MYB2 Is Important for Tapetal PCD and Pollen Development by Directly Activating Protease Expression in Arabidopsis
Abstract
:1. Introduction
2. Results
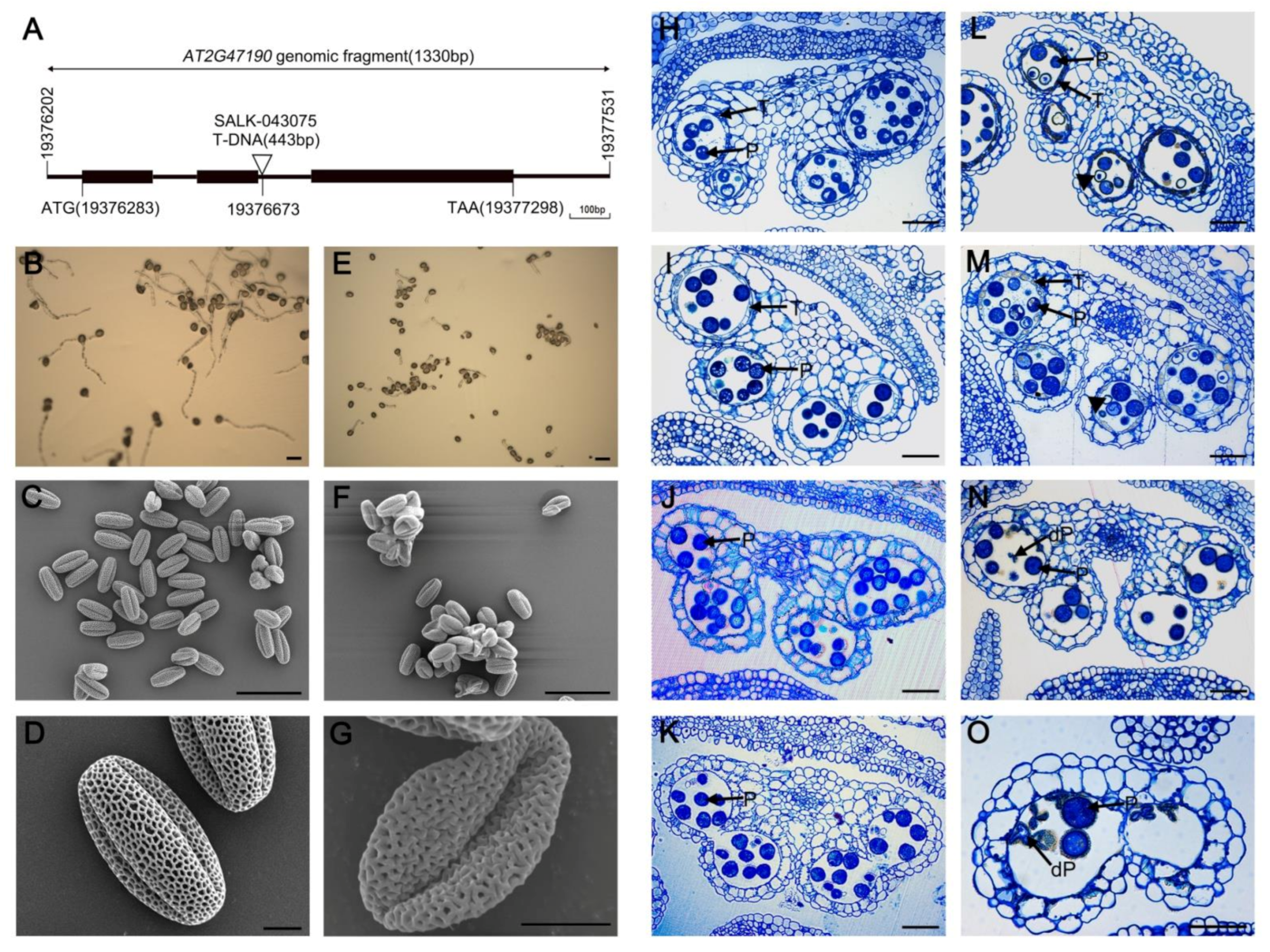
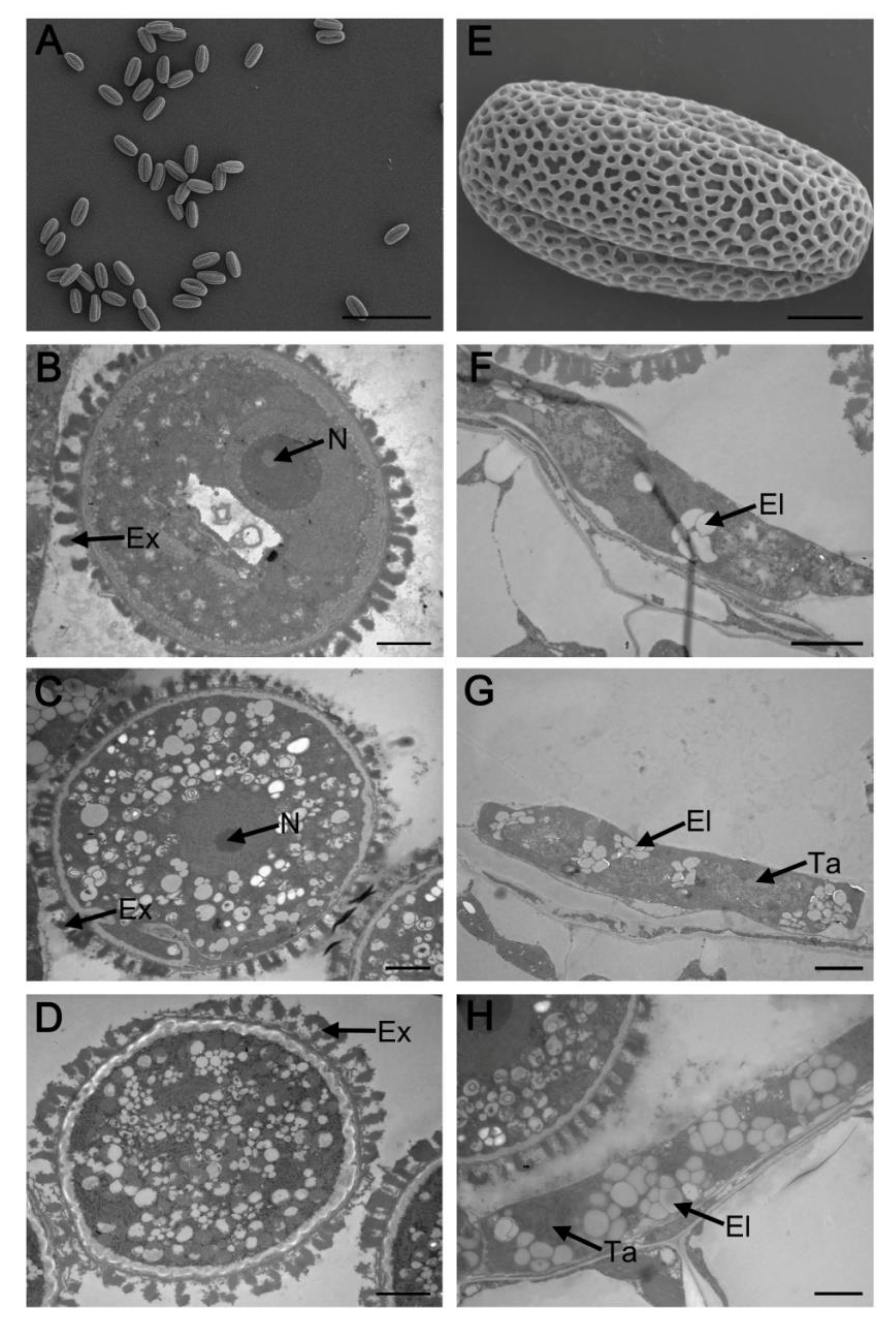
2.1. Phenotypic Characterization of myb2 Mutant
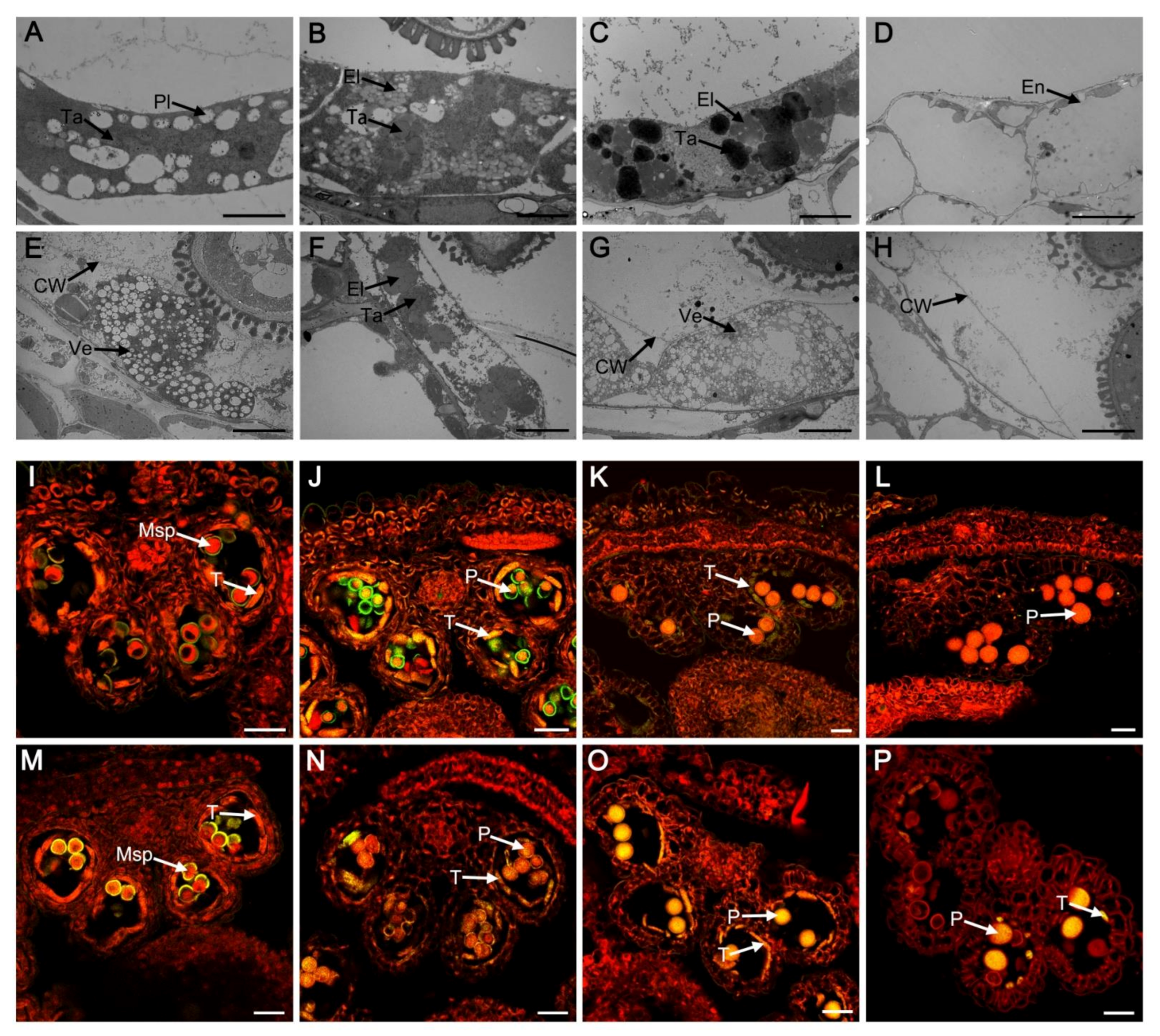
2.2. Tapetal PCD Was Retarded in myb2 Mutant
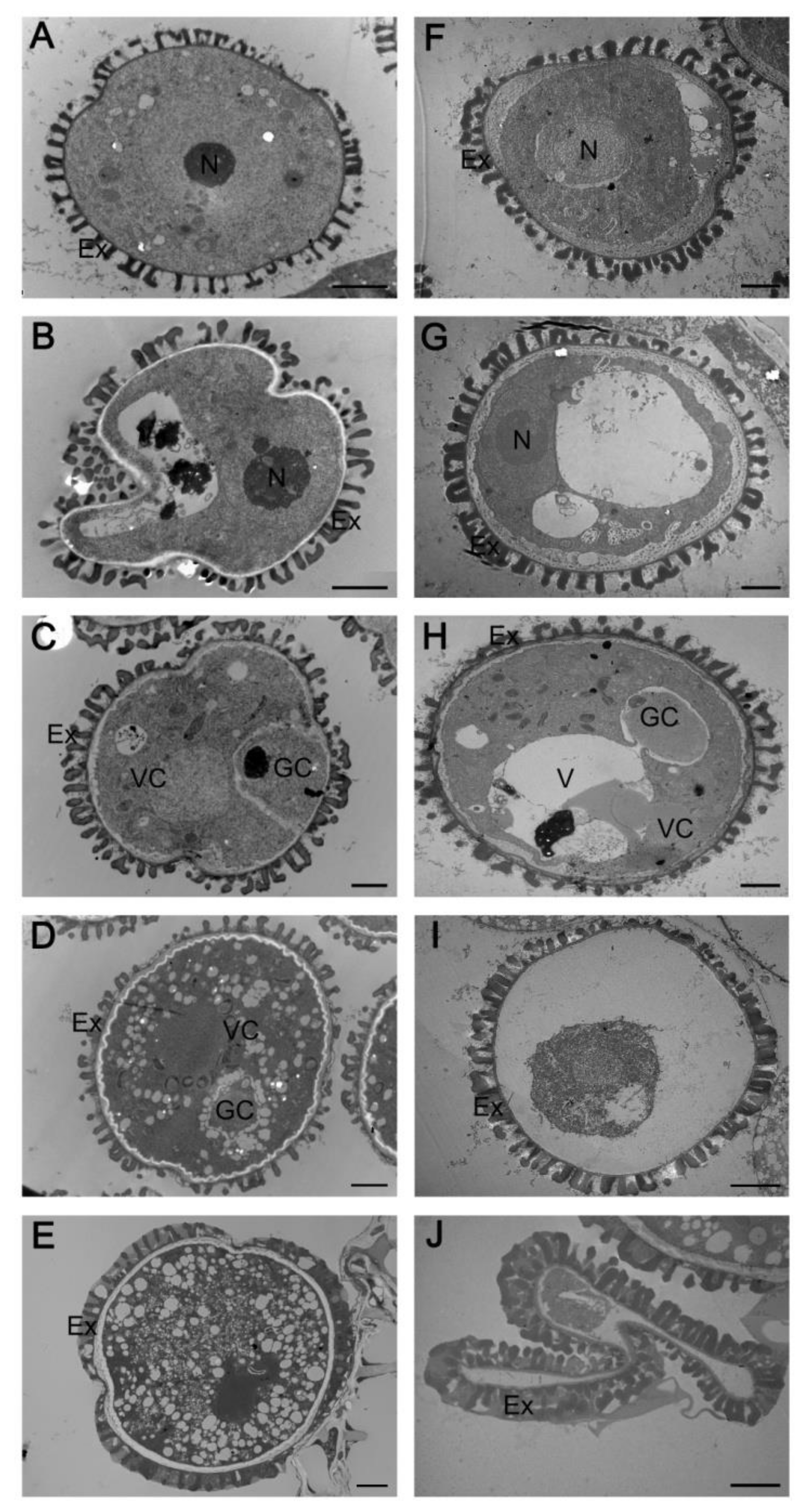
2.3. Abnormal Pollen Development in the myb2 Mutant
2.4. Complementation Analysis
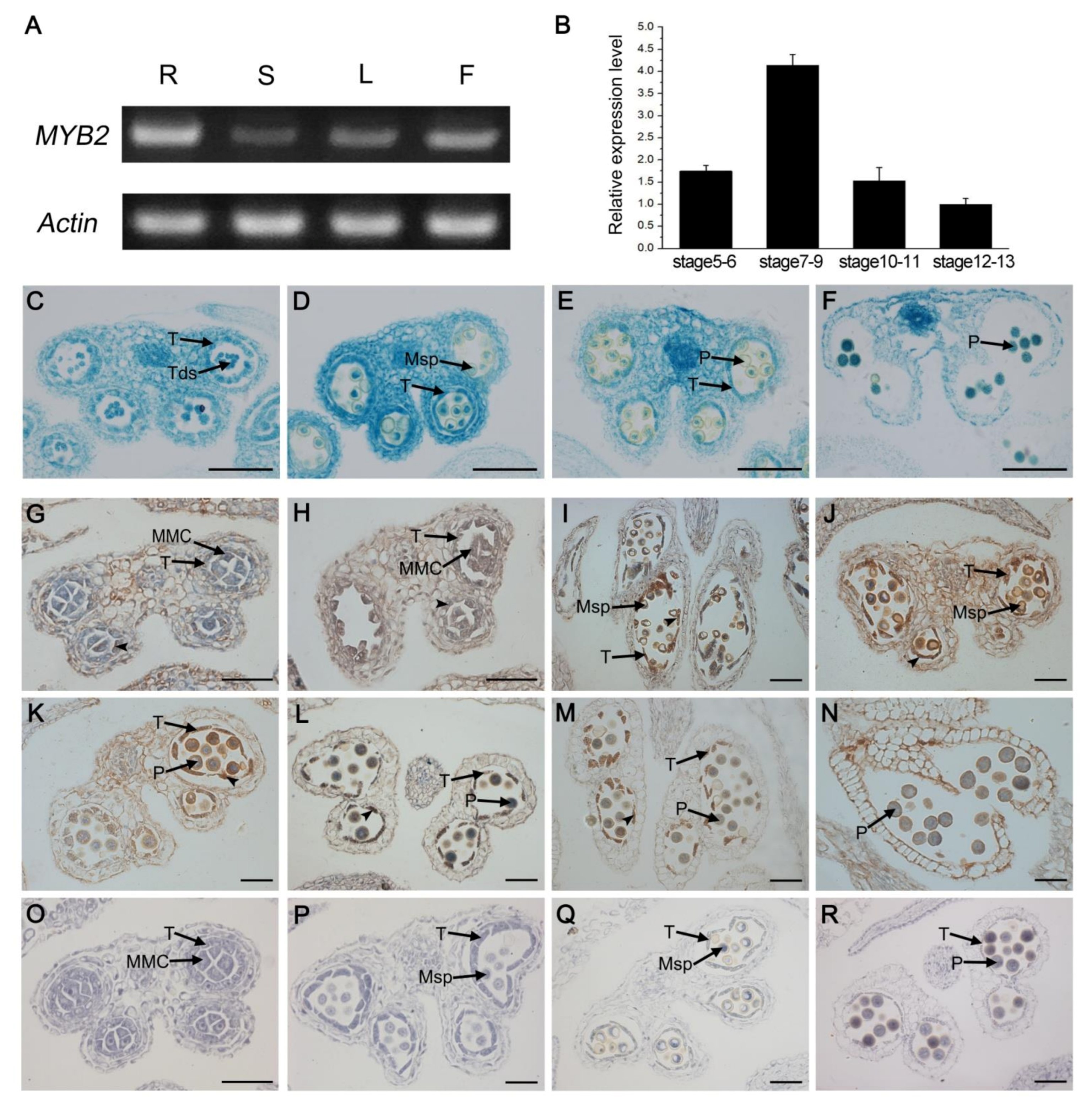
2.5. Spatio-Temporal Expression Pattern of MYB2
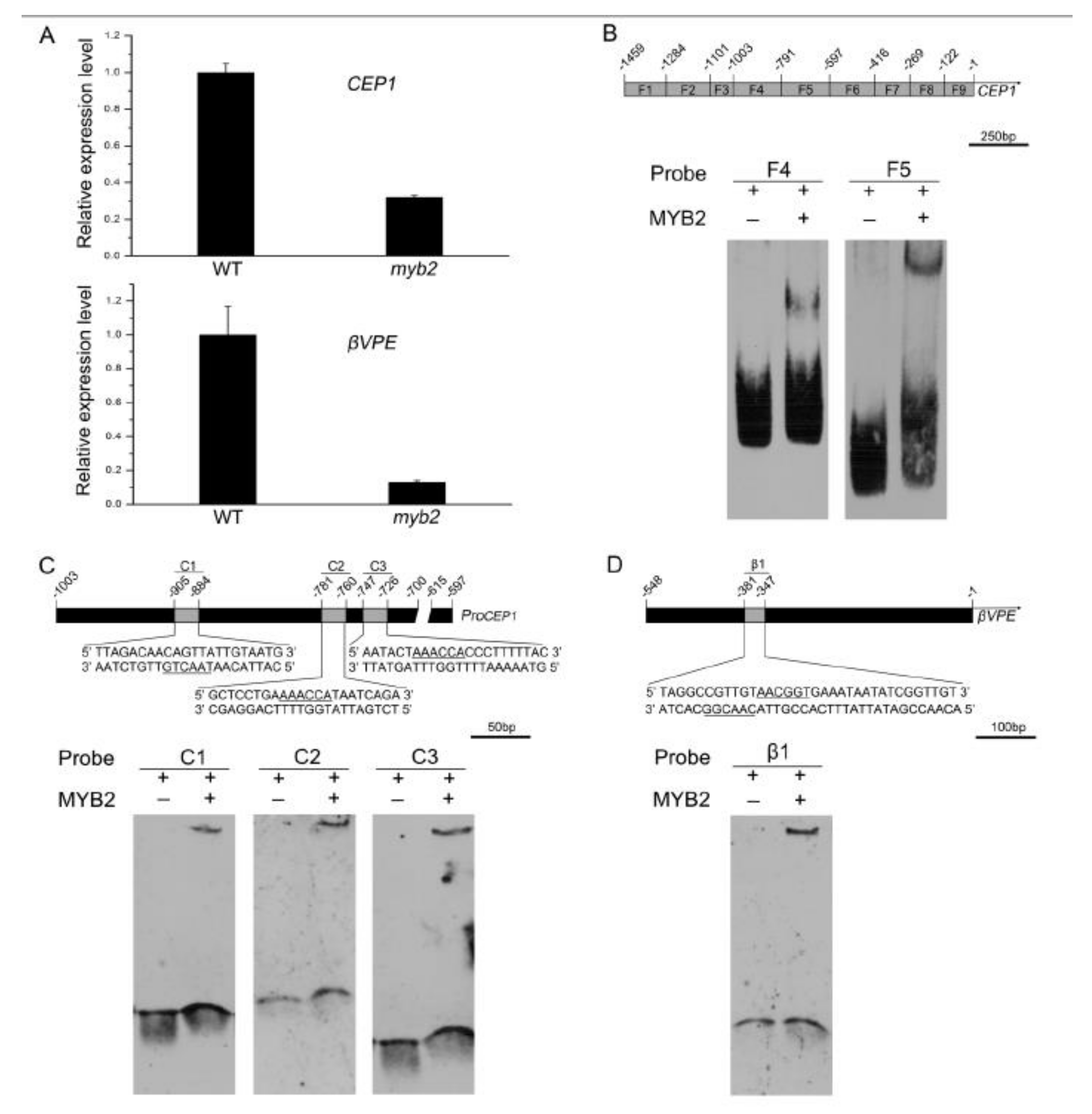
2.6. MYB2 Directly Binds to Cysteine Proteases CEP1 and βVPE Promoter and Activates Their Expression
2.7. The Deficiency of Cysteine Proteases Is Responsible for myb2 Mutant Phenotype
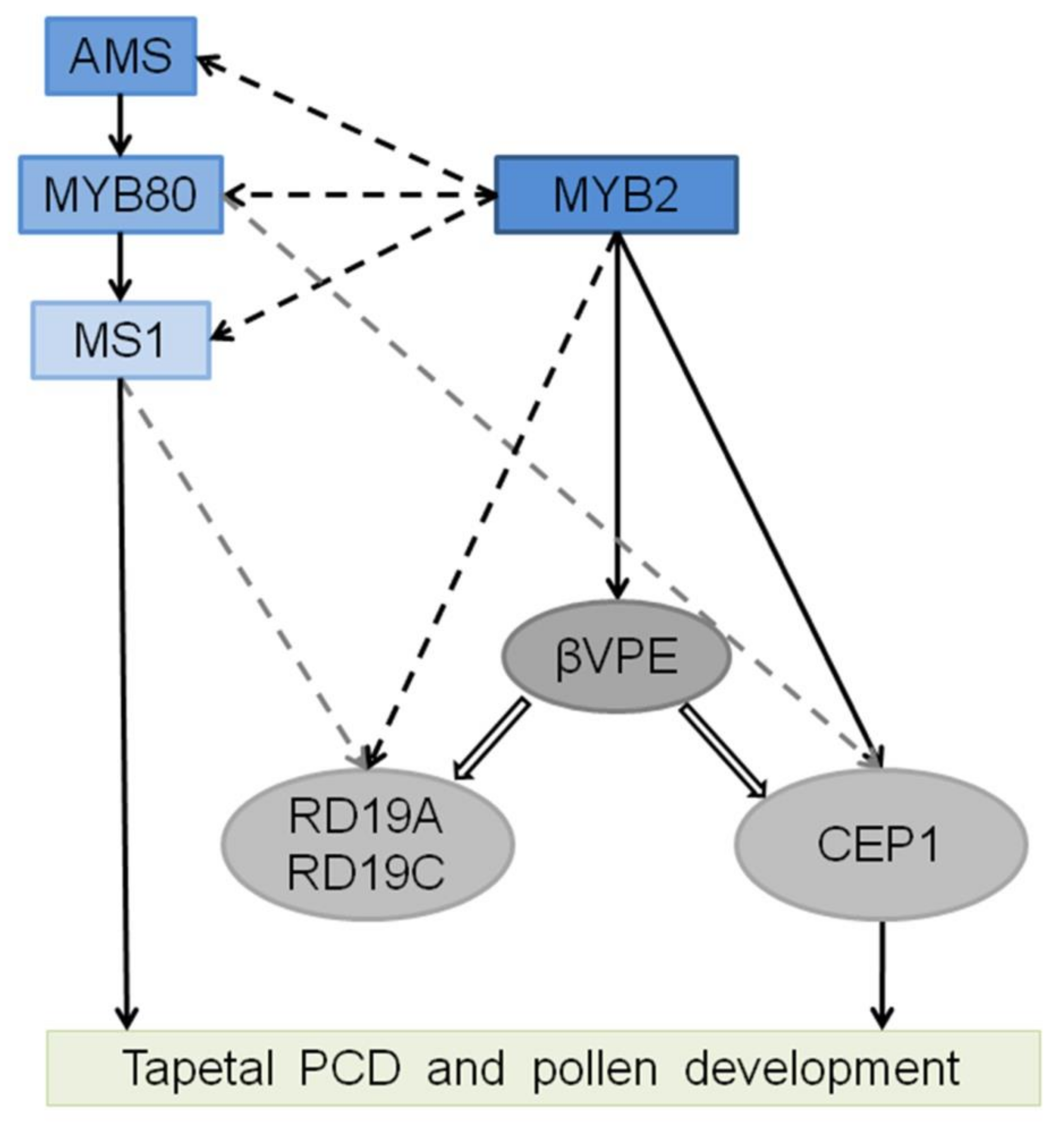
3. Discussion
3.1. MYB2 Is a Key Component during Tapetal PCD and Pollen Development
3.2. MYB2 Directly Regulates Expression of the Protease to Supervise Tapetal PCD
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Characterization of the Plants Phenotype
4.3. Semi-Thin Sections
4.4. TEM
4.5. Scanning Electron Microscopy
4.6. TUNEL
4.7. GUS Assay
4.8. In Situ Hybridization
4.9. RNA Extraction and RT-qPCR Analysis
4.10. EMSA
4.11. Transactivation Assay
4.12. Complementation of the myb2 Mutant
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Accession Numbers
References
- Ma, H. Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 2005, 56, 393–434. [Google Scholar] [CrossRef] [PubMed]
- Parish, R.W.; Li, S.F. Death of a tapetum: A programme of developmental altruism. Plant Sci. 2010, 178, 73–89. [Google Scholar] [CrossRef]
- Hanamata, S.; Sawada, J.; Ono, S.; Ogawa, K.; Fukunaga, T.; Nonomura, K.I.; Kimura, S.; Kurusu, T.; Kuchitsu, K. Impact of Autophagy on Gene Expression and Tapetal Programmed Cell Death during Pollen Development in Rice. Front. Plant Sci. 2020, 11, 172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chichkova, N.V.; Galiullina, R.A.; Beloshistov, R.E.; Balakireva, A.V.; Vartapetian, A.B. Phytaspases: Aspartate-specific proteases involved in plant cell death. Russ. J. Bioorganic Chem. 2014, 40, 658–664. [Google Scholar] [CrossRef]
- Cai, Y.-M.; Gallois, P. Programmed Cell Death Regulation by Plant Proteases with Caspase-Like Activity. In Plant Programmed Cell Death; Plant Physiol; Springer International Publishing: Cham, Switzerland, 2015; pp. 191–202. [Google Scholar] [CrossRef]
- Zamyatnin, A.A., Jr. Plant Proteases Involved in Regulated Cell Death. Biochemistry 2015, 80, 1701–1715. [Google Scholar] [CrossRef]
- Daneva, A.; Gao, Z.; Van Durme, M.; Nowack, M.K. Functions and Regulation of Programmed Cell Death in Plant Development. Annu. Rev. Cell Dev. Biol. 2016, 32, 441–468. [Google Scholar] [CrossRef]
- Buono, R.A.; Hudecek, R.; Nowack, M.K. Plant proteases during developmental programmed cell death. J. Exp. Bot. 2019, 70, 2097–2112. [Google Scholar] [CrossRef]
- Shukla, P.; Singh, N.K.; Kumar, D.; Vijayan, S.; Ahmed, I.; Kirti, P.B. Expression of a pathogen-induced cysteine protease (AdCP) in tapetum results in male sterility in transgenic tobacco. Funct. Integr. Genom. 2014, 14, 307–317. [Google Scholar] [CrossRef]
- Gautam, R.; Shukla, P.; Kirti, P.B. Targeted expression of a cysteine protease (AdCP) in tapetum induces male sterility in Indian mustard, Brassica juncea. Funct. Integr. Genom. 2019, 19, 703–714. [Google Scholar] [CrossRef]
- Zhang, X.M.; Wang, Y.; Lv, X.M.; Li, H.; Sun, P.; Lu, H.; Li, F.L. NtCP56, a new cysteine protease in Nicotiana tabacum L., involved in pollen grain development. J. Exp. Bot. 2009, 60, 1569–1577. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Dong, C.; Yu, J.; Shi, L.; Tong, C.; Li, Z.; Huang, J.; Liu, S. Cysteine Protease 51 (CP51), an anther-specific cysteine protease gene, is essential for pollen exine formation in Arabidopsis. Plant Cell Tissue Organ Cult. (PCTOC) 2014, 119, 383–397. [Google Scholar] [CrossRef]
- Song, L.; Zhou, Z.; Tang, S.; Zhang, Z.; Plant, J.T.J.; Physiology, C. Ectopic Expression of BnaC.CP20.1 Results in Premature Tapetal Programmed Cell Death in Arabidopsis. Plant Cell Physiol. 2016, 57, 1972–1984. [Google Scholar] [CrossRef]
- Zhang, D.; Liu, D.; Lv, X.; Wang, Y.; Xun, Z.; Liu, Z.; Li, F.; Lu, H. The cysteine protease CEP1, a key executor involved in tapetal programmed cell death, regulates pollen development in Arabidopsis. Plant Cell 2014, 26, 2939–2961. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Z.; Guo, X.; Zhang, J.; Liu, Y.; Wang, B.; Li, H.; Lu, H. betaVPE is involved in tapetal degradation and pollen development by activating proprotease maturation in Arabidopsis thaliana. J. Exp. Bot. 2020, 71, 1943–1955. [Google Scholar] [CrossRef]
- Zhu, E.; You, C.; Wang, S.; Cui, J.; Niu, B.; Wang, Y.; Qi, J.; Ma, H.; Chang, F. The DYT1-interacting proteins bHLH010, bHLH089 and bHLH091 are redundantly required for Arabidopsis anther development and transcriptome. Plant J. 2015, 83, 976–990. [Google Scholar] [CrossRef]
- Lou, Y.; Zhou, H.S.; Han, Y.; Zeng, Q.Y.; Zhu, J.; Yang, Z.N. Positive regulation of AMS by TDF1 and the formation of a TDF1-AMS complex are required for anther development in Arabidopsis thaliana. New Phytol. 2018, 217, 378–391. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Ding, Z.; Vizcay-Barrena, G.; Shi, J.; Liang, W.; Yuan, Z.; Werck-Reichhart, D.; Schreiber, L.; Wilson, Z.A.; Zhang, D. Aborted Microspores Acts as a Master Regulator of Pollen Wall Formation in Arabidopsis. Plant Cell 2014, 26, 1544–1556. [Google Scholar] [CrossRef] [Green Version]
- Ferguson, A.C.; Pearce, S.; Band, L.R.; Yang, C.; Ferjentsikova, I.; King, J.; Yuan, Z.; Zhang, D.; Wilson, Z.A. Biphasic regulation of the transcription factor ABORTED MICROSPORES (AMS) is essential for tapetum and pollen development in Arabidopsis. New Phytol. 2017, 213, 778–790. [Google Scholar] [CrossRef]
- Yang, C.; Vizcay-Barrena, G.; Conner, K.; Wilson, Z.A. MALE STERILITY1 is required for tapetal development and pollen wall biosynthesis. Plant Cell 2007, 19, 3530–3548. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Iacuone, S.; Li, S.; Parish, R.W. MYB80 homologues in Arabidopsis, cotton and Brassica: Regulation and functional conservation in tapetal and pollen development. BMC Plant Biol. 2014, 14, 278. [Google Scholar]
- Cai, C.-F.; Zhu, J.; Lou, Y.; Guo, Z.-L.; Xiong, S.-X.; Wang, K.; Yang, Z.-N. The functional analysis of OsTDF1 reveals a conserved genetic pathway for tapetal development between rice and Arabidopsis. Sci. Bull. 2015, 60, 1073–1082. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Zhou, S.D.; Fan, J.J.; Zhou, L.; Shi, Q.S.; Zhang, Y.F.; Liu, X.L.; Chen, X.; Zhu, J.; Yang, Z.N. OsMS188 Is a Key Regulator of Tapetum Development and Sporopollenin Synthesis in Rice. Rice 2021, 14, 4. [Google Scholar] [CrossRef]
- Moon, S.; Hong, W.-J.; Kim, Y.-J.; Chandran, A.K.N.; Gho, Y.-S.; Yoo, Y.-H.; Nguyen, V.N.T.; An, G.; Park, S.K.; Jung, K.-H. Comparative Transcriptome Analysis Reveals Gene Regulatory Mechanism of UDT1 on Anther Development. J. Plant Biol. 2020, 63, 289–296. [Google Scholar] [CrossRef]
- Niu, N.; Liang, W.; Yang, X.; Jin, W.; Wilson, Z.A.; Hu, J.; Zhang, D. EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nat. Commun. 2013, 4, 1445. [Google Scholar] [CrossRef]
- Li, H.; Yuan, Z.; Vizcay-Barrena, G.; Yang, C.; Liang, W.; Zong, J.; Wilson, Z.A.; Zhang, D. PERSISTENT TAPETAL CELL1 encodes a PHD-finger protein that is required for tapetal cell death and pollen development in rice. Plant Physiol. 2011, 156, 615–630. [Google Scholar] [CrossRef] [Green Version]
- Uzair, M.; Xu, D.; Schreiber, L.; Shi, J.; Liang, W.; Jung, K.H.; Chen, M.; Luo, Z.; Zhang, Y.; Yu, J.; et al. PERSISTENT TAPETAL CELL2 Is Required for Normal Tapetal Programmed Cell Death and Pollen Wall Patterning. Plant Physiol. 2020, 182, 962–976. [Google Scholar] [CrossRef] [Green Version]
- Phan, H.A.; Iacuone, S.; Li, S.F.; Parish, R.W. The MYB80 transcription factor is required for pollen development and the regulation of tapetal programmed cell death in Arabidopsis thaliana. Plant Cell 2011, 23, 2209–2224. [Google Scholar] [CrossRef] [Green Version]
- Verma, N.; Burma, P.K. Regulation of tapetum-specific A9 promoter by transcription factors AtMYB80, AtMYB1 and AtMYB4 in Arabidopsis thaliana and Nicotiana tabacum. Plant J. 2017, 92, 481–494. [Google Scholar] [CrossRef]
- Sanders, P.M.; Bui, A.Q.; Weterings, K.; McIntire, K.N.; Hsu, Y.-C.; Lee, P.Y.; Truong, M.T.; Beals, T.P.; Goldberg, R.B. Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 1999, 11, 297–322. [Google Scholar] [CrossRef]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef] [Green Version]
- Baek, D.; Park, H.C.; Kim, M.C.; Yun, D.J. The role of Arabidopsis MYB2 in miR399f-mediated phosphate-starvation response. Plant Signal. Behav. 2013, 8, e23488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baek, D.; Chun, H.J.; Kang, S.; Shin, G.; Park, S.J.; Hong, H.; Kim, C.; Kim, D.H.; Lee, S.Y.; Kim, M.C.; et al. A Role for Arabidopsis miR399f in Salt, Drought, and ABA Signaling. Mol. Cells 2016, 39, 111–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.; Gan, S. AtMYB2 regulates whole plant senescence by inhibiting cytokinin-mediated branching at late stages of development in Arabidopsis. Plant Physiol. 2011, 156, 1612–1619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Huang, J.; Parameswaran, S.; Ito, T.; Seubert, B.; Auer, M.; Rymaszewski, A.; Jia, G.; Owen, H.A.; Zhao, D. The SPOROCYTELESS/NOZZLE gene is involved in controlling stamen identity in Arabidopsis. Plant Physiol. 2009, 151, 1401–1411. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Lou, Y.; Xu, X.; Yang, Z.N. A genetic pathway for tapetum development and function in Arabidopsis. J. Integr. Plant Biol. 2011, 53, 892–900. [Google Scholar] [CrossRef]
- Cui, J.; You, C.; Zhu, E.; Huang, Q.; Ma, H.; Chang, F. Feedback Regulation of DYT1 by Interactions with Downstream bHLH Factors Promotes DYT1 Nuclear Localization and Anther Development. Plant Cell 2016, 28, 1078–1093. [Google Scholar] [CrossRef] [Green Version]
- Li, D.D.; Xue, J.S.; Zhu, J.; Yang, Z.N. Gene Regulatory Network for Tapetum Development in Arabidopsis thaliana. Front. Plant Sci. 2017, 8, 1559. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.Y.; Xiong, S.X.; Yin, W.; Teng, X.D.; Lou, Y.; Zhu, J.; Zhang, C.; Gu, J.N.; Wilson, Z.A.; Yang, Z.N. MS1, a direct target of MS188, regulates the expression of key sporophytic pollen coat protein genes in Arabidopsis. J. Exp. Bot. 2020, 71, 4877–4889. [Google Scholar] [CrossRef]
- Pelayo, M.A.; Yamaguchi, N.; Ito, T. One factor, many systems: The floral homeotic protein AGAMOUS and its epigenetic regulatory mechanisms. Curr. Opin. Plant Biol. 2021, 61, 102009. [Google Scholar] [CrossRef]
- Wijeratne, A.J.; Zhang, W.; Sun, Y.; Liu, W.; Albert, R.; Zheng, Z.; Oppenheimer, D.G.; Zhao, D.; Ma, H. Differential gene expression in Arabidopsis wild-type and mutant anthers: Insights into anther cell differentiation and regulatory networks. Plant J. 2007, 52, 14–29. [Google Scholar] [CrossRef]
- Feng, B.; Lu, D.; Ma, X.; Peng, Y.; Sun, Y.; Ning, G.; Ma, H. Regulation of the Arabidopsis anther transcriptome by DYT1 for pollen development. Plant J. 2012, 72, 612–624. [Google Scholar] [CrossRef]
- Ma, X.; Feng, B.; Ma, H. AMS-dependent and independent regulation of anther transcriptome and comparison with those affected by other Arabidopsis anther genes. BMC Plant Biol. 2012, 12, 23. [Google Scholar] [CrossRef] [Green Version]
- Ito, T.; Nagata, N.; Yoshiba, Y.; Ohme-Takagi, M.; Ma, H.; Shinozaki, K. Arabidopsis MALE STERILITY1 encodes a PHD-type transcription factor and regulates pollen and tapetum development. Plant Cell 2007, 19, 3549–3562. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Song, J.; Ferguson, A.C.; Klisch, D.; Simpson, K.; Mo, R.; Taylor, B.; Mitsuda, N.; Wilson, Z.A. Transcription Factor MYB26 Is Key to Spatial Specificity in Anther Secondary Thickening Formation. Plant Physiol. 2017, 175, 333–350. [Google Scholar] [CrossRef]
- Preston, J.; Wheeler, J.; Heazlewood, J.; Li, S.F.; Parish, R.W. AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant J. 2004, 40, 979–995. [Google Scholar] [CrossRef]
- Battat, M.; Eitan, A.; Rogachev, I.; Hanhineva, K.; Fernie, A.; Tohge, T.; Beekwilder, J.; Aharoni, A. A MYB Triad Controls Primary and Phenylpropanoid Metabolites for Pollen Coat Patterning. Plant Physiol. 2019, 180, 87–108. [Google Scholar] [CrossRef] [Green Version]
- Millar, A.A.; Gubler, F. The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 2005, 17, 705–721. [Google Scholar] [CrossRef] [Green Version]
- Gu, J.N.; Zhu, J.; Yu, Y.; Teng, X.D.; Lou, Y.; Xu, X.F.; Liu, J.L.; Yang, Z.N. DYT1 directly regulates the expression of TDF1 for tapetum development and pollen wall formation in Arabidopsis. Plant J. 2014, 80, 1005–1013. [Google Scholar] [CrossRef]
- Liu, L.; Fan, X.D. Tapetum: Regulation and role in sporopollenin biosynthesis in Arabidopsis. Plant Mol. Biol. 2013, 83, 165–175. [Google Scholar] [CrossRef]
- Trobacher, C.P.; Senatore, A.; Greenwood, J.S. Masterminds or minions? Cysteine proteinases in plant programmed cell deathThis review is one of a selection of papers published in the Special Issue on Plant Cell Biology. Can. J. Bot. 2006, 84, 651–667. [Google Scholar] [CrossRef]
- Liu, H.; Hu, M.; Wang, Q.; Cheng, L.; Zhang, Z. Role of Papain-Like Cysteine Proteases in Plant Development. Front. Plant Sci. 2018, 9, 1717. [Google Scholar] [CrossRef] [Green Version]
- Misas-Villamil, J.C.; Toenges, G.; Kolodziejek, I.; Sadaghiani, A.M.; Kaschani, F.; Colby, T.; Bogyo, M.; van der Hoorn, R.A. Activity profiling of vacuolar processing enzymes reveals a role for VPE during oomycete infection. Plant J. 2013, 73, 689–700. [Google Scholar] [CrossRef]
- Hatsugai, N.; Yamada, K.; Goto-Yamada, S.; Hara-Nishimura, I. Vacuolar processing enzyme in plant programmed cell death. Front. Plant Sci. 2015, 6, 234. [Google Scholar] [CrossRef] [Green Version]
- Kuroyanagi, M.; Nishimura, M.; Hara-Nishimura, I. Activation of Arabidopsis Vacuolar Processing Enzyme by Self-Catalytic Removal of an Auto-Inhibitory Domain of the C-Terminal Propeptide. Plant Cell Physiol. 2002, 43, 143–151. [Google Scholar] [CrossRef] [Green Version]
- He, F.; Li, H.G.; Wang, J.J.; Su, Y.; Wang, H.L.; Feng, C.H.; Yang, Y.; Niu, M.X.; Liu, C.; Yin, W.; et al. PeSTZ1, a C2H2-type zinc finger transcription factor from Populus euphratica, enhances freezing tolerance through modulation of ROS scavenging by directly regulating PeAPX2. Plant Biotechnol. J. 2019, 17, 2169–2183. [Google Scholar] [CrossRef] [Green Version]


| Plants | Normal Pollen Rate | Pollen Germination Rate |
|---|---|---|
| WT | 92.96% (317/341) | 87.50% ± 1.68 |
| myb2 | 39.18% (125/319) | 29.55% ± 2.10 |
| 35S: CEP1/myb2 | 61.24% (237/387) | 40.67% ± 2.49 |
| 35S: βVPE/myb2 | 70.92% (217/306) | 54.00% ± 3.27 |
| 35S: CEP1 + 35S: βVPE/myb2 | 83.74% (273/326) | 79.75% ± 2.15 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, X.; Li, L.; Liu, X.; Zhang, C.; Yao, X.; Xun, Z.; Zhao, Z.; Yan, W.; Zou, Y.; Liu, D.; et al. MYB2 Is Important for Tapetal PCD and Pollen Development by Directly Activating Protease Expression in Arabidopsis. Int. J. Mol. Sci. 2022, 23, 3563. https://doi.org/10.3390/ijms23073563
Guo X, Li L, Liu X, Zhang C, Yao X, Xun Z, Zhao Z, Yan W, Zou Y, Liu D, et al. MYB2 Is Important for Tapetal PCD and Pollen Development by Directly Activating Protease Expression in Arabidopsis. International Journal of Molecular Sciences. 2022; 23(7):3563. https://doi.org/10.3390/ijms23073563
Chicago/Turabian StyleGuo, Xiaorui, Lihong Li, Xiatong Liu, Chong Zhang, Xiaoyun Yao, Zhili Xun, Zhijing Zhao, Wenwen Yan, Yirong Zou, Di Liu, and et al. 2022. "MYB2 Is Important for Tapetal PCD and Pollen Development by Directly Activating Protease Expression in Arabidopsis" International Journal of Molecular Sciences 23, no. 7: 3563. https://doi.org/10.3390/ijms23073563
APA StyleGuo, X., Li, L., Liu, X., Zhang, C., Yao, X., Xun, Z., Zhao, Z., Yan, W., Zou, Y., Liu, D., Li, H., & Lu, H. (2022). MYB2 Is Important for Tapetal PCD and Pollen Development by Directly Activating Protease Expression in Arabidopsis. International Journal of Molecular Sciences, 23(7), 3563. https://doi.org/10.3390/ijms23073563






