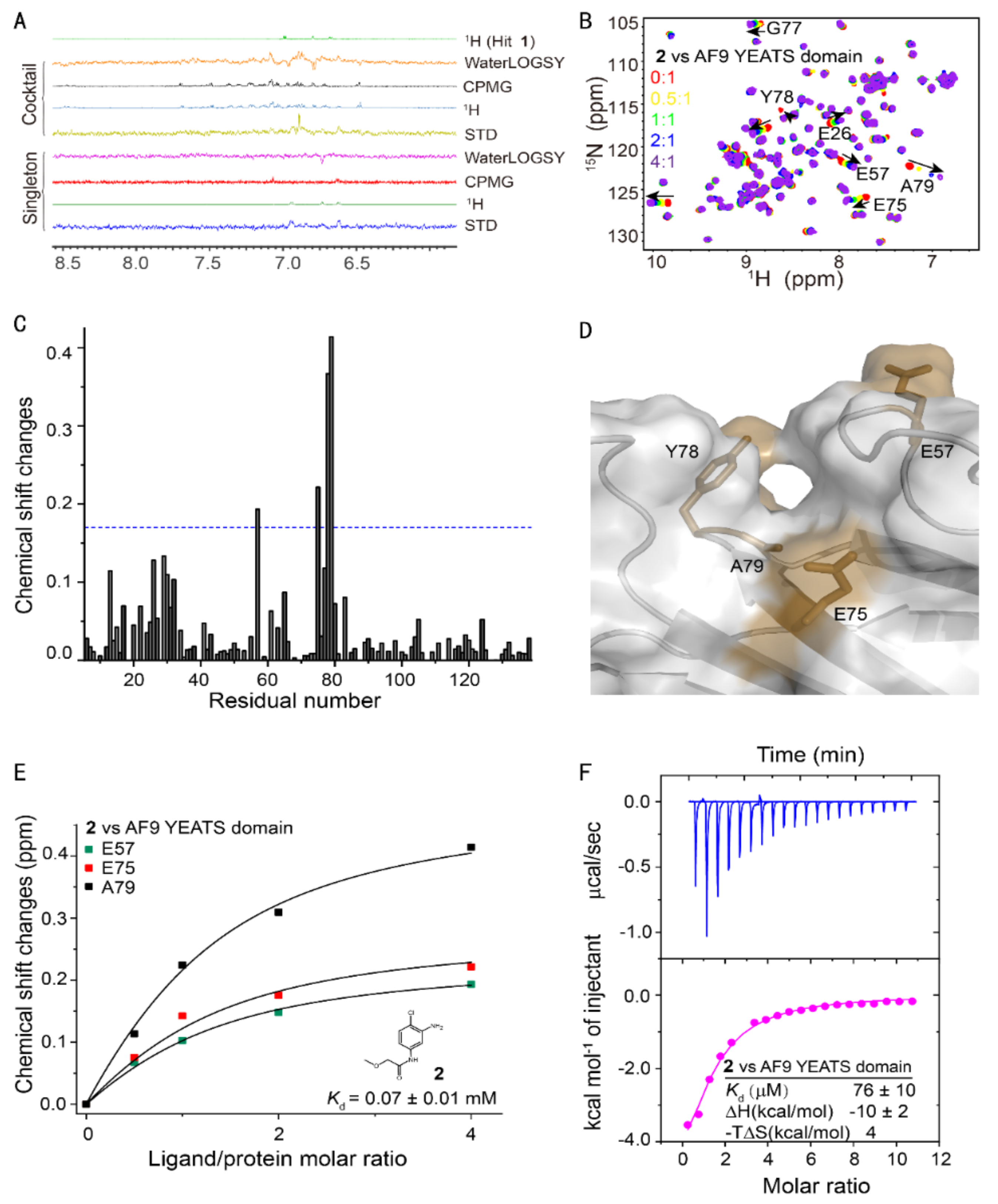
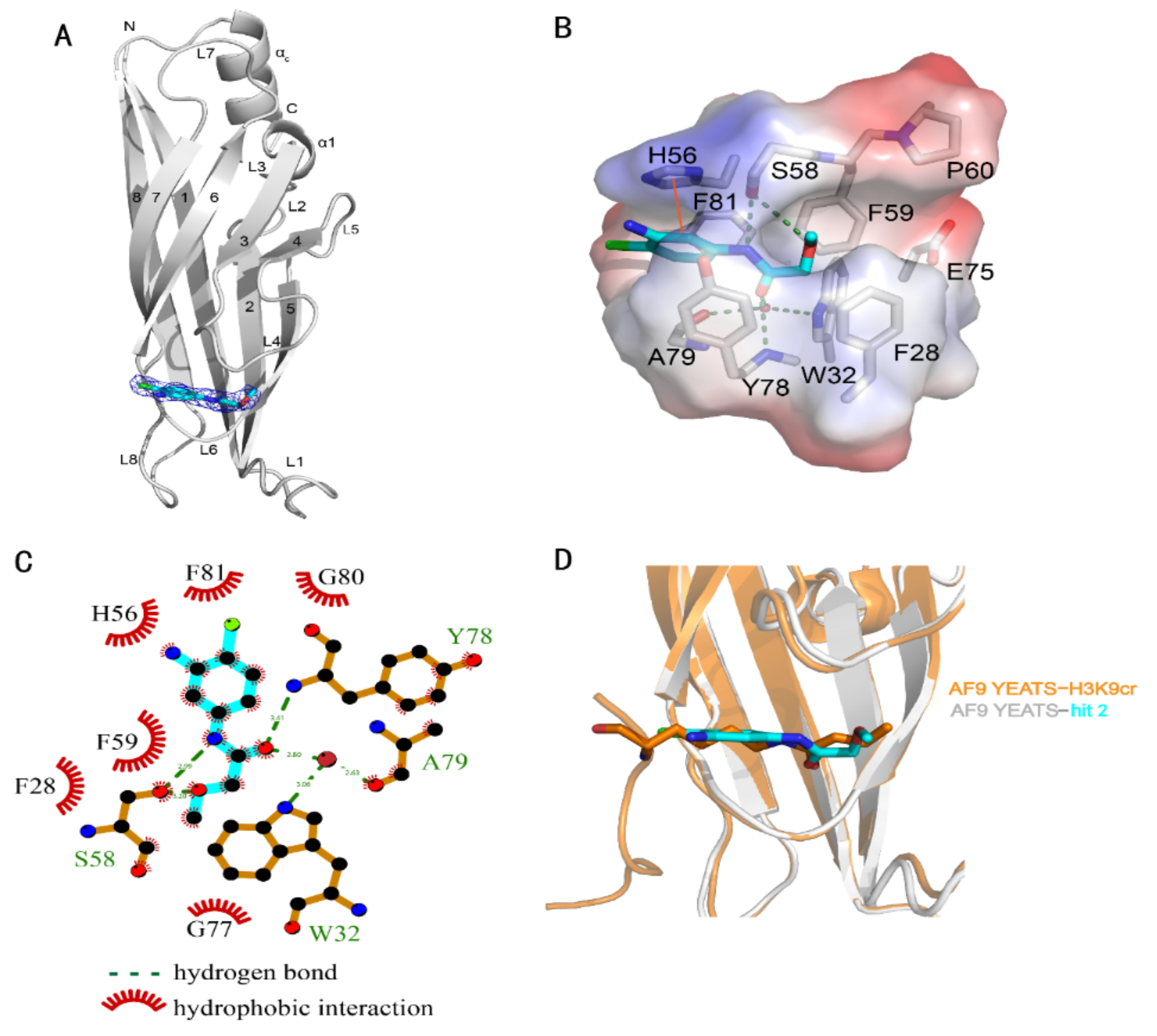
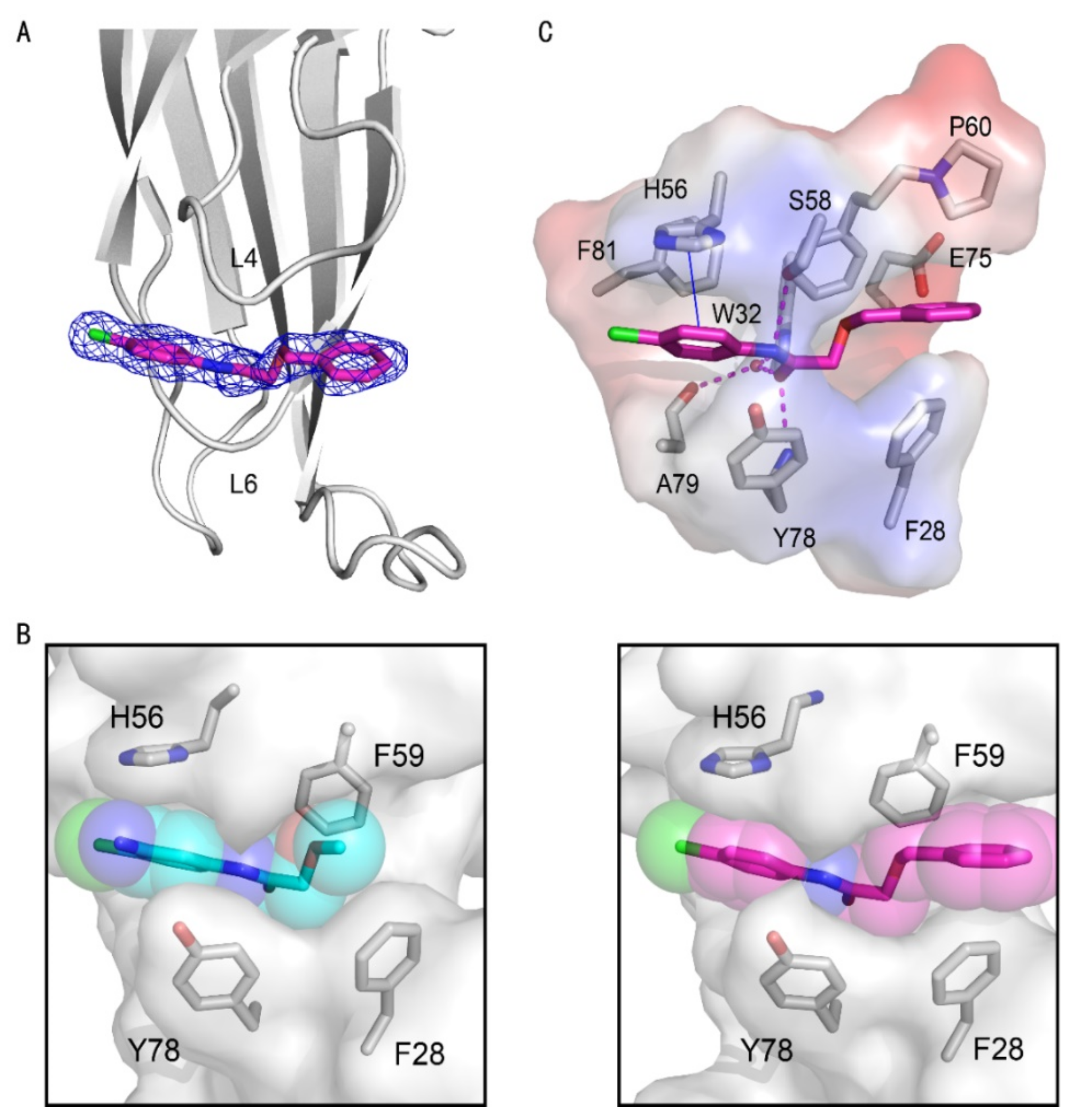
2. Results
3. Conclusions
4. Materials and Methods
4.1. Cloning, Protein Expression, and Purification
4.2. Ligand-Observed Fragment Screening
4.3. NMR Chemical Shift Perturbation
4.4. Isothermal Titration Calorimetry
4.5. Crystallization, Data collection
4.6. Structure Determination and Refinement
4.7. Cell Viability Assay
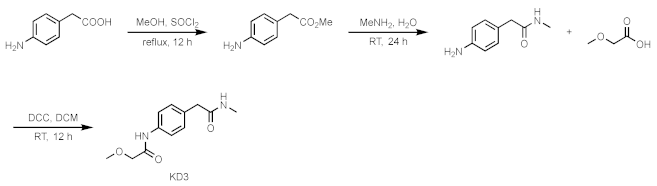
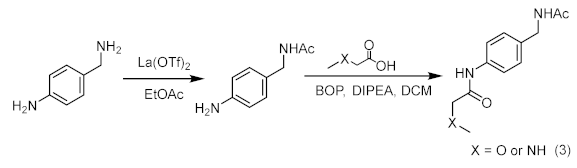
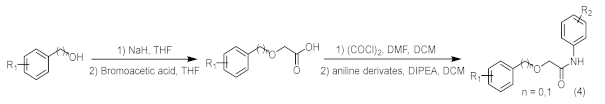
4.8. Chemical Synthesis and Characterization

Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| YEATS | YAF9, ENL, AF9, TAF14, SAS5 |
| STD | Saturation Transfer Difference |
| CPMG | Carr–Purcell–Meiboom–Gill |
| CSPs | Chemical shift perturbations |
| HSQC | Heteronuclear single-quantum coherence |
| NMR | Nuclear Magnetic Resonance |
References
- Tolsma, T.O.; Hansen, J.C. Post-translational modifications and chromatin dynamics. Essays Biochem. 2019, 63, 89–96. [Google Scholar] [PubMed]
- Zeng, L.; Zhang, Q.; Li, S.; Plotnikov, A.N.; Walsh, M.J.; Zhou, M.-M. Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature 2010, 466, 258–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhalluin, C.; Carlson, J.E.; Zeng, L.; He, C.; Aggarwal, A.K.; Zhou, M.M. Structure and ligand of a histone acetyltransferase bromodomain. Nature 1999, 399, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Panchenko, T.; Yang, S.; Zhao, S.; Yan, P.; Zhang, W.; Xie, P.Y.W.Z.W.; Li, Y.; Zhao, Y.; Allis, T.P.C.D.; et al. Selective recognition of histone crotonylation by double PHD fingers of MOZ and DPF2. Nat. Chem. Biol. 2016, 12, 1111–1118. [Google Scholar] [CrossRef] [Green Version]
- Zhao, D.; Li, Y.; Xiong, X.; Chen, Z.; Li, H. YEATS Domain—A Histone Acylation Reader in Health and Disease. J. Mol. Biol. 2017, 429, 1994–2002. [Google Scholar] [CrossRef]
- Li, Y.; Sabari, B.R.; Panchenko, T.; Wen, H.; Zhao, D.; Guan, H.; Wan, L.; Huang, H.; Tang, Z.; Zhao, Y.; et al. Molecular Coupling of Histone Crotonylation and Active Transcription by AF9 YEATS Domain. Mol. Cell 2016, 62, 181–193. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Zeng, L.; Zhao, C.; Ju, Y.; Konuma, T.; Zhou, M.-M. Structural Insights into Histone Crotonyl-Lysine Recognition by the AF9 YEATS Domain. Structure 2016, 24, 1606–1612. [Google Scholar] [CrossRef] [Green Version]
- Zhao, D.; Guan, H.; Zhao, S.; Mi, W.; Wen, H.; Li, Y.; Zhao, Y.; Allis, C.D.; Shi, X.; Li, H. YEATS2 is a selective histone crotonylation reader. Cell Res. 2016, 26, 629–632. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Jin, J.; Chung, M.W.; Feng, L.; Sun, H.; Hao, Q. Identification of the YEATS domain of GAS41 as a pH-dependent reader of histone succinylation. Proc. Natl. Acad. Sci. USA 2018, 115, 2365–2370. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Wen, H.; Xi, Y.; Tanaka, K.; Wang, H.; Peng, D.; Ren, Y.; Jin, Q.; Dent, S.; Li, W.; et al. AF9 YEATS Domain Links Histone Acetylation to DOT1L-Mediated H3K79 Methylation. Cell 2014, 159, 558–571. [Google Scholar] [CrossRef] [Green Version]
- Andrews, F.H.; Shanle, E.K.; Strahl, B.D.; Kutateladze, T.G. The essential role of acetyllysine binding by the YEATS domain in transcriptional regulation. Transcription 2016, 7, 14–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shanle, E.K.; Andrews, F.H.; Meriesh, H.; Mc Daniel, S.L.; Dronamraju, R.; Di Fiore, J.V.; Jha, D.; Wozniak, G.G.; Bridgers, J.B.; Kerschner, J.L.; et al. Association of Taf14 with acetylated histone H3 directs gene transcription and the DNA damage response. Genes Dev. 2015, 29, 1795–1800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Zhao, D.; Chen, Z.; Li, H. YEATS domain: Linking histone crotonylation to gene regulation. Transcription 2017, 8, 9–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulze, J.M.; Wang, A.Y.; Kobor, M.S. YEATS domain proteins: A diverse family with many links to chromatin modification and transcription. Biochem. Cell Biol. 2009, 87, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Frattini, V.; Trifonov, V.; Chan, J.; Castano, A.; Lia, M.; Abate, F.; Keir, S.T.; Ji, A.X.; Zoppoli, P.; Niola, F.; et al. The integrated landscape of driver genomic alterations in glioblastoma. Nat. Genet. 2013, 45, 1141–1149. [Google Scholar] [CrossRef] [Green Version]
- Morin, R.; Assouline, S.; Alcaide, M.; Mohajeri, A.; Johnston, R.; Chong, L.; Grewal, J.; Yu, S.; Fornika, D.; Bushell, K.; et al. Genetic Landscapes of Relapsed and Refractory Diffuse Large B-Cell Lymphomas. Clin. Cancer Res. 2016, 22, 2290–2300. [Google Scholar] [CrossRef] [Green Version]
- Stavropoulou, V.; Kaspar, S.; Brault, L.; Sanders, M.A.; Juge, S.; Morettini, S.; Tzankov, A.; Iacovino, M.; Lau, I.-J.; Milne, T.; et al. MLL-AF9 Expression in Hematopoietic Stem Cells Drives a Highly Invasive AML Expressing EMT-Related Genes Linked to Poor Outcome. Cancer Cell 2016, 30, 43–58. [Google Scholar] [CrossRef] [Green Version]
- Zeisig, D.T.; Bittner, C.B.; Zeisig, B.B.; García-Cuéllar, M.-P.; Hess, J.L.; Slany, R.K. The eleven-nineteen-leukemia protein ENL connects nuclear MLL fusion partners with chromatin. Oncogene 2005, 24, 5525–5532. [Google Scholar] [CrossRef] [Green Version]
- Erb, M.A.; Scott, T.G.; Li, B.E.; Xie, H.; Paulk, J.; Seo, H.-S.; Souza, A.; Roberts, J.M.; Dastjerdi, S.; Buckley, D.L.; et al. Transcription control by the ENL YEATS domain in acute leukaemia. Nature 2017, 543, 270–274. [Google Scholar] [CrossRef] [Green Version]
- Wan, L.; Wen, H.; Li, Y.L.H.; Lyu, J.; Xi, Y.; Hoshii, T.; Joseph, J.K.; Wang, X.; Loh, Y.-H.E.; Erb, M.; et al. ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia. Nature 2017, 543, 265–269. [Google Scholar] [CrossRef] [Green Version]
- Perlman, E.J.; Gadd, S.; Arold, S.T.; Radhakrishnan, A.; Gerhard, D.S.; Jennings, L.J.; Huff, V.; Auvil, J.M.G.; Davidsen, T.M.; Dome, J.S.; et al. MLLT1 YEATS domain mutations in clinically distinctive Favourable Histology Wilms tumours. Nat. Commun. 2015, 6, 10013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moustakim, M.; Christott, T.; Monteiro, O.P.; Bennett, J.; Giroud, C.; Ward, J.; Rogers, C.M.; Smith, P.; Panagakou, I.; Díaz-Sáez, L.; et al. Discovery of an MLLT1/3 YEATS Domain Chemical Probe. Angew. Chem. Int. Ed. Engl. 2018, 57, 16302–16307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christott, T.; Bennett, J.; Coxon, C.; Monteiro, O.; Giroud, C.; Beke, V.; Felce, S.L.; Gamble, V.; Gileadi, C.; Poda, G.; et al. Discovery of a Selective Inhibitor for the YEATS Domains of ENL/AF9. SLAS Discov. Adv. Sci. Drug Discov. 2018, 24, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Asiaban, J.N.; Milosevich, N.; Chen, E.; Bishop, T.R.; Wang, J.; Zhang, Y.; Ackerman, C.J.; Hampton, E.N.; Young, T.S.; Hull, M.V.; et al. Cell-Based Ligand Discovery for the ENL YEATS Domain. ACS Chem. Biol. 2020, 15, 895–903. [Google Scholar] [CrossRef]
- Li, X.; Li, X.M.; Jiang, Y.; Liu, Z.; Cui, Y.; Fung, K.Y.; van der Beelen, S.H.; Tian, G.; Wan, L.; Shi, X.; et al. Structure-guided development of YEATS domain inhibitors by targeting pi-pi-pi stacking. Nat. Chem. Biol. 2018, 14, 1140–1149. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, G.; Li, X.-M.; Liu, S.; Tian, G.; Li, Y.; Li, X.; Li, H.; Li, X.D. Selective Targeting of AF9 YEATS Domain by Cyclopeptide Inhibitors with Preorganized Conformation. J. Am. Chem. Soc. 2020, 142, 21450–21459. [Google Scholar] [CrossRef]
- Ma, X.R.; Xu, L.; Xu, S.; Klein, B.J.; Wang, H.; Das, S.; Li, K.; Yang, K.S.; Sohail, S.; Chapman, A.; et al. Discovery of Selective Small-Molecule Inhibitors for the ENL YEATS Domain. J. Med. Chem. 2021, 64, 10997–11013. [Google Scholar] [CrossRef]
- Wu, F.; Nie, S.; Yao, Y.; Huo, T.; Li, X.; Wu, X.; Zhao, J.; Lin, Y.L.; Zhang, Y.; Mo, Q.; et al. Small-molecule inhibitor of AF9/ENL-DOT1L/AF4/AFF4 interactions suppresses malignant gene expression and tumor growth. Theranostics 2021, 11, 8172–8184. [Google Scholar] [CrossRef]
- Behan, F.M.; Iorio, F.; Picco, G.; Gonçalves, E.; Beaver, C.M.; Migliardi, G.; Santos, R.; Rao, Y.; Sassi, F.; Pinnelli, M.; et al. Prioritization of cancer therapeutic targets using CRISPR–Cas9 screens. Nature 2019, 568, 511–516. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, S.; Liu, M.; Liu, Y.; Nshogoza, G.; Gao, J.; Ma, R.; Yang, Y.; Wu, J.; Zhang, J.; et al. Structural plasticity of the TDRD3 Tudor domain probed by a fragment screening hit. FEBS J. 2018, 285, 2091–2103. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Gao, J.; Ma, R.; Liu, Y.; Liu, M.; Zhong, F.; Hu, J.; Li, S.; Wu, J.; Jiang, H.; et al. Recent progress in fragment-based drug discovery facilitated by NMR spectroscopy. Magn. Reson. Lett. 2021, 100025, 100025. [Google Scholar] [CrossRef]
- Gao, J.; Ma, R.; Wang, W.; Wang, N.; Sasaki, R.; Snyderman, D.; Wu, J.; Ruan, K. Automated NMR Fragment Based Screening Identified a Novel Interface Blocker to the LARG/RhoA Complex. PLoS ONE 2014, 9, e88098. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Li, F.; Bao, H.; Li, J.; Wu, J.; Ruan, K. NMR Fragment Screening Hit Induces Plasticity of BRD7/9 Bromodomains. Chembiochem 2016, 17, 1456–1463. [Google Scholar] [CrossRef] [PubMed]
- Meyer, M.M.a.B. Group Epitope Mapping by Saturation Transfer Difference NMR To Identify Segments of a Ligand in Direct Contact with a Protein Receptor. J. Am. Chem. Soc. 2001, 123, 6108–6117. [Google Scholar] [CrossRef] [PubMed]
- Dalvit, C.; Fogliatto, G.; Stewart, A.; Veronesi, M.; Stockman, B. WaterLOGSY as a method for primary NMR screening: Practical aspects and range of applicability. J. Biomol. NMR 2001, 21, 349–359. [Google Scholar] [CrossRef]
- Johnson, J.A.; Olson, N.M.; Tooker, M.J.; Bur, S.K.; Pomerantz, W.C.K. Combined Protein- and Ligand-Observed NMR Workflow to Screen Fragment Cocktails against Multiple Proteins: A Case Study Using Bromodomains. Molecules 2020, 25, 3949. [Google Scholar] [CrossRef]
- Chen, J.; Wang, Z.; Guo, X.; Li, F.; Wei, Q.; Chen, X.; Gong, D.; Xu, Y.; Chen, W.; Liu, Y.; et al. TRIM66 reads unmodified H3R2K4 and H3K56ac to respond to DNA damage in embryonic stem cells. Nat. Commun. 2019, 10, 4273. [Google Scholar] [CrossRef] [Green Version]
- Hajduk, P.J.; Olejniczak, E.T.; Fesik, S.W. One-Dimensional Relaxation- and Diffusion-Edited NMR Methods for Screening Compounds That Bind to Macromolecules. J. Am. Chem. Soc. 1997, 119, 12257–12261. [Google Scholar] [CrossRef]
- Liu, J.; Li, F.; Bao, H.; Jiang, Y.; Zhang, S.; Ma, R.; Gao, J.; Wu, J.; Ruan, K. The polar warhead of a TRIM 24 bromodomain inhibitor rearranges a water-mediated interaction network. FEBS J. 2017, 284, 1082–1095. [Google Scholar] [CrossRef]
- Williamson, M.P. Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Magn. Reson. Spectrosc. 2013, 73, 1–16. [Google Scholar] [CrossRef]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar] [PubMed]
- Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994, 50, 760–763. [CrossRef] [PubMed]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Struct. Biol. Crystallogr. 2004, D60, 2126–2132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; Liu, J.; Hong, W.; Fei, X.; Liu, R. Arctigenin Inhibits Glioblastoma Proliferation through the AKT/mTOR Pathway and Induces Autophagy. Biomed. Res. Int. 2020, 2020, 3542613. [Google Scholar] [CrossRef] [PubMed]

| ID | Structure | Kd (mM) a | LE | ID | Structure | Kd (mM) | LE |
|---|---|---|---|---|---|---|---|
| 1 |  | 0.137 ± 0.024 b | 0.38 | 8 | 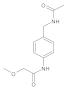 | 0.31 ± 0.004 | 0.36 |
| 2 | 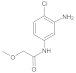 | 0.07 ± 0.01 | 0.41 | 9 |  | 0.66 ± 0.07 | 0.26 |
| 3 | 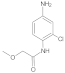 | 0.10 ± 0.01 | 0.39 | 10 |  | 0.038 ± 0.006 | 0.42 |
| 4 | 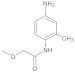 | 0.44 ± 0.10 | 0.33 | 11 | 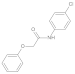 | 0.088 ± 0.03 | 0.31 |
| 5 |  | 0.08 ± 0.06 | 0.43 | 12 | 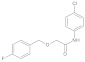 | 0.087 ± 0.011 | 0.28 |
| 6 | 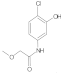 | 0.061 ± 0.004 | 0.41 | 13 | 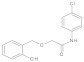 | 0.07 ± 0.009 | 0.28 |
| 7 | 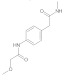 | 0.27 ± 0.02 | 0.29 | 14 | 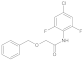 | 0.026 ± 0.004 | 0.30 |
| PDB ID | 7VKH | 7VKG |
|---|---|---|
| Data Collection | ||
| Space group | P1 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | 31.652, 41.426, 59.725 | 41.381, 31.553, 89.378 |
| α, β, γ (°) | 102, 90.87, 90.357 | 90, 102.04, 90 |
| Wavelength (Å) | 0.9774 | 0.9785 |
| Resolution (Å) | 40.00–2.25 (2.29–2.25) * | 40.00–1.83 (1.86–1.83) * |
| Completeness (%) | 97.6 (93.6) | 99.1 (97.4) |
| Redundancy | 3.3 (2.5) | 6.6 (5.9) |
| Rsym or Rmerge (%) | 11.1 (38.1) | 7.9 (65.2) |
| I/σI | 9.26 (2.05) | 20.44 (2.67) |
| Refinement | ||
| No. reflections used/free | 13,767/703 | 13,843/649 |
| Rwork/Rfree (%) | 20.61/24.45 | 20.20/24.54 |
| R.m.s.deviations | ||
| Bond lengths (Å) | 0.003 | 0.004 |
| Bond angles (°) | 0.669 | 0.767 |
| B-factors (Å2) | ||
| Protein | 28.08 | 26.00 |
| Ligand | 33.16 | 31.26 |
| Water | 26.91 | 32.43 |
| No. atoms | ||
| Protein | 2228 | 1116 |
| Ligand | 28 | 19 |
| Water | 35 | 61 |
| Ramachandran plot | ||
| Favored/allowed/outlier (%) | 98.87/1.13/0 | 100/0/0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Jin, R.; Lu, H.; Bian, K.; Wang, R.; Wang, L.; Gao, R.; Zhang, J.; Wu, J.; Yao, X.; et al. Fragment-Based Discovery of AF9 YEATS Domain Inhibitors. Int. J. Mol. Sci. 2022, 23, 3893. https://doi.org/10.3390/ijms23073893
Liu Y, Jin R, Lu H, Bian K, Wang R, Wang L, Gao R, Zhang J, Wu J, Yao X, et al. Fragment-Based Discovery of AF9 YEATS Domain Inhibitors. International Journal of Molecular Sciences. 2022; 23(7):3893. https://doi.org/10.3390/ijms23073893
Chicago/Turabian StyleLiu, Yaqian, Ruoxing Jin, Hui Lu, Kangjie Bian, Rui Wang, Lei Wang, Rui Gao, Jiahai Zhang, Jihui Wu, Xuebiao Yao, and et al. 2022. "Fragment-Based Discovery of AF9 YEATS Domain Inhibitors" International Journal of Molecular Sciences 23, no. 7: 3893. https://doi.org/10.3390/ijms23073893
APA StyleLiu, Y., Jin, R., Lu, H., Bian, K., Wang, R., Wang, L., Gao, R., Zhang, J., Wu, J., Yao, X., Liu, X., Liu, D., Wang, X., Zhang, Z., & Ruan, K. (2022). Fragment-Based Discovery of AF9 YEATS Domain Inhibitors. International Journal of Molecular Sciences, 23(7), 3893. https://doi.org/10.3390/ijms23073893







