MicroRNAs Expression in Response to rhNGF in Epithelial Corneal Cells: Focus on Neurotrophin Signaling Pathway
Abstract
:1. Introduction
2. Results
2.1. MiRNA Expression in HCEpiC Cells in Response to rhNGF
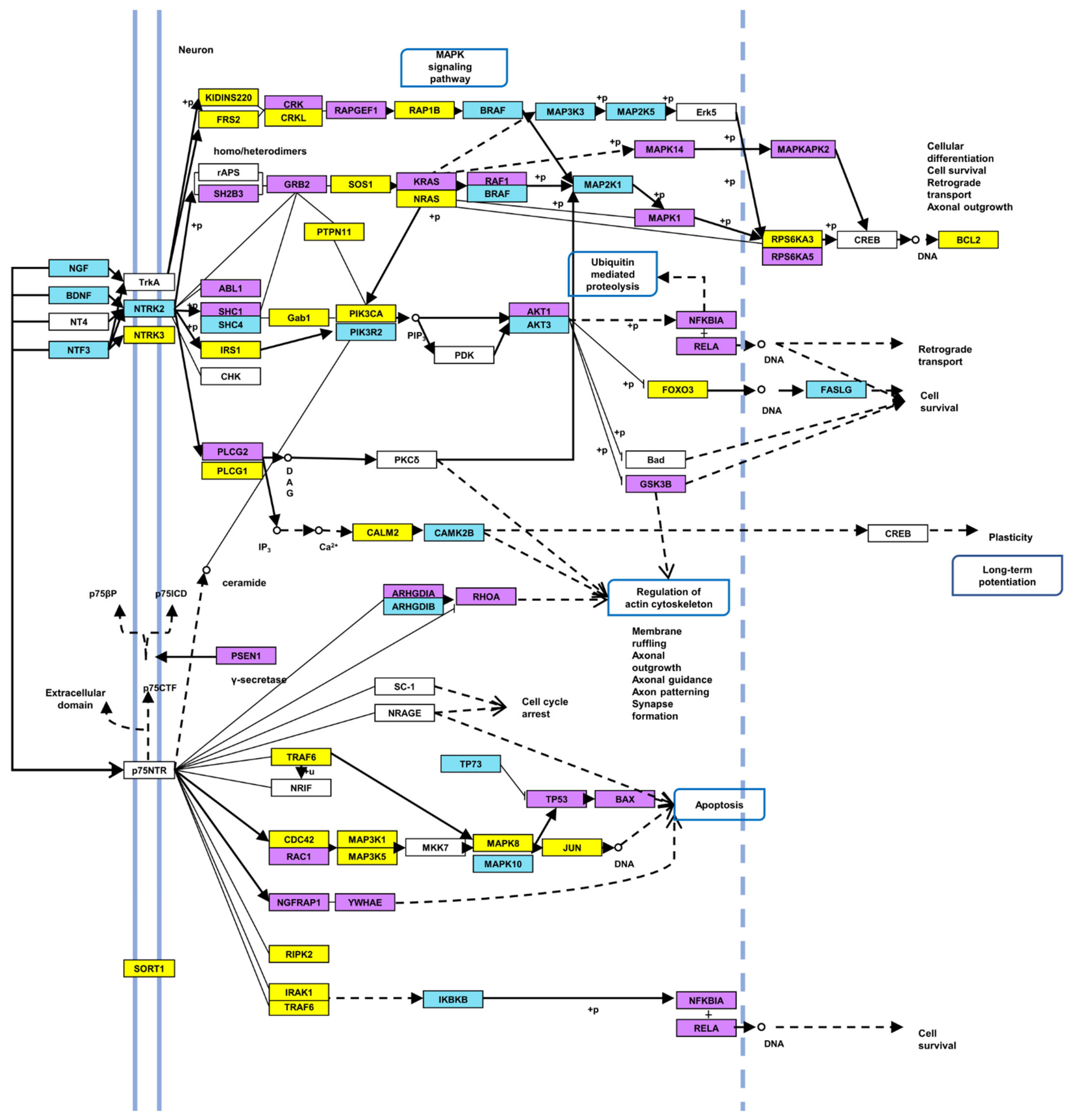
2.2. Identification of Target Genes and Pathways
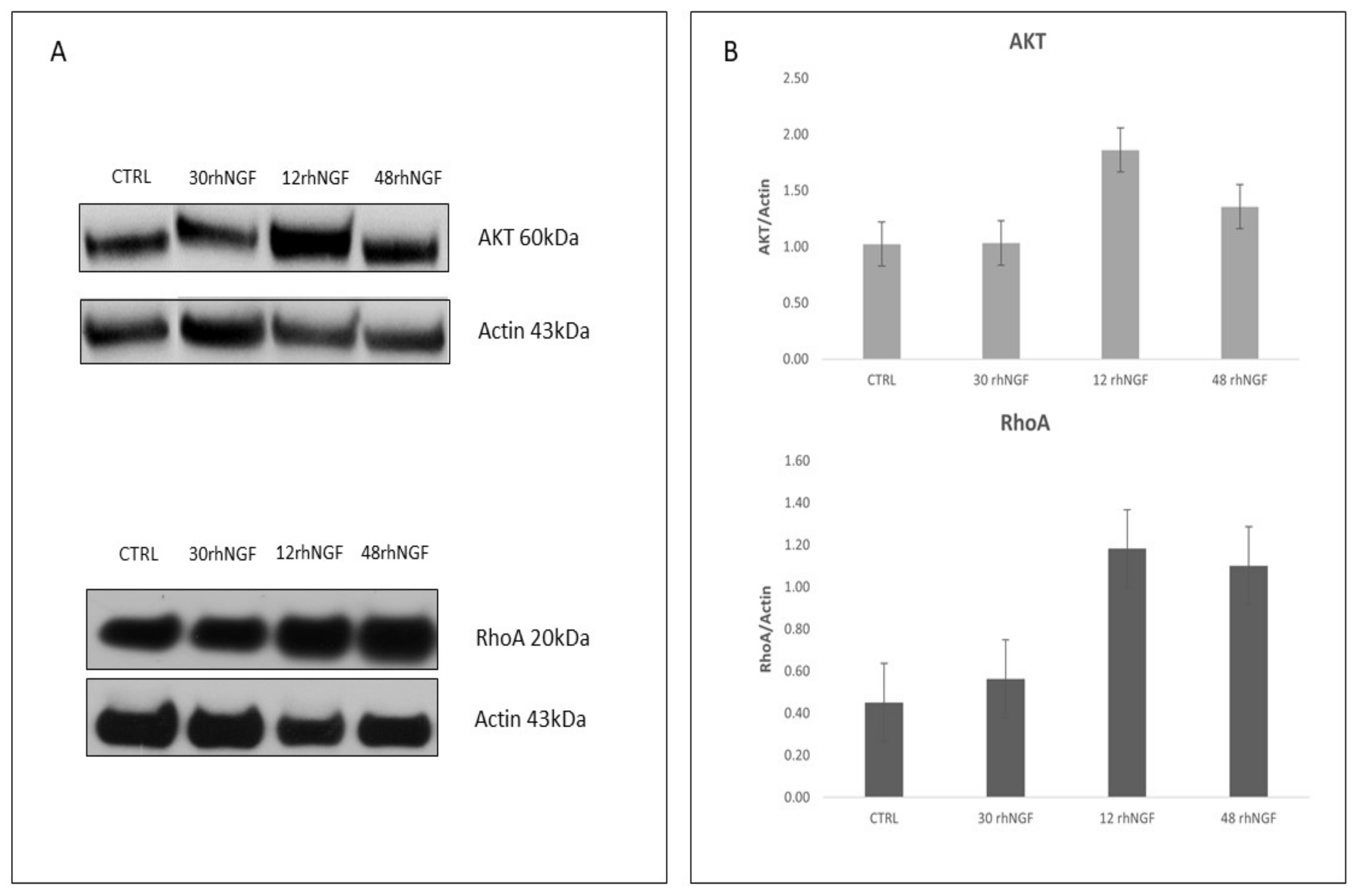
2.3. Protein Expression Levels of Target AKT and RhoA
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Treatment
4.2. Total RNA Isolation
4.3. Reverse Transcription and TaqMan miRNA Array
4.4. Statistics and Target Genes/Pathways Analysis
4.5. Western Blot Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wiesmann, C.; de Vos, A.M. Nerve growth factor: Structure and Function. Cell Mol. Life Sci. 2001, 58, 748–759. [Google Scholar] [CrossRef] [PubMed]
- Aloe, L.; Rocco, M.L.; Bianchi, P.; Manni, L. Nerve growth factor: From the Early Discoveries to the Potential Clinical Use. J. Transl. Med. 2012, 10, 239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernabei, R.; Landi, F.; Bonini, S.; Onder, G.; Lambiase, A.; Pola, R.; Aloe, L. Effect of topical application of nerve-growth factor on pressure ulcers. Lancet 1999, 354, 307. [Google Scholar] [CrossRef]
- Tuveri, M.; Generini, S.; Matucci-Cerinic, M.; Aloe, L. NGF, a useful tool in the treatment of chronic vasculitic ulcers in rheumatoid arthritis. Lancet 2000, 356, 1739–1740. [Google Scholar] [CrossRef]
- Chiaretti, A.; Piastra, M.; Caresta, E.; Nanni, L.; Aloe, L. Improving ischaemic skin revascularisation by nerve growth factor in a child with crush syndrome. Arch. Dis. Child. 2002, 87, 446–448. [Google Scholar] [PubMed] [Green Version]
- Landi, F.; Aloe, L.; Russo, A.; Cesari, M.; Onder, G.; Bonini, S.; Carbonin, P.U.; Bernabei, R. Topical treatment of pressure ulcers with nerve growth factor: A Randomized Clinical Trial. Ann. Intern. Med. 2003, 139, 635–641. [Google Scholar] [CrossRef]
- Generini, S.; Tuveri, M.A.; Matucci Cerinic, M.; Mastinu, F.; Manni, L.; Aloe, L. Topical application of nerve growth factor in human diabetic foot ulcers. A study of three cases. Exp. Clin. Endocrinol. Diabetes 2004, 112, 542–544. [Google Scholar] [CrossRef]
- Liu, Z.; Wu, H.; Huang, S. Role of NGF and its receptors in wound healing (Review). Exp. Ther. Med. 2021, 21, 599. [Google Scholar] [CrossRef]
- Lambiase, A.; Rama, P.; Bonini, S.; Caprioglio, G.; Aloe, L. Topical treatment with nerve growth factor for corneal neurotrophic ulcers. N. Engl. J. Med. 1998, 338, 1174–1180. [Google Scholar]
- Lambiase, A.; Manni, L.; Bonini, S.; Rama, P.; Micera, A.; Aloe, L. Nerve growth factor promotes corneal healing: Structural, Biochemical, and Molecular Analyses of Rat and Human Corneas. Investig. Ophthalmol. Vis. Sci. 2000, 41, 1063–1069. [Google Scholar]
- Bonini, S.; Lambiase, A.; Rama, P.; Caprioglio, G.; Aloe, L. Topical treatment with nerve growth factor for neurotrophic keratitis. Ophthalmology 2000, 107, 1347–1351. [Google Scholar] [CrossRef]
- Lambiase, A.; Mantelli, F.; Sacchetti, M.; Rossi, S.; Aloe, L.; Bonini, S. Clinical applications of NGF in ocular diseases. Arch. Ital. Biol. 2011, 149, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Lambiase, A.; Sacchetti, M.; Bonini, S. Nerve growth factor therapy for corneal disease. Curr. Opin. Ophthalmol. 2012, 23, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Versura, P.; Giannaccare, G.; Pellegrini, M.; Sebastiani, S.; Campos, E.C. Neurotrophic keratitis: Current Challenges and Future Prospects. Eye Brain 2018, 10, 37–45. [Google Scholar] [CrossRef] [Green Version]
- Bonini, S.; Lambiase, A.; Rama, P.; Filatori, I.; Allegretti, M.; Chao, W.; Mantelli, F.; REPARO Study Group. Phase I Trial of Recombinant Human Nerve Growth Factor for Neurotrophic Keratitis. Ophthalmology 2018, 125, 1468–1471. [Google Scholar] [CrossRef] [Green Version]
- Bonini, S.; Lambiase, A.; Rama, P.; Sinigaglia, F.; Allegretti, M.; Chao, W.; Mantelli, F.; REPARO Study Group. Phase II Randomized, Double-Masked, Vehicle-Controlled Trial of Recombinant Human Nerve Growth Factor for Neurotrophic Keratitis. Ophthalmology 2018, 125, 1332–1343. [Google Scholar] [CrossRef] [Green Version]
- Eftimiadi, G.; Soligo, M.; Manni, L.; Di Giuda, D.; Calcagni, M.L.; Chiaretti, A. Topical delivery of nerve growth factor for treatment of ocular and brain disorders. Neural Regen. Res. 2021, 16, 1740–1750. [Google Scholar] [CrossRef]
- Sacchetti, M.; Lambiase, A.; Schmidl, D.; Schmetterer, L.; Ferrari, M.; Mantelli, F.; Allegretti, M.; Garhoefer, G. Effect of recombinant human nerve growth factor eye drops in patients with dry eye: A Phase IIa, Open Label, Multiple-Dose Study. Br. J. Ophthalmol. 2020, 104, 127–135. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Kowdley, K.V. MicroRNAs in common human diseases. Genom. Proteom. Bioinform. 2012, 10, 246–253. [Google Scholar] [CrossRef] [Green Version]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Etheridge, A.; Lee, I.; Hood, L.; Galas, D.; Wang, K. Extracellular microRNA: A New Source of Biomarkers. Mutat. Res. 2011, 717, 85–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell. Biol. 2011, 13, 423–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Zhu, D.; Huang, L.; Zhang, J.; Bian, Z.; Chen, X.; Liu, Y.; Zhang, C.Y.; Zen, K. Argonaute 2 complexes selectively protect the circulating microRNAs in cell-secreted microvesicles. PLoS ONE 2012, 7, e46957. [Google Scholar] [CrossRef]
- Sanz-Rubio, D.; Martin-Burriel, I.; Gil, A.; Cubero, P.; Forner, M.; Khalyfa, A.; Marin, J.M. Stability of Circulating Exosomal miRNAs in Healthy Subjects. Sci. Rep. 2018, 8, 10306. [Google Scholar] [CrossRef] [Green Version]
- Pouw, A.E.; Greiner, M.A.; Coussa, R.G.; Jiao, C.; Han, I.C.; Skeie, J.M.; Fingert, J.H.; Mullins, R.F.; Sohn, E.H. Cell-Matrix Interactions in the Eye: From Cornea to Choroid. Cells 2021, 10, 687. [Google Scholar] [CrossRef]
- Kimura, K.; Kawano, S.; Mori, T.; Inoue, J.; Hadachi, H.; Saito, T.; Nishida, T. Quantitative analysis of the effects of extracellular matrix proteins on membrane dynamics associated with corneal epithelial cell motility. Investig. Ophthalmol. Vis. Sci. 2010, 51, 4492–4499. [Google Scholar] [CrossRef]
- Barbariga, M.; Vallone, F.; Mosca, E.; Bignami, F.; Magagnotti, C.; Fonteyne, P.; Chiappori, F.; Milanesi, L.; Rama, P.; Andolfo, A.; et al. The role of extracellular matrix in mouse and human corneal neovascularization. Sci. Rep. 2019, 9, 14272. [Google Scholar] [CrossRef] [Green Version]
- Kowtharapu, B.S.; Murín, R.; Jünemann, A.G.M.; Stachs, O. Role of Corneal Stromal Cells on Epithelial Cell Function during Wound Healing. Int. J. Mol. Sci. 2018, 19, 464. [Google Scholar] [CrossRef] [Green Version]
- McKay, T.B.; Schlötzer-Schrehardt, U.; Pal-Ghosh, S.; Stepp, M.A. Integrin: Basement membrane adhesion by corneal epithelial and endothelial cells. Exp. Eye Res. 2020, 198, 108138. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Fu, W.; Liu, Y.; Wang, Q.; Wang, L.; Huang, Y. Agrin Promotes Limbal Stem Cell Proliferation and Corneal Wound Healing Through Hippo-Yap Signaling Pathway. Investig. Ophthalmol. Vis. Sci. 2020, 61, 7. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ge, L.; Chen, X.; Mao, Y.; Gu, X.; Ren, B.; Zeng, Y.; Chen, M.; Chen, S.; Liu, J.; et al. The common YAP activation mediates corneal epithelial regeneration and repair with different-sized wounds. NPJ Regen. Med. 2021, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.G.; Li, J.S.; Liu, Y.F.; Xu, Q. MicroRNA-200b suppresses the invasion and migration of hepatocellular carcinoma by downregulating RhoA and circRNA_000839. Tumour. Biol. 2017, 39, 1010428317719577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mastropasqua, L.; Massaro-Giordano, G.; Nubile, M.; Sacchetti, M. Understanding the Pathogenesis of Neurotrophic Keratitis: The Role of Corneal Nerves. J. Cell. Physiol. 2017, 232, 717–724. [Google Scholar] [CrossRef]
- Aloe, L.; Rocco, M.L.; Balzamino, B.O.; Micera, A. Nerve Growth Factor: A Focus on Neuroscience and Therapy. Curr. Neuropharmacol. 2015, 13, 294–303. [Google Scholar] [CrossRef] [Green Version]
- Rassi, D.M.; De Paiva, C.S.; Dias, L.C.; Módulo, C.M.; Adriano, L.; Fantucci, M.Z.; Rocha, E.M. Review: MicroRNAS in Ocular Surface and Dry Eye Diseases. Ocul. Surf. 2017, 15, 660–669. [Google Scholar] [CrossRef]
- Funari, V.A.; Winkler, M.; Brown, J.; Dimitrijevich, S.D.; Ljubimov, A.V.; Saghizadeh, M. Differentially expressed wound healing-related microRNAs in the human diabetic cornea. PLoS ONE 2013, 8, e84425. [Google Scholar] [CrossRef] [Green Version]
- Winkler, M.A.; Dib, C.; Ljubimov, A.V.; Saghizadeh, M. Targeting miR-146a to treat delayed wound healing in human diabetic organ-cultured corneas. PLoS ONE 2014, 9, e114692. [Google Scholar] [CrossRef] [Green Version]
- Pauley, K.M.; Stewart, C.M.; Gauna, A.E.; Dupre, L.C.; Kuklani, R.; Chan, A.L.; Pauley, B.A.; Reeves, W.H.; Chan, E.K.; Cha, S. Altered miR-146a expression in Sjögren’s syndrome and its functional role in innate immunity. Eur. J. Immunol. 2011, 41, 2029–2039. [Google Scholar] [CrossRef] [Green Version]
- Zilahi, E.; Tarr, T.; Papp, G.; Griger, Z.; Sipka, S.; Zeher, M. Increased microRNA-146a/b, TRAF6 gene and decreased IRAK1 gene expressions in the peripheral mononuclear cells of patients with Sjögren’s syndro. Immunol. Lett. 2012, 141, 165–168. [Google Scholar] [CrossRef] [PubMed]
- Movahedan, A.; Majdi, M.; Afsharkhamseh, N.; Sagha, H.M.; Saadat, N.S.; Shalileh, K.; Milani, B.Y.; Ying, H.; Djalilian, A.R. Notch inhibition during corneal epithelial wound healing promotes migration. Invest. Ophthalmol. Vis. Sci. 2012, 53, 7476–7483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poe, A.J.; Kulkarni, M.; Leszczynska, A.; Tang, J.; Shah, R.; Jami-Alahmadi, Y.; Wang, J.; Kramerov, A.A.; Wohlschlegel, J.; Punj, V.; et al. Integrated Transcriptome and Proteome Analyses Reveal the Regulatory Role of miR-146a in Human Limbal Epithelium via Notch Signaling. Cells 2020, 9, 2175. [Google Scholar] [CrossRef] [PubMed]
- Kuang, W.; Tan, J.; Duan, Y.; Duan, J.; Wang, W.; Jin, F.; Jin, Z.; Yuan, X.; Liu, Y. Cyclic stretch induced miR-146a upregulation delays C2C12 myogenic differentiation through inhibition of Numb. Biochem. Biophys. Res. Commun. 2009, 378, 259–263. [Google Scholar] [CrossRef] [PubMed]
- Hung, P.S.; Liu, C.J.; Chou, C.S.; Kao, S.Y.; Yang, C.C.; Chang, K.W.; Chiu, T.H.; Lin, S.C. miR-146a enhances the oncogenicity of oral carcinoma by concomitant targeting of the IRAK1, TRAF6 and NUMB genes. PLoS ONE 2013, 8, e79926. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Li, J.; Yin, L.; Pan, J.; Zhang, Y.; Jiang, Z. Role of microRNA 146a on the healing of cornea alkali burn treated with mesenchymal stem cells. Mol. Med. Rep. 2018, 18, 3203–3210. [Google Scholar] [CrossRef] [Green Version]
- Alexander, G.; Carlsen, H.; Blomhoff, R. Corneal NF-kappaB activity is necessary for the retention of transparency in the cornea of UV-B-exposed transgenic reporter mice. Exp. Eye Res. 2006, 82, 700–709. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Meng, Q.; Kao, W.; Xia, Y. IκB kinase β regulates epithelium migration during corneal wound healing. PLoS ONE 2011, 6, e16132. [Google Scholar] [CrossRef] [Green Version]
- Yao, B.; Wang, S.; Xiao, P.; Wang, Q.; Hea, Y.; Zhang, Y. MAPK signaling pathways in eye wounds: Multifunction and Cooperation. Exp. Cell. Res. 2017, 359, 10–16. [Google Scholar] [CrossRef]
- Li, J.; Qi, X.; Wang, X.; Li, W.; Li, Y.; Zhou, Q. PTEN Inhibition Facilitates Diabetic Corneal Epithelial Regeneration by Reactivating Akt Signaling Pathway. Transl. Vis. Sci. Technol. 2020, 9, 5. [Google Scholar] [CrossRef] [Green Version]
- Hong, J.; Qian, T.; Le, Q.; Sun, X.; Wu, J.; Chen, J.; Yu, X.; Xu, J. NGF promotes cell cycle progression by regulating D-type cyclins via PI3K/Akt and MAPK/Erk activation in human corneal epithelial cells. Mol. Vis. 2012, 18, 758–764. [Google Scholar] [PubMed]
- Liu, Y.; Di, G.; Wang, Y.; Chong, D.; Cao, X.; Chen, P. Aquaporin 5 Facilitates Corneal Epithelial Wound Healing and Nerve Regeneration by Reactivating Akt Signaling Pathway. Am. J. Pathol. 2021, 191, 1974–1985. [Google Scholar] [CrossRef] [PubMed]
- Sabater, A.L.; Andreu, E.J.; García-Guzmán, M.; López, T.; Abizanda, G.; Perez, V.L.; Moreno-Montañés, J.; Prósper, F. Combined PI3K/Akt and Smad2 Activation Promotes Corneal Endothelial Cell Proliferation. Investig. Ophthalmol. Vis. Sci. 2017, 58, 745–754. [Google Scholar] [CrossRef]
- Park, J.H.; Kang, S.S.; Kim, J.Y.; Tchah, H. Nerve Growth Factor Attenuates Apoptosis and Inflammation in the Diabetic Cornea. Investig. Ophthalmol. Vis. Sci. 2016, 57, 6767–6775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.T.; Han, B.; Li, F.; Chen, S.Y.; Tighe, S.; Zhang, S.; Tseng, S.C. Knockdown of both p120 catenin and Kaiso promotes expansion of human corneal endothelial monolayers via RhoA-ROCK-noncanonical BMP-NFκB pathway. Investig. Ophthalmol. Vis. Sci. 2014, 55, 1509–1518. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.H.; Hu, Z.X.; Gao, Z.X.; Song, X.L.; Feng, Q.Y.; Yang, G.; Li, Z.J.; Pan, H.W. Airborne particulate matter impairs corneal epithelial cells migration via disturbing FAK/RhoA signaling pathway and cytoskeleton organization. Nanotoxicology 2018, 12, 312–324. [Google Scholar] [CrossRef] [PubMed]
- Ortega, M.C.; Santander-García, D.; Marcos-Ramiro, B.; Barroso, S.; Cox, S.; Jiménez-Alfaro, I.; Millán, J. Activation of Rac1 and RhoA Preserve Corneal Endothelial Barrier Function. Investig. Ophthalmol. Vis. Sci. 2016, 57, 6210–6222. [Google Scholar] [CrossRef] [Green Version]
- Kimura, K.; Kawamoto, K.; Teranishi, S.; Nishida, T. Role of Rac1 in fibronectin-induced adhesion and motility of human corneal epithelial cells. Investig. Ophthalmol. Vis. Sci. 2006, 47, 4323–4329. [Google Scholar] [CrossRef] [Green Version]
- Pothula, S.; Bazan, H.E.; Chandrasekher, G. Regulation of Cdc42 expression and signaling is critical for promoting corneal epithelial wound healing. Investig. Ophthalmol. Vis. Sci. 2013, 54, 5343–5352. [Google Scholar] [CrossRef] [Green Version]
- Rogge, M.; Yin, X.T.; Godfrey, L.; Lakireddy, P.; Potter, C.A.; Del Rosso, C.R.; Stuart, P.M. Therapeutic Use of Soluble Fas Ligand Ameliorates Acute and Recurrent Herpetic Stromal Keratitis in Mice. Investig. Ophthalmol. Vis. Sci. 2015, 56, 6377–6386. [Google Scholar] [CrossRef] [Green Version]
- Aluru, S.V.; Shweta, A.; Bhaskar, S.; Geetha, K.; Sivakumar, R.M.; Utpal, T.; Padmanabhan, P.; Angayarkanni, N. Tear Fluid Protein Changes in Dry Eye Syndrome Associated with Rheumatoid Arthritis: A Proteomic Approach. Ocul. Surf. 2017, 15, 112–129. [Google Scholar] [CrossRef] [PubMed]
- Tessitore, A.; Cicciarelli, G.; Mastroiaco, V.; Vecchio, F.D.; Capece, D.; Verzella, D.; Fischietti, M.; Vecchiotti, D.; Zazzeroni, F.; Alesse, E. Therapeutic Use of MicroRNAs in Cancer. Anticancer Agents Med. Chem. 2016, 16, 7–19. [Google Scholar] [CrossRef] [PubMed]
- Lam, J.K.; Chow, M.Y.; Zhang, Y.; Leung, S.W. siRNA Versus miRNA as Therapeutics for Gene Silencing. Mol. Ther. Nucleic Acids 2015, 4, e252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a New Era for the Management of Cancer and Other Diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Zhou, S.S.; Jin, J.P.; Wang, J.Q.; Zhang, Z.G.; Freedman, J.H.; Zheng, Y.; Cai, L. miRNAS in cardiovascular diseases: Potential Biomarkers, Therapeutic Targets and Challenges. Acta Pharmacol. Sin. 2018, 39, 1073–1084. [Google Scholar] [CrossRef] [Green Version]
- Shah, V.; Shah, J. Recent trends in targeting miRNAs for cancer therapy. J. Pharm. Pharmacol. 2020, 72, 1732–1749. [Google Scholar] [CrossRef]
- Forterre, A.; Komuro, H.; Aminova, S.; Harada, M. A Comprehensive Review of Cancer MicroRNA Therapeutic Delivery Strategies. Cancers 2020, 12, 1852. [Google Scholar] [CrossRef]
- Askou, A.L.; Alsing, S.; Holmgaard, A.; Bek, T.; Corydon, T.J. Dissecting microRNA dysregulation in age-related macular degeneration: New Targets for Eye Gene Therapy. Acta Ophthalmol. 2018, 96, 9–23. [Google Scholar] [CrossRef] [Green Version]
- Mukwaya, A.; Jensen, L.; Peebo, B.; Lagali, N. MicroRNAs in the cornea: Role and implications for treatment of corneal neovascularization. Ocul. Surf. 2019, 17, 400–411. [Google Scholar] [CrossRef]
- Liu, C.H.; Huang, S.; Britton, W.R.; Chen, J. MicroRNAs in Vascular Eye Diseases. Int. J. Mol. Sci. 2020, 21, 649. [Google Scholar] [CrossRef] [Green Version]
- You, L.; Kruse, F.E.; Völcker, H.E. Neurotrophic factors in the human cornea. Investig. Ophthalmol. Vis. Sci. 2000, 41, 692–702. [Google Scholar] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath v3.0: Deciphering microRNA Function with Experimental Support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef] [PubMed]
- Paraskevopoulou, M.D.; Georgakilas, G.; Kostoulas, N.; Vlachos, I.S.; Vergoulis, T.; Reczko, M.; Filippidis, C.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-microT web server v5.0: Service Integration into miRNA Functional Analysis Workflows. Nucleic Acids Res. 2013, 41, W169–W173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karagkouni, D.; Paraskevopoulou, M.D.; Chatzopoulos, S.; Vlachos, I.S.; Tastsoglou, S.; Kanellos, I.; Papadimitriou, D.; Kavakiotis, I.; Maniou, S.; Skoufos, G.; et al. DIANA-TarBase v8: A Decade-Long Collection of Experimentally Supported miRNA-Gene Interactions. Nucleic Acids Res. 2018, 46, D239–D245. [Google Scholar] [CrossRef] [Green Version]
- Robinson, I.; Webber, J.; Eifrem, E. Graph Databases, 2nd ed.; O’Reilly Media, Inc.: Sebastopol, CA, USA, 2013. [Google Scholar]
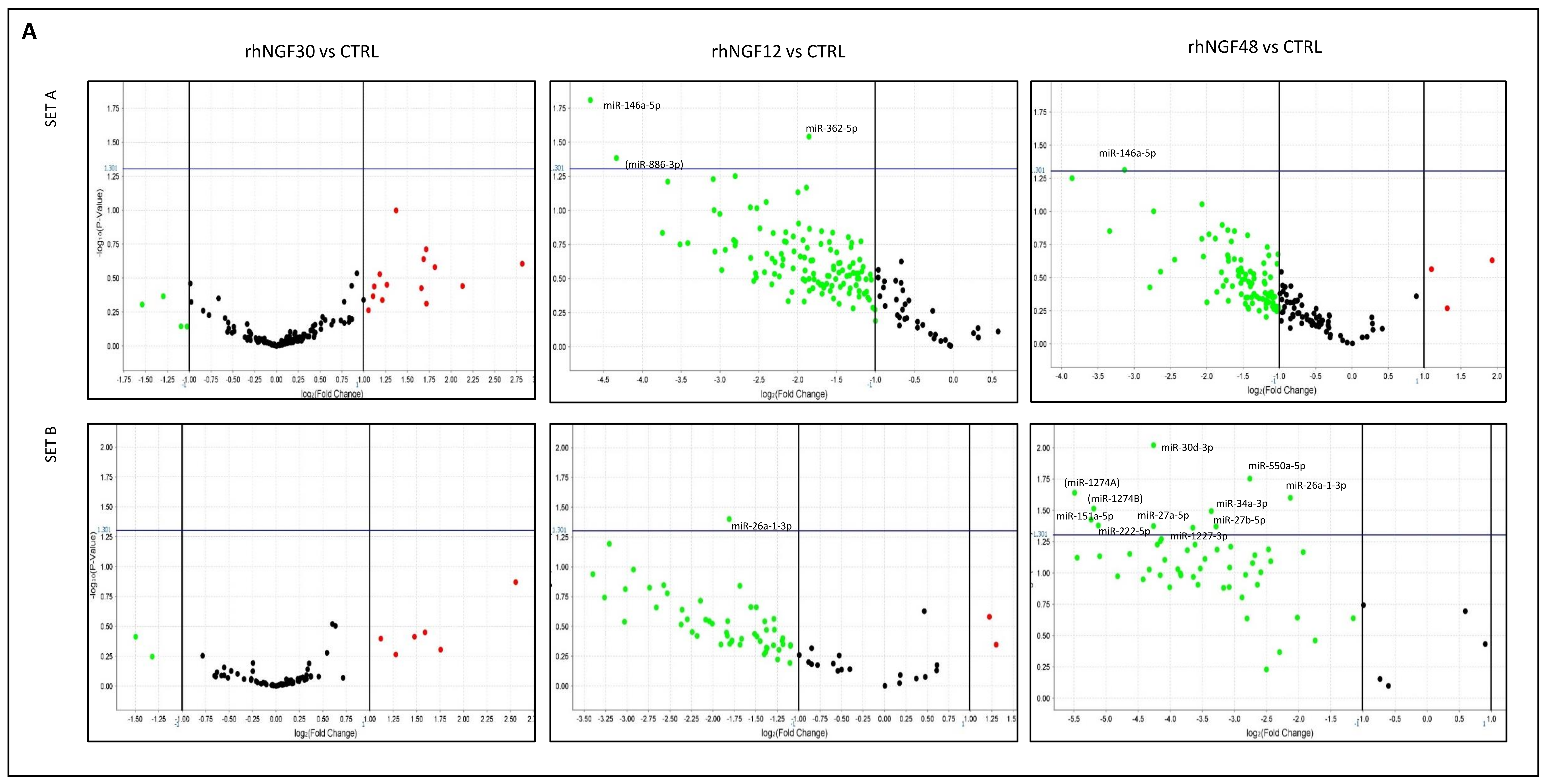
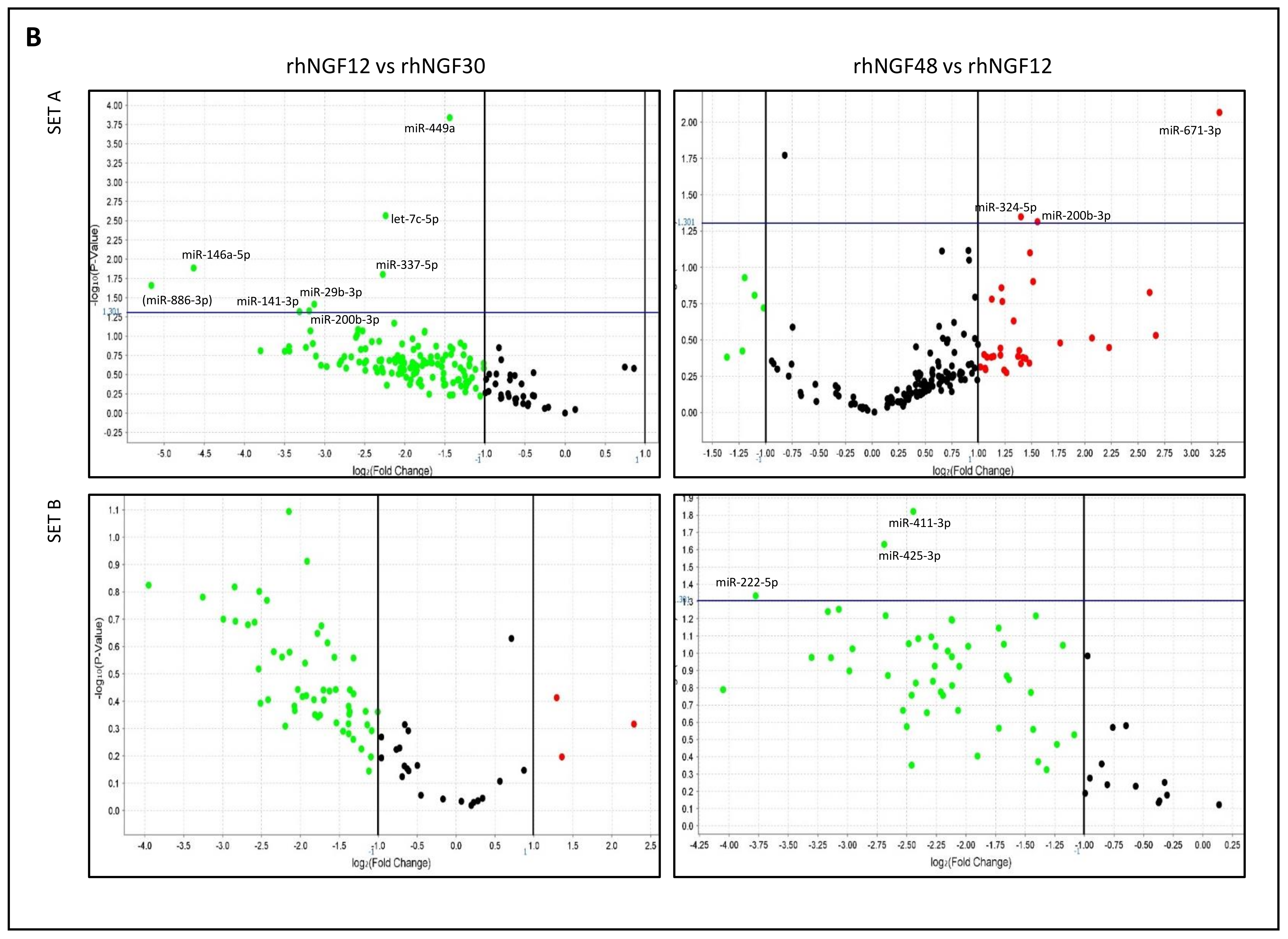

| miRNA miRbase ID | NGF30 | p-Value | NGF12 | p-Value | NGF48 | p-Value | NGF12vs30 | p-Value | NGF48vs12 | p-Value |
|---|---|---|---|---|---|---|---|---|---|---|
| hsa-let7c-5p | 7.035 | 0.248 | 1.491 | 0.773 | 2.485 | 0.538 | 0.212 | 0.003 | 1.629 | 0.330 |
| hsa-mir-29b-3p | 4.379 | 0.362 | 0.5 | 0.645 | 0.955 | 0.977 | 0.114 | 0.038 | 1.917 | 0.536 |
| hsa-miR-449a | 3.279 | 0.194 | 1.199 | 0.797 | 1.85 | 0.436 | 0.368 | 0.000 | 1.548 | 0.255 |
| hsa-miR-337-5p | 3.215 | 0.229 | 0.661 | 0.617 | 2.105 | 0.980 | 0.207 | 0.016 | 3.414 | 0.980 |
| hsa-miR-671-3p | 1.819 | 0.634 | 0.399 | 0.374 | 3.819 | 0.234 | 0.219 | 0.207 | 9.616 | 0.009 |
| hsa-mir-1227-3p | 1.292 | 0.823 | 0.44 | 0.397 | 0.08 | 0.044 | 0.343 | 0.361 | 0.179 | 0.088 |
| hsa-miR-26a-1-3p | 1.264 | 0.724 | 0.285 | 0.04 | 0.228 | 0.025 | 0.226 | 0.080 | 0.799 | 0.560 |
| hsa-miR-27b-5p | 1.175 | 0.870 | 0.34 | 0.218 | 0.102 | 0.043 | 0.291 | 0.225 | 0.303 | 0.071 |
| hsa-miR-141-3p | 1.175 | 0.867 | 0.118 | 0.059 | 0.514 | 0.578 | 0.101 | 0.048 | 4.192 | 0.306 |
| hsa-miR-200b-3p | 1.139 | 0.903 | 0.125 | 0.106 | 0.373 | 0.340 | 0.109 | 0.047 | 2.936 | 0.049 |
| hsa-mir-425-3p | 1.043 | 0.973 | 0.661 | 0.649 | 0.104 | 0.065 | 0.631 | 0.687 | 0.155 | 0.023 |
| hsa-mir-550a-5p | 1.038 | 0.961 | 0.311 | 0.144 | 0.147 | 0.018 | 0.301 | 0.211 | 0.471 | 0.296 |
| hsa-mir-222-5p | 1.013 | 0.993 | 0.388 | 0.482 | 0.029 | 0.042 | 0.384 | 0.523 | 0.073 | 0.046 |
| hsa-mir-34a-3p | 0.974 | 0.976 | 0.168 | 0.143 | 0.097 | 0.032 | 0.173 | 0.158 | 0.572 | 0.578 |
| hsa-miR-146a-5p | 0.973 | 0.976 | 0.039 | 0.015 | 0.114 | 0.049 | 0.04 | 0.013 | 2.854 | 0.125 |
| hsa-mir-151a-5p | 0.968 | 0.984 | 0.236 | 0.278 | 0.027 | 0.038 | 0.244 | 0.361 | 0.111 | 0.057 |
| hsa-mir-27a-5p | 0.863 | 0.909 | 0.226 | 0.193 | 0.052 | 0.042 | 0.261 | 0.289 | 0.23 | 0.105 |
| hsa-mir-411-3p | 0.725 | 0.980 | 1.391 | 0.840 | 0.203 | 0.430 | 1.927 | 0.980 | 0.184 | 0.015 |
| hsa-miR-324-5p | 0.712 | 0.722 | 0.173 | 0.097 | 0.446 | 0.186 | 0.245 | 0.221 | 2.639 | 0.045 |
| hsa-miR-30d-3p | 0.682 | 0.682 | 0.194 | 0.304 | 0.052 | 0.01 | 0.285 | 0.446 | 0.268 | 0.394 |
| hsa-miR-362-5p | 0.504 | 0.347 | 0.276 | 0.029 | 0.271 | 0.160 | 0.548 | 0.418 | 0.939 | 0.940 |
| KEGG Pathway | p-Value | #genes | #miRNAs |
|---|---|---|---|
| ECM-receptor interaction | 1.94 × 10−31 | 39 | 15 |
| Adherens junction | 1.37 × 10−11 | 47 | 16 |
| Proteoglycans in cancer | 1.75 × 10−9 | 85 | 19 |
| Prion diseases | 3.82 × 10−9 | 12 | 15 |
| Viral carcinogenesis | 5.99 × 10−9 | 91 | 17 |
| Focal adhesion | 1.90 × 10−7 | 102 | 18 |
| Protein processing in endoplasmic reticulum | 2.99 × 10−7 | 87 | 18 |
| Pathways in cancer | 1.04 × 10−6 | 163 | 19 |
| Fatty acid biosynthesis | 1.76 × 10−6 | 4 | 5 |
| Chronic myeloid leukemia | 3.63 × 10−6 | 42 | 17 |
| Cell cycle | 5.44 × 10−6 | 62 | 17 |
| Glioma | 5.44 × 10−6 | 34 | 18 |
| Renal cell carcinoma | 5.44 × 10−6 | 38 | 19 |
| Hepatitis B | 5.79 × 10−6 | 66 | 17 |
| Oocyte meiosis | 6.01 × 10−6 | 55 | 18 |
| Bacterial invasion of epithelial cells | 6.63 × 10−6 | 41 | 16 |
| Ubiquitin mediated proteolysis | 7.71 × 10−6 | 70 | 17 |
| Prostate cancer | 1.30 × 10−5 | 47 | 17 |
| Neurotrophin signaling pathway | 2.11 × 10−5 | 60 | 18 |
| Small cell lung cancer | 3.09 × 10−5 | 46 | 17 |
| PI3K-Akt signaling pathway | 4.53 × 10−5 | 141 | 18 |
| Transcriptional misregulation in cancer | 7.92 × 10−5 | 70 | 19 |
| Hippo signaling pathway | 9.95 × 10−5 | 59 | 18 |
| Central carbon metabolism in cancer | 0.000127 | 34 | 16 |
| p53 signaling pathway | 0.000134 | 37 | 17 |
| Shigellosis | 0.000227 | 33 | 14 |
| Colorectal cancer | 0.000227 | 33 | 16 |
| Lysine degradation | 0.000344 | 20 | 14 |
| FoxO signaling pathway | 0.000344 | 62 | 17 |
| Endocytosis | 0.000446 | 84 | 17 |
| Acute myeloid leukemia | 0.000496 | 29 | 17 |
| Endometrial cancer | 0.000515 | 27 | 16 |
| Pancreatic cancer | 0.000515 | 34 | 17 |
| TGF-beta signaling pathway | 0.000952 | 36 | 17 |
| Epstein–Barr virus infection | 0.001042 | 87 | 17 |
| Sulfur metabolism | 0.004825 | 5 | 5 |
| Fatty acid elongation | 0.005702 | 6 | 6 |
| Non-small cell lung cancer | 0.005702 | 27 | 17 |
| Bladder cancer | 0.005702 | 22 | 18 |
| NF-kappa B signaling pathway | 0.006256 | 32 | 17 |
| Arrhythmogenic right ventricular cardiomyopathy (ARVC) | 0.006413 | 26 | 16 |
| HIF-1 signaling pathway | 0.006698 | 47 | 18 |
| Spliceosome | 0.007512 | 56 | 18 |
| Amoebiasis | 0.011039 | 41 | 14 |
| Sphingolipid signaling pathway | 0.011162 | 49 | 18 |
| Regulation of actin cytoskeleton | 0.019688 | 79 | 16 |
| AMPK signaling pathway | 0.019688 | 53 | 17 |
| Thyroid hormone signaling pathway | 0.019688 | 52 | 19 |
| Circadian rhythm | 0.023121 | 16 | 13 |
| mRNA surveillance pathway | 0.024669 | 40 | 16 |
| RNA transport | 0.024669 | 64 | 18 |
| Pathogenic Escherichia coli infection | 0.024739 | 26 | 13 |
| Dorso-ventral axis formation | 0.024739 | 15 | 15 |
| Insulin signaling pathway | 0.024739 | 57 | 16 |
| Hepatitis C | 0.024739 | 53 | 17 |
| N-Glycan biosynthesis | 0.037600 | 19 | 14 |
| Prolactin signaling pathway | 0.039423 | 32 | 17 |
| Thyroid cancer | 0.048754 | 14 | 16 |
| Melanoma | 0.049562 | 29 | 17 |
| KEGG Pathway | p-Value | #genes | #miRNAs |
|---|---|---|---|
| ECM-receptor interaction | 1.74 × 10−15 | 34 | 15 |
| Prion diseases | 1.12 × 10−8 | 9 | 9 |
| ErbB signaling pathway | 1.12 × 10−8 | 46 | 13 |
| Glioma | 3.13 × 10−6 | 32 | 13 |
| Focal adhesion | 3.13 × 10−6 | 92 | 16 |
| Proteoglycans in cancer | 3.13 × 10−6 | 84 | 19 |
| Renal cell carcinoma | 3.35 × 10−6 | 36 | 12 |
| Glycosaminoglycan biosynthesis—heparan sulfate/heparin | 5.01 × 10−6 | 14 | 8 |
| Choline metabolism in cancer | 1.32 × 10−5 | 51 | 14 |
| FoxO signaling pathway | 0.000104 | 58 | 14 |
| PI3K-Akt signaling pathway | 0.000104 | 127 | 20 |
| Amoebiasis | 0.000116 | 43 | 14 |
| Adherens junction | 0.000182 | 37 | 14 |
| Lysine degradation | 0.000186 | 19 | 10 |
| mTOR signaling pathway | 0.000192 | 33 | 14 |
| Thyroid hormone signaling pathway | 0.000211 | 49 | 18 |
| Ras signaling pathway | 0.000345 | 85 | 17 |
| Rap1 signaling pathway | 0.000661 | 84 | 16 |
| Axon guidance | 0.000768 | 51 | 15 |
| Glycosaminoglycan biosynthesis—keratan sulfate | 0.001064 | 8 | 6 |
| Pathways in cancer | 0.001065 | 142 | 18 |
| Adrenergic signaling in cardiomyocytes | 0.001896 | 54 | 18 |
| p53 signaling pathway | 0.004244 | 31 | 14 |
| TGF-beta signaling pathway | 0.004617 | 33 | 16 |
| Glycosaminoglycan biosynthesis—chondroitin sulfate/dermatan sulfate | 0.004672 | 8 | 6 |
| Small cell lung cancer | 0.005864 | 38 | 12 |
| Neurotrophin signaling pathway | 0.005864 | 50 | 16 |
| Hippo signaling pathway | 0.006055 | 49 | 15 |
| HIF-1 signaling pathway | 0.006421 | 45 | 14 |
| Glycosphingolipid biosynthesis—lacto and neolacto series | 0.006829 | 11 | 8 |
| AMPK signaling pathway | 0.007480 | 49 | 18 |
| Prostate cancer | 0.008563 | 38 | 13 |
| Prolactin signaling pathway | 0.008642 | 27 | 12 |
| Circadian rhythm | 0.008642 | 17 | 15 |
| cGMP-PKG signaling pathway | 0.100938 | 62 | 17 |
| Regulation of actin cytoskeleton | 0.012869 | 76 | 15 |
| Phosphatidylinositol signaling system | 0.014451 | 32 | 13 |
| Biotin metabolism | 0.014567 | 1 | 1 |
| Long-term depression | 0.014567 | 26 | 13 |
| Sphingolipid signaling pathway | 0.014740 | 44 | 16 |
| Non-small cell lung cancer | 0.019923 | 24 | 10 |
| MAPK signaling pathway | 0.021152 | 90 | 17 |
| Pancreatic cancer | 0.024846 | 26 | 10 |
| Melanoma | 0.024846 | 30 | 12 |
| Estrogen signaling pathway | 0.029488 | 35 | 15 |
| Wnt signaling pathway | 0.035309 | 55 | 16 |
| Chagas disease (American trypanosomiasis) | 0.038162 | 38 | 15 |
| Bacterial invasion of epithelial cells | 0.042141 | 28 | 13 |
| Adipocytokine signaling pathway | 0.043705 | 27 | 13 |
| miR-222-5p | miR-151a-5p | miR-29b-3p | miR-26a-1-3p | miR-30d-3p | miR- 449a | miR-550a-5p | miR-141-3p | miR-27a-5p | miR-200b-3p | miR-146a-5p | miR-324-5p | miR-425-3p | miR-362-5p | miR-411-3p | miR-27b-5p | miR-671-3p | let-7c-5p |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CALM3 | ARHGDIA | ABL1 | PTPN11 | AKT3 | ARHGDIB | MAPK10 | ABL1 | ABL1 | AKT3 | ABL1 | AKT1 | NGFRAP1 | AKT3 | CAMK4 | FASLG | GRB2 | MAP3K1 |
| CRKL | GRB2 | AKT1 | GRB2 | BCL2 | MAPKAPK2 | BAX | AKT1 | BCL2 | AKT2 | ARHGDIA | PLCG2 | CAMK2B | GSK3B | JUN | MAPK8 | ||
| KIDINS220 | MAPK1 | AKT2 | MAP3K5 | CAMK4 | PLCG1 | BCL2 | CALM2 | BDNF | BRAF | MAP3K1 | RAP1A | GSK3B | RAC1 | NRAS | NGF | ||
| MAP3K5 | NTRK2 | BAX | MAPKAPK2 | IRAK3 | RAF1 | CALM2 | CALM3 | CAMK4 | CAMK2D | NTRK3 | RAP1B | RPS6KA3 | NRAS | ||||
| MAPK1 | RAPGEF1 | BCL2 | PTPN11 | IRS1 | RPS6KA3 | CDC42 | GRB2 | CRK | GSK3B | SOS1 | TP53 | NTRK3 | |||||
| SORT1 | RIPK2 | CALM3 | SOS1 | MAP2K1 | CRK | MAPK3 | CRKL | IRAK1 | |||||||||
| SOS2 | CAMK4 | YWHAE | MAPKAPK2 | CRKL | PLCG1 | FRS2 | IRAK2 | ||||||||||
| YWHAE | CDC42 | NFKB1 | FOXO3 | RHOA | GAB1 | JUN | |||||||||||
| FOXO3 | PIK3CA | FRS2 | SH2B3 | IKBKB | MAP3K5 | ||||||||||||
| FRS2 | PIK3CB | GAB1 | SHC1 | JUN | NRAS | ||||||||||||
| GSK3B | PLCG1 | GRB2 | KIDINS220 | SORT1 | |||||||||||||
| JUN | PTPN11 | IRS1 | KRAS | TRAF6 | |||||||||||||
| KIDINS220 | RPS6KA3 | KIDINS220 | MAP2K5 | ||||||||||||||
| MAPK8 | MAP3K1 | MAP3K1 | |||||||||||||||
| NRAS | MAP3K3 | NTF3 | |||||||||||||||
| PIK3R1 | MAPK14 | PIK3CA | |||||||||||||||
| PIK3R2 | NFKBIA | PLCG1 | |||||||||||||||
| PIK3R3 | PIK3R1 | PTPN11 | |||||||||||||||
| RELA | PSEN1 | RAP1B | |||||||||||||||
| SOS1 | RAC1 | RHOA | |||||||||||||||
| TP53 | SHC1 | RIPK2 | |||||||||||||||
| YWHAE | RPS6KA3 | ||||||||||||||||
| RPS6KA5 | |||||||||||||||||
| SHC1 | |||||||||||||||||
| SHC4 | |||||||||||||||||
| SORT1 | |||||||||||||||||
| SOS1 | |||||||||||||||||
| TP73 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Compagnoni, C.; Zelli, V.; Bianchi, A.; Di Marco, A.; Capelli, R.; Vecchiotti, D.; Brandolini, L.; Cimini, A.M.; Zazzeroni, F.; Allegretti, M.; et al. MicroRNAs Expression in Response to rhNGF in Epithelial Corneal Cells: Focus on Neurotrophin Signaling Pathway. Int. J. Mol. Sci. 2022, 23, 3597. https://doi.org/10.3390/ijms23073597
Compagnoni C, Zelli V, Bianchi A, Di Marco A, Capelli R, Vecchiotti D, Brandolini L, Cimini AM, Zazzeroni F, Allegretti M, et al. MicroRNAs Expression in Response to rhNGF in Epithelial Corneal Cells: Focus on Neurotrophin Signaling Pathway. International Journal of Molecular Sciences. 2022; 23(7):3597. https://doi.org/10.3390/ijms23073597
Chicago/Turabian StyleCompagnoni, Chiara, Veronica Zelli, Andrea Bianchi, Antinisca Di Marco, Roberta Capelli, Davide Vecchiotti, Laura Brandolini, Anna Maria Cimini, Francesca Zazzeroni, Marcello Allegretti, and et al. 2022. "MicroRNAs Expression in Response to rhNGF in Epithelial Corneal Cells: Focus on Neurotrophin Signaling Pathway" International Journal of Molecular Sciences 23, no. 7: 3597. https://doi.org/10.3390/ijms23073597
APA StyleCompagnoni, C., Zelli, V., Bianchi, A., Di Marco, A., Capelli, R., Vecchiotti, D., Brandolini, L., Cimini, A. M., Zazzeroni, F., Allegretti, M., Alesse, E., & Tessitore, A. (2022). MicroRNAs Expression in Response to rhNGF in Epithelial Corneal Cells: Focus on Neurotrophin Signaling Pathway. International Journal of Molecular Sciences, 23(7), 3597. https://doi.org/10.3390/ijms23073597







