Abstract
Microbe–host communication is essential to maintain vital functions of a healthy host, and its disruption has been associated with several diseases, including Crohn’s disease and ulcerative colitis, the two major forms of inflammatory bowel disease (IBD). Although individual members of the intestinal microbiota have been associated with experimental IBD, identifying microorganisms that affect disease susceptibility and phenotypes in humans remains a considerable challenge. Currently, the lack of a definition between what is healthy and what is a dysbiotic gut microbiome limits research. Nevertheless, although clear proof-of-concept of causality is still lacking, there is an increasingly evident need to understand the microbial basis of IBD at the microbial strain, genomic, epigenomic, and functional levels and in specific clinical contexts. Recent information on the role of diet and novel environmental risk factors affecting the gut microbiome has direct implications for the immune response that impacts the development of IBD. The complexity of IBD pathogenesis, involving multiple distinct elements, suggests the need for an integrative approach, likely utilizing computational modeling of molecular datasets to identify more specific therapeutic targets.
1. Introduction
In the last two decades, the gut microbiota has become a focus of major interest in the study of inflammatory bowel disease (IBD) pathogenesis. Technological advancements have allowed the characterization of several gut microbiome abnormalities in patients with Crohn’s disease (CD) and ulcerative colitis (UC), the two major forms of IBD. Abnormal immune reactivity against commensal microorganisms [1,2] and defects in innate and adaptive immunity have long been described in studies on IBD [3,4,5]. Nonetheless, the most consistent association between IBD and bacteria has been derived from animal models. For example, germ-free mice do not develop colitis, and inflammatory changes can be induced after colonization with commensal bacteria [6]. In a study using interleukin (IL)-10-deficient mice, which are genetically susceptible to colitis development, antibiotic administration early in life increased the risk of colitis [7].
Regarding genetic predisposition, several studies have identified altered regulators of the complex network underlying IBD in patients, many of which control the immune response to microbes. Nucleotide-binding oligomerization domain 2 (NOD2) polymorphisms, which encode an intracellular pattern recognition receptor and regulate the production of defensins by Paneth cells, have been associated with the risk of CD [8]. Several other gene variants related to bacterial clearance or protection against epithelial invasion have also been associated with IBD [9]. Currently, healthy immune homeostasis is linked to a state of tolerance towards resident microbiota, and disequilibrium of normal homeostatic conditions has been proposed as a necessary event for the initiation and/or maintenance of inflammation in IBD [10].
Although there is not yet a clear definition of what a healthy gut microbiome is, dysbiosis has generally been defined as an altered balance between the microbiota and its host [11]. Hence, commensal microbiota play a critical role in the early education of the immune system and microorganisms, and their products modulate immune responses through the induction of immune cells, signaling pathways, and inflammatory mediators [9]. In contrast, exposure to environmental triggers, such as dietary components, gastrointestinal infections, medications, psychological stress, and smoking, in genetically susceptible individuals leads to dysbiosis-associated mucosal immune dysfunction, which delineates IBD. Prolonged dysbiotic conditions, characterized by increased aggressive bacterial strains and decreased regulatory species, result in dysfunction of the mucosal immune response (Figure 1). Together with defective intestinal barrier function, gut dysbiosis is likely to sustain mucosal inflammation and potentially lead to IBD [12]. In this review, we aimed to discuss and summarize the basic mechanisms and potential associations between dysbiosis and IBD development, as well as exposome influences, methodological approaches to the microbiome, epigenetic changes, and recent developments in therapeutics.
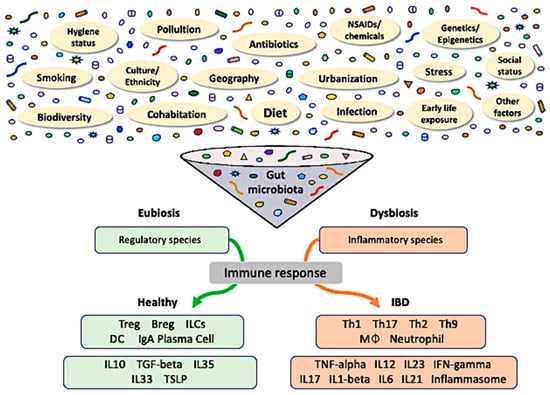
Figure 1.
The role of gut dysbiosis in the pathogenesis of inflammatory bowel disease. Gut microbiota reflect an interaction of host genetics with dynamic exposure to innumerable stimuli from the exposome. Crosstalk amongst these factors results in long-standing consequences to the gut microbiota and epigenetic modifications in a multidirectional fashion, potentially affecting susceptibility to diseases. The prevalence of either regulatory (eubiosis) or inflammatory (dysbiosis) species within the gut microbial community determines the respective predominant immune response. Treg, regulatory T-cell; Breg, regulatory B-cell; ILC, innate lymphoid cell; IgA, immunoglobulin A; MØ, macrophage; TSLP, thymic stromal lymphopoietin.
2. Intestinal Microbial Dysbiosis
The gut microbiota is an important physical, chemical, and immunological interface between the environment and host; thus, any dysregulation or breakdown of this barrier can contribute to disease states. For example, altered physical epithelial barrier function, a thinner mucus layer, and altered responses to endoplasmic reticulum stress (via mutations in MUC19, ITLN1, FUT2, and XBP1) have all been identified as risk factors for IBD [13,14,15]. Currently, the pathogenesis of human IBD is believed to involve inappropriate activation of the immune system when genetically susceptible individuals are exposed to gut antigens, such as microbiome components [16]. Although alterations in the gut microbiome have been proposed to be critical in IBD pathogenesis, it is not yet clear how this process occurs and whether dysbiosis is a central cause or a common consequence of the disease [17].
In healthy individuals, 99% of gut bacterial phyla are Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria. Firmicutes and Bacteroidetes account for approximately 90% of the total microbiome composition. These phyla are critically important in maintaining gut homeostasis and produce short-chain fatty acids (SCFAs), especially butyrate and propionate, from the fermentation of dietary components such as indigestible fibers. SCFAs are important energy sources for colonic mucosa cells but have also been shown to play key roles in regulating immune homeostasis [18]. Dysbiosis is defined as an alteration in gut microbiota composition and diversity and a shift in the balance between commensal and potentially pathogenic microorganisms [19]. Several pieces of evidence support the role of the microbiome and dysbiosis in IBD development. For example, experimental mice subjected to germ-free conditions develop attenuated colitis [20]. In studies using mouse models, the transfer of bacterial strains associated with IBD induces intestinal inflammation in genetically susceptible mice [21]. Similarly, fecal transplantation from human IBD donors to germ-free mice stimulates proinflammatory responses, with increased Th17 cell infiltration and proinflammatory mediators compared with transplants from healthy human donors [22]. Britton et al. colonized groups of adult wild-type or Rag1-deficient mice in germ-free conditions with human microbiota and assessed the mucosal immune response. Microbiota from healthy human donors induced, on average, higher frequencies of RORγt + Foxp3 + Treg cells in the intestinal lamina propria and prevented disease exacerbation. In contrast, microbiota from IBD donors resulted in enhanced RORγt + Th17 effector cell frequencies and enhanced disease severity in colitis-susceptible mice [23].
Determining the groups of microbes that are related to the development of intestinal inflammation has been a focus of extensive research. Patients with IBD tend to present several changes, not only in composition, but also in the diversity of their microbiome populations when compared to healthy individuals (Table 1). Evidence shows that alterations in microbiome components can also be involved in different IBD phenotypes [24]. The IBD microbiota has been characterized by an increase in the abundance of Bacteroidetes and Proteobacteria and a decrease in Firmicutes compared to control individuals. Specifically, levels of Faecalibacterium prausnitzii, a highly metabolically active commensal bacterium, are reduced in individuals with IBD [25]. Patients with IBD have reduced microbiome diversity (mostly a decrease in the relative abundance of Firmicutes) and an increase in the presence of Proteobacteria, such as Enterobacteriaceae and Bilophila, and certain members of Bacteroidetes [26]. Dysbiosis can potentially lead to a reduction in key functions necessary for maintaining intestinal barrier integrity and gut homeostasis. Therefore, alterations in the immune response and proinflammatory activity could be due to a dysbiotic microenvironment.

Table 1.
Association between the gut microbiome and inflammatory bowel disease.
Clooney et al. showed that among the microbial species found to be significantly increased in CD compared to controls, there was an increased presence of Ruminococcus gnavus and Fusobacterium nucleatum. Conversely, the presence of Ruminococcus albus, Eubacterium rectale, and Faecalibacterium prausnitzii were decreased in CD. Eubacterium and Roseburia were among the most important species in classifying either CD or UC compared with controls [24,47]. Furthermore, some species are particularly associated with certain subgroups of patients. For instance, Bacteroides vulgatus, Akkermansia muciniphila, and Escherichia/Shigella were increased in patients with a history of prior surgical resection [44]. In the largest pediatric Crohn’s cohort to date, including >400 patients and 200 controls, microbiomes from new-onset CD cases in multiple gastrointestinal locations were analyzed by Gevers et al. An axis defined by an increased abundance of bacteria such as Enterobacteriaceae, Pasteurellacaea, and Fusobacteriaceae and decreased abundance of Erysipelotrichales, Bacteroidales, and Clostridiales was strongly correlated with disease status. Moreover, microbiome comparison between patients with CD with and without antibiotic exposure indicated that antibiotic use amplified the microbial dysbiosis associated with CD in a new-onset pediatric cohort. The microbial dysbiosis index, which is characterized by the differential relative abundance of specific taxa, was associated with disease severity. Additionally, the rectal mucosa-associated microbiome, but not the fecal microbiome, has been shown to be a robust disease predictor [30]. Similarly, another study of a pediatric IBD cohort showed significant correlation between microbiota composition and disease severity, with resolution of dysbiosis in patients responding to anti-tumor necrosis factor (TNF) therapy [48]. Although changes in the gut microbiota profile in new-onset and treatment-naive pediatric patients with IBD were further corroborated in a recent systematic review, no clear conclusion can be drawn at the moment due to inconsistent results and heterogeneous methodologies [49].
In one of the largest longitudinal analyses of the IBD microbiome, Halfvarson et al. dissected the long-term dynamic behavior of the gut microbiome by comparing patients with IBD with healthy controls. Phylogenetic analysis of fecal samples showed that gut microbiomes of different IBD subtypes displayed different species distributions relative to controls. They identified potential microbial indicators of IBD subtypes, including genera such as Lachnospira, Clostridium, Oscillospira, and many unidentified Ruminococcaceae. Furthermore, they found that the microbiomes of patients with IBD fluctuate more than those of healthy individuals, based on deviation from a newly defined healthy plane. In addition, patients with ileal CD deviated the most from the healthy plane, especially those who underwent surgical resection. Interestingly, the microbiomes of some patients with IBD periodically visited the healthy plane and then deviated away from it [17]. In a study of 132 participants with IBD and controls, Lloyd-Price et al. assessed host and microbial data from colon biopsies, blood, and stool, for one year each. Principal coordinate analysis based on species-level Bray–Curtis dissimilarity showed that most variation was driven by a trade-off between the phyla Bacteroidetes and Firmicutes. Samples from individuals with IBD (particularly CD) had lower alpha diversity. Moreover, in patients with CD, taxonomic perturbations during dysbiosis were observed, such as a depletion of obligate anaerobes, including F. prausnitzii and Roseburia hominis, and an enrichment of facultative anaerobes such as Escherichia coli. In the metabolome, SCFAs were generally reduced during IBD dysbiosis. Overall, metabolite pools were less diverse in individuals with IBD, paralleling the observations of microbial diversity [50]. The investigation of whether the response to treatment with biologic agents could be associated with alterations in the composition of the intestinal microbiota of patients with CD was performed in a prospective study. The therapeutic intervention based on adalimumab was associated with the restoration of a eubiotic environment after six months of treatment. Particularly, in the cohort of patients with CD, those receiving adalimumab displayed a reduction in Proteobacteria and an increase in Lachnospiraceae. These results characteristically predominated among those patients who achieved therapeutic success, suggesting that dysbiosis could be directly involved with the response to treatment [51].
The role of pathogenic components in IBD has also been studied. For example, adherent-invasive E. coli (AIEC) is more prevalent in the mucosa of patients with IBD than in healthy individuals. Overall, the prevalence of AIEC in the mucosa of adult patients with CD ranges from 21–63%, and it is more associated with ileal CD than with colonic disease [41]. Mycobacterium avium, especially the subspecies paratuberculosis, has also been implicated in IBD pathogenesis. The abundance of these bacteria is higher in patients with IBD than in controls, and they are associated with increased production of proinflammatory cytokines [31]. Similarly, Listeria monocytogenes, which induces a Th1-type immune response [52], has also been shown to be increased in patients with IBD [33].
Archaea members have also been identified as part of the human microbiome, especially methane-producing species, such as Methanobrevibacter smithii, Methanosphaera stadtmanae, and Methanomassiliicoccus luminyensis in human stools, and Methanobrevibacter oralis in the oral mucosa [53,54]. Dysbiosis appears to affect the relative abundance of archaea members and, particularly in patients with IBD, M. smithii had a reduced abundance, whereas the immunogenic M. stadtmanae was remarkably increased compared to healthy controls [55,56].
Viruses and fungi are widely present in the gut and may play important roles in homeostasis. Changes in the enteric group of viruses in the gastrointestinal tract can have consequences on the bacterial microbiome and its diversity, as viruses are drivers of bacterial resistance. For instance, viruses can be responsible for the horizontal transfer of genetic material among bacterial communities, which implies a change in the balance of different ecosystems. An increase in the abundance of Caudovirales bacteriophages has been observed in patients with CD [57,58]. In addition, results from another study demonstrated an increased abundance of phages infecting Clostridiales, Alteromonadales, and Clostridium acetobutylicum, as well as viruses from the Retroviridae family, in patients with IBD [59,60].
Similarly, fungal components regulate and trigger immune responses. However, the gut mycobiome is less stable than the bacterial microbiome; therefore, a mycobiome signature has not yet been described [61]. Candida is the most prominent component of the fungal microbiota in humans. Its cell wall constituents, such as beta-glucans, chitin, and mannoses, can activate components of the innate immune system, such as Toll-like receptors (TLRs) 2 and 4, dectin-1, CD5, CD36, and SCARF1, and complement system components. The activation of these molecules leads to immune signaling and proinflammatory responses. Some studies have proposed a correlation between the gut mycobiome and gut bacterial components. Mason et al. showed that the colonization of C. albicans strain CHN1 in the stomach and cecum of C57BL6 mice prompted an overgrowth of Lactobacillus spp. and Enterococcus spp. during antibiotic treatment [62]. However, Bernardes et al. demonstrated that bacteria may influence fungal colonization. They showed that the colonization of bacteria together with fungi increased the relative abundance of C. parapsilosis and Issatchenkia orientalis, and a lack of co-colonization with bacteria or elimination of bacteria by antibiotics led to an overgrowth of C. albicans [63].
3. Influence of the Exposome
The first organisms on earth were microbes, and they have evolved and adapted to live in extreme environments all over the planet. All biological entities appearing later on Earth, including mammals, have evolved in a microbially dominant world. Therefore, humans have coevolved with microbes immersed within two complex ecological communities: the external and internal microbial environments. In this dualistic world of microbes, the exposome and gut microbiome impact each other as well as all other “–omes” in a reciprocal manner [64,65].
3.1. Geosocial Factors
Environmental factors have been associated with complex health conditions, including chronic immune-mediated inflammatory diseases (IMIDs), which have been increasing in incidence during the last century. Features are shared among the different IMIDs, including IBD, such as an inflammatory basis, multifactorial nature, and yet-unknown causes. In addition, epidemiological data revealing the co-occurrence of IMIDs and geographic expansion reinforce a common pathophysiological background that has evolved over the past several decades to reach a worldwide distribution [66]. The emergence of IMIDs and IBD has been linked to societal transformations, most commonly socioeconomic development or industrialization [67,68]. Such changes have been almost invariably accompanied by increasing urbanization [69], which, in turn, has been associated with distinct gut microbiota compositions different to those found in rural areas that are supposedly protective against IBD development [70,71].
Socioeconomic development and social behavior are crucial elements fueling the emergence of IBD [72]. Such changes have been associated with improvements in sanitation, quality of water supply in distribution systems, and a resulting decrease in infectious diseases, which constitute the basis of the hygiene hypothesis [73]. Nonetheless, these changes bring about several other simultaneous environmental modifications that need to be considered. It is important to highlight, for example, changes in homes, family structures, workplaces, dietary habits, the widespread use and production of chemicals, and the use of medications, including antibiotics. Growing urbanization has led to a continuous increase in population density in cities that have become progressively more polluted, competitive, and stressful, causing dramatic changes in peoples’ lifestyles. Moreover, the attraction between manpower and industry, or other economic activities, resulted in more human agglomeration, whether that be in households or factories; it also gathered people from different backgrounds, be they genetic, geographic, or cultural. These observations are in accordance with previous studies showing that individuals who migrate from low to high prevalence IBD areas, that is, from less developed to more developed areas, are more susceptible to developing IBD, predominantly affecting the first- and second-generation offspring of these immigrants [74,75,76,77]. In addition, among the potentially relevant stimuli from the exposome, cohabitation has been shown to strongly affect immune responses [78]. Transmission of microbial strains, predominantly detected among first-degree relatives sharing a household, has recently been demonstrated [79], helping to explain the link between the exposome and the immune response. This also reinforces a microbial basis for IMIDs.
While several human diseases have been associated with abnormalities in host-associated microbial communities, and the human body is seen as an ecosystem [80], defining a healthy microbiome continues to represent a complex challenge due to the formidable variability shown in population-based studies [81,82]. This is also true for IBD, as a large study confirmed a reduction in microbial diversity in patients with CD and UC but did not explain the increased variability compared to controls [24]. Whether such a variance is stochastic or due to environmental factors has not yet been established [83]. Nevertheless, the microbiome reflects a complex combination of endogenous and exogenous elements, particularly environmental and lifestyle factors. Previous studies have shown that the gut microbiome of Western populations is characteristically less diverse [84,85,86]. As the intestine represents the largest surface of contact with the external environment, IBD could be facilitated by a combination of both a cleaner external milieu (as in the hygiene hypothesis) and an impoverished biome influencing the internal milieu to become less diverse, resulting in inappropriate immune system education and responses.
The hypothesis that loss of biodiversity is an important environmental factor has been supported by data showing that reduced contact of people with the natural environment may negatively impact the commensal microbiota and its immunomodulatory properties [87,88]. In addition to its worldwide distribution and progressive increase as a result of diverse human activities, loss of biodiversity has been regarded as a critical factor in the rise of allergic diseases [89], among which asthma, in particular, has been intimately associated with IBD [90]. Loss of biodiversity has recently been proposed as a novel factor in the pathogenesis and prevention of IBD, based on the non-uniform disease distribution in large developing countries, showing pronounced regional dissimilarities and disease hotspots associated with specific geosocial and ecosystem factors [91].
3.2. Antibiotics
Exposure to antibiotics has been associated with increased risk for developing IBD, especially CD [92,93]. Evidence from different studies has shown that patients diagnosed with IBD during childhood were more likely to have been exposed to antibiotics early in life [94]. In a pediatric prospective study, the strongest association between antibiotic use and future development of IBD was in the first 3 months following the use of antibiotics and among children who had more courses of antibiotics [95]. Contrarily, a recent study found that exposure to antibiotics during pregnancy, but not in infancy, is associated with an increased risk of early onset IBD [96]. Although current evidence does not confirm a consistent causal link with IBD, early exposure to antibiotics has been suggested to affect the development of tolerance to the gut microbiota, consequently raising inappropriate immune reactivity that underlies chronic intestinal inflammation [97]. Additionally, recent evidence from a study investigating the microbiome of humans, domestic animals, and their environment, in relation to antibiotic use, suggested the exchange of antimicrobial-resistant strains between reservoirs [98]. Together, these data appear to support the idea that the risk of developing IBD associated with intestinal dysbiosis may occur at both the individual and community levels. This also includes crosstalk with nonhuman components, reinforcing the existence of dynamic interactions between the environment and host regarding the exchange and sharing of microorganisms.
3.3. Dietary Factors
Several studies have investigated diet, arguably the most ubiquitous environmental factor, and its potential to shape the gut microbiota. For instance, evidence has shown that a high-calorie diet, consisting of fat- and carbohydrate-based foods, determines a preferential expansion of the genera Bacteroides and Prevotella and the Bacteroidetes phylum in adults, with shifts occurring in a relatively rapid fashion [99]. In another study, strictly animal-based food increased the relative abundance of bile-tolerant microorganisms, reducing the presence of microorganisms capable of metabolizing dietary plant polysaccharides. These results showed shifts between carbohydrate and protein fermentation, confirming that the microbiota can rapidly adjust to changes in dietary patterns. Moreover, changes in microbial composition were followed by changes in the molecular output of the microbiome with dietary interventions. SCFAs, products of bacterial digestion of fibers with critical homeostatic functions in the mucosa and anti-inflammatory properties, were shown to increase with plant-based diets. This may explain why a reduction in SCFAs in a typical Western-style diet (animal-based, high-calorie, high-fat, and low-dietary fiber) has been associated with the risk of IBD [100]. In fact, a Western-style diet, rich in sugar and fat, has been the predominant profile associated with a higher risk of developing IBD. While individuals who consume higher proportions of red meat and fats have a higher risk of IBD, others who predominantly consume fibers from vegetables and fruits have a lower risk [101,102]. Regarding dietary fat content, particularly polyunsaturated fatty acids, recent data indicate that a high omega-6 to omega-3 ratio, typical of Western-style diets, is associated with proinflammatory effects [103]. Furthermore, polyunsaturated fatty acids have been shown to exert not only effects on the immune response, directly acting on immune cells, but also influence the composition of the gut microbiota, thereby affecting host–microbiome interactions at different levels [104]. The consumption of processed foods, usually low in omega-3 fatty acids and micronutrients such as zinc and vitamins D and E, another common feature of Westernized diets, has also been associated with the development of chronic inflammatory diseases [105,106,107,108]. Globally, major shifts in dietary patterns towards progressively more Westernized diets, together with socioeconomic and demographic changes, represent a global transition that may explain the widespread increase in the rates of several metabolic and IMIDs [109], potentially involving changes in the gut microbiome and its interaction with the host.
4. Genetic Susceptibility
A clear connection with genetic predisposition has long been demonstrated in IBD, more so in CD than in UC [110]. There are over 200 genetic loci associated with IBD susceptibility, most of which regulate host–microbe interactions and immune-related pathways [110,111]. Some of the more studied genes include those involved in IL-23 receptors and Janus-activated kinase signaling, and those in innate mucosal defense, cytokine production, lymphocyte activation, epithelial barrier integrity, and multiple proteins involved in autophagy [112]. Genome-wide association studies have highlighted higher IBD genetic risk in individuals with NOD2 receptors, autophagy-related protein 16-like 1 (ATG16L1), immunity-related GTPase family, M (IRGM), IL-23 receptor gene, protein tyrosine phosphatase, non-receptor type 2 (PTPN2), X-box binding protein 1 (XBP1), and leucine-rich repeat kinase 2 (LRRK2) variants [111,113]. Genetic risk variants are also associated with changes in microbiota composition; for example, Roseburia spp., an acetate-to-butyrate converter, was less abundant in patients with IBD with these high-risk mutations [114].
Mutations in autophagy-related genes alter anti-bacterial, fungal, and viral responses and impair the clearance of various invading pathogens such as Mycobacterium tuberculosis, Group A Streptococcus, L. monocytogenes, and E. coli [115,116,117]. An ATG16L1 single nucleotide polymorphism (SNP) confers susceptibility to CD and is a common genetic variation present in 40–50% of the population, although most individuals with this SNP do not develop IBD [118,119]. The role of autophagy variants in Salmonella clearance is not well established. Messer et al. observed that ATG16L1 deficiency promoted cell resistance to Salmonella, while Conway et al. observed autophagy induction after Salmonella infection with the participation of ATG16L1 in intestines [120,121]. These variants also affect antimicrobial peptide production by Paneth cells, cytokine production, antigen presentation, and response to endoplasmic reticulum stress [122]. Atg16L1-deficient mice exhibited elevated inflammasome activation and IL-1β production when stimulated with lipopolysaccharides (LPS) and abnormalities in ileal Paneth cells, such as the escape of antimicrobial peptides into the cytoplasm [123]. There is crosstalk between NOD2 and ATG16L1, as NOD2 activation triggers autophagy in dendritic cells with the participation of ATG16L1, and deficiency in ATG16L1 heightens cytokine production via NOD [124,125]. Patients with CD with risk variants of ATG16L1 or NOD2 present with abnormal Paneth cell morphology [126]. In mouse models, decreased expression levels of Atg5, Atg7, or Atg4B generated abnormal Paneth cell functions, and in CD-like ileitis, deficiency of Atg16L1 also altered Paneth cell morphology [127]. In addition, norovirus infection in Atg16L1-deficient animals increased their susceptibility to dextran sodium sulfate (DSS) in a TNF-dependent phenotype resembling aspects of IBD [58]. Complete knockout of Atg3, Atg5, Atg7, or Atg16L1 is lethal in mice, and impairment of either Atg7 or Atg16L1 results in severe CD-like transmural ileitis [128]. Autophagic defects also worsen goblet cell function, production of mucus membrane defenses, and absorptive functions of the microvilli [129].
NOD2 recognizes bacterial peptidoglycan (muramyl dipeptide) in the cell walls of Gram-negative and Gram-positive bacteria and triggers the production of intestinal antimicrobial peptides to protect cells and immune responses in the gut [130]. NOD2 activation leads to NF-κB activation and production of IL-1b, TNF-α, IL-6, IL-8, and α-defensins [130,131]. NOD2 interacts with autophagy-related proteins to help destroy intracellular pathogens, and mutations in NOD2, also known as the caspase recruitment domain family, member 15 gene (CARD15), disrupt Paneth cells’ ability to recognize and eliminate invading pathogens [132,133]. In IBD, NOD2 mutations are associated with decreased release of defensins [134]. NOD2-mediated autophagy is important for the generation of major histocompatibility complex (MHC)-II-restricted CD4+ T cell responses in dendritic cells, and patients with CD with high-risk NOD2 or ATG16L1 variants exhibit impaired MHC II antigen presentation [124].
IL-23 signaling affects both the innate and adaptive immune systems in mice and is required for colitis development in several models [135,136,137]. The dominant IL23R SNP protects against IBD and generates a soluble receptor antagonist of IL-23 [138]. Variants in the autophagy-associated IRGM gene interfere with Paneth cell morphology and function, and are associated with abnormal secretory granule development, decreased antimicrobial peptide production, and higher susceptibility to colitis in a DSS-induced model [139]. Mutations in PTPN2 lead to defective autophagosome formation and bacterial elimination and promote T cell differentiation into Th1 and Th17 types [140,141,142]. Patients with IBD with PTPN2 variants demonstrate increased levels of interferon (IFN)-γ, IL-17, and IL-22 in the serum and intestinal mucosa [143]. LRRK2 is involved in the activation of dendritic cells (DCs) and production of IL-2 and TNF-α in CD [144].
5. Epigenetic Modifications
Recent data have provided the basis for the hypothesis that epigenetic modifications, resulting from interactions between the host and exposome, determine the phenotypic expression of IBD. For instance, the relatively high discordance rate among monozygotic twins [145] and an increased risk of developing the disease among people migrating from low- to high-incidence regions of IBD [146] constitute important epidemiological information to support the pathogenic role of epigenetic changes. Consequently, epigenetic factors have been suggested to mediate critical interactions between the exposome and genome, offering new insights into the pathogenesis of several diseases, including IBD [147].
Epigenetic changes related to the gut microbiome include modifications to DNA or histones, as well as the regulation of non-coding RNAs [148]. For example, recent studies have shown that microorganisms can bind to lysine on histones and regulate host chromatin by modifying histone proteins [149]. In turn, post-translational modifications of histones induced by microorganisms lead to changes in transcriptional gene activity [150,151]. Other studies investigating microRNAs (miRNAs) have suggested their participation in the immune response to microorganisms, resulting in the regulation of inflammatory mediators [152]. For example, miR-10a has been shown to suppress CD4+ T cell production of IL-10, favoring the induction of more severe colitis in genetically predisposed Rag1−/− mice [153]. In addition, miR-155 has been shown to promote Th17 differentiation and upregulate Th17-related cytokines [154]. Moreover, the induction of miR-155 and miR-146 family members has been implicated in the regulation of inflammatory responses triggered by microorganisms [155,156].
Dietary components also promote epigenetic modifications either directly or through the action of the gut microbiome, as some metabolites may modulate gene expression, chromatin remodeling, and DNA methylation. For example, polyphenols in green tea or soybeans, such as epigallocatechin-3-gallate and genistein, have been shown to inhibit DNA methyltransferase activity. Additionally, the gut microbiome generates a variety of SCFAs, such as acetate, butyrate, and propionate, which are essential for epithelial cell homeostasis but can also epigenetically regulate the immune response [157]. Bacteria from Clostridium, Eubacterium, and Butyrivibrio genera can synthesize butyrate, which inhibits histone deacetylases, from non-digestible fibers in the gut lumen [158]. In addition to being a nutrient for epithelial cells, SCFAs can also induce intracellular signaling pathways through the activation of G-protein-coupled receptors, regulating cell metabolism, inflammation, and oxidative stress [159,160]. Furthermore, the gut microbiome also contributes to the absorption and secretion of minerals, such as iodine, zinc, selenium, cobalt, and other cofactors that participate in epigenetic processes. Additionally, other key metabolites of the gut microbiota, including S-adenosylmethionine, acetyl-coenzyme A, nicotinamide adenine dinucleotide, α-ketoglutarate, and adenosine triphosphate, serve as essential cofactors for epigenetic enzymes that regulate DNA methylation and histone modifications [161,162].
6. Inappropriate Immune Response
The epithelium and its specialized cell types act as a barrier, separating the microbiota in the lumen from the immune cells in the lamina propria. In this reciprocal relationship, the microbiota also produces the metabolites necessary for epithelial cells, such as SCFAs and bacteriocins [163]. Immune cells in the lamina propria constitute the mucosa-associated lymphoid tissue that responds to microbiota stimuli, along with the epithelium. Some innate immune cells such as DCs, macrophages, natural killer cells, and innate lymphoid cells (ILCs) sample lumen antigens and induce a tolerogenic immune response. Under homeostatic conditions, this immune surveillance does not initiate a proinflammatory response; in contrast, it induces Treg cells. These immune cells capture antigens and migrate to lymphoid tissues to activate lymphocytes and link innate and adaptive responses [163,164]. The interaction between the immune system and microbiota, especially segmented filamentous bacteria, symbiotically aids the maturation of the immune system [164,165].
The dysregulated immune response observed in IBD is thought to result from crosstalk among genetic susceptibility, environmental factors, and gut microbiota [166,167]. Dysbiosis changes the composition of the gut microbiota, resulting in the loss of commensal bacteria and growth of pathogenic microorganisms [17,168]. In IBD, dysbiosis is characterized by a decrease in alpha diversity, with a decrease in abundance of Bacteroidetes and Firmicutes and an increase in that of Gamma-proteobacteria, especially AIEC [114,169]. Moreover, dysbiosis in IBD leads to a shift towards a proinflammatory environment with activated immune cells. In this context, cells increase the expression levels of pattern recognition receptors (PRRs) and the production of proinflammatory mediators in response to pathogens [170]. PRRs recognize microbe-, pathogen-, and/or danger-associated molecular patterns (MAMPs, PAMPs, and DAMPs, respectively, e.g., ATP and high mobility group box 1 protein (HMGB1)) released by cells during inflammation [171]. Examples of MAMPs include LPS (a TLR-4 ligand) and flagellin (a TLR-5 ligand) from bacteria, β-1,3-glucans (a dectin-1, C-type lectin receptor (CLR) ligand) from fungi, and viral nucleic acid molecules [172]. TLRs and CLRs are distributed on the surface of immune cells, epithelial cells, and other cell types, whereas NOD-like receptors (NLRs) and RIG-I-like receptors are present in the cytoplasm [173,174].
Tissue-resident macrophages comprise a large population of macrophages in the gut and control either tolerance or defense against microorganisms. Therefore, macrophages exhibit specialized gene expression related to their localization within the intestinal mucosa [175,176]. ILCs represent a group of innate immune cells, mostly localized at mucosal sites, with important participation in immune-mediated diseases such as IBD. In addition to increasing in density in the inflamed mucosa, ILC-1-producing IFN-γ cells characteristically accumulate in CD [177]. Because of their proximity to the gut microbiome, mucosal ILCs are thought to participate in a dichotomous regulatory mechanism, in which ILCs interfere with the microbial composition of the gut, and the gut resident microbes shape the plasticity and physiological functions of ILCs [178].
Inflammasomes have recently been regarded as central and specifically attractive in IBD immunopathogenesis because of their participation in complex crosstalk between the host mucosal immune system and environment, particularly the microbiota. Inflammasomes are multiprotein platforms formed in the cytoplasm that cleave and activate caspase-1, leading to the production of inflammatory cytokines, including IL-1b and IL-18. Inflammasomes comprise intracellular sensors formed by NLR proteins NLRP1, NLRP3, NLRC4, NLRP6, and NAIP5, or by the DNA-sensing complex AIM2, and can be activated by extracellular and intracellular pathogens in the presence of DAMPs [179]. Inflammasomes participate in responses against several bacteria such as L. monocytogenes, M. tuberculosis, and Fusobacterium [180]. In patients with CD, increased NLRP3 and AIM2 activity has been reported, and an SNP in NLRP3 is associated with CD susceptibility [181,182]. In contrast, NLRP3 inflammasome activation and production of IL-1b and IL-18 appear to be protective in experimental IBD [183].
Together, through these mechanisms, innate immune cells sense PAMPs and DAMPs, induce an inflammatory response, and shape adaptive immunity, promoting lymphoid tissue expansion and T- (Th1, Th2, Th9, Th17, and Treg cells) and B-cell responses [184]. Additional direct and indirect participation of gut microbiota through the metabolism of dietary vitamins and SCFAs, such as butyrate, also influences the immune response, promoting Treg differentiation and tolerance [185,186]. For example, appropriate Th17 and Th1 responses are important for the clearance of Citrobacter rodentium and non-typhoidal Salmonella enterica infections [187,188]. Although, under normal conditions, the gut constitutes a microenvironment controlled by balanced T-cell responses, prolonged dysbiosis favors an inappropriate persistent proinflammatory response [189].
7. Microbial-Based Therapies
Antibiotics have long been considered in the treatment of IBD because of the prevalence of microbial abnormalities and the presence of known pathogens. Although the use of antibiotics has been clearly supported for the treatment of infectious complications related to IBD, several pieces of evidence have failed to find consistent beneficial associations between antibiotic treatment and IBD remission [190]. Selby et al. did not find beneficial outcomes for CD with treatment with clarithromycin, rifabutin, and clofazimine aimed at eradicating M. avium subspecies paratuberculosis in a two-year randomized clinical trial [191]. Nevertheless, some data support the clinical application of antibiotics such as ciprofloxacin, with or without metronidazole, for treating active fistulizing perianal CD [21,192].
Other methods of bacterial manipulation have provided additional evidence supporting the role of the microbiota in the pathogenesis of IBD. One method of microbiota manipulation in IBD is the introduction of dietary probiotics to control the growth of pathological components and/or switch the global composition towards a healthier one. E. coli Nissle 1917, a nonpathogenic strain clinically used as a probiotic, has been shown to be effective in inducing the remission of patients with UC. In addition, E. coli Nissle 1917 has been associated with maintenance of remission in patients with UC for at least one year [193,194]. Similarly, the probiotic VSL#3, a set of eight bacterial strains (Bifidobacterium breve, B. longum, B. infantis, Lactobacillus acidophilus, L. plantarum, L. paracasei, L. bulgaricus, and Streptococcus thermophilus) has significantly reduced scores of disease severity and induced remission in patients with UC compared to a placebo [37,195,196]. Other probiotics, such as Lactobacillus GG, have been shown to be effective when associated with IBD oral therapy, such as mesalamine [38,197]. Nevertheless, so far, data regarding the effectiveness of probiotics for treating patients with CD have failed to reach substantial association with the induction of remission.
Although currently available probiotics potentially modulate dysbiosis in IBD, their effects are transient and limited in most IBD subsets. In fact, most existing probiotics encounter colonization resistance in the host intestine and are present only for a limited period, even after long-term administration. Therefore, new alternatives have been investigated, including the use of genetically modified organisms with recombinant bacteria as vectors to deliver therapeutic molecules at target sites in the gut. For example, modified strains of Lactobacillus casei BL23 [198] and Streptococcus thermophilus CRL 807 [199] were engineered to produce superoxide dismutase, which has anti-inflammatory properties. Lactococcus lactis-secreting IL-10 [200], elafin (a human protease inhibitor) [201], and IL-27 (an immunosuppressive cytokine) [202] reportedly have anti-inflammatory effects in colitis models and may therefore represent potential candidates for future clinical trials.
Another method of microbiota manipulation of increasing interest for potential therapeutic applications in various diseases is fecal microbiota transplantation (FMT). Recently, randomized clinical trials have assessed the benefits of the use of FMT in the treatment of patients with IBD. Moayyedi et al., for example, found that patients with recently diagnosed UC could be induced to remission after treatment with FMT [203]. Data from another study showed that for patients in remission, treatment with FMT was able to maintain clinical remission in 87.1% of patients compared to 66.7% receiving a placebo. These results indicate that the long-term beneficial effect of FMT in patients with UC in clinical remission could help sustain endoscopic, histological, and clinical remission [204]. A recent meta-analysis showed that FMT was effective in promoting clinical remission (OR = 3.47, 95% CI = 1.93–6.25) and clinical response (OR = 2.48, 95% CI = 1.18–5.21) to patients with active UC when compared to placebo [205]. Studies on the effectiveness of therapeutic microbiota manipulation in IBD are still in progress, and the results are expected to further understanding and guide the potential application in clinical practice.
8. Complex Genetic and Molecular Network
As previously mentioned, by using modern sequencing methodologies, several studies have compared and described the microbiota composition of healthy individuals versus patients with IBD [17,50,168]. For example, recent data revealed a loss of microbial diversity in patients with IBD, with a clear separation between CD and healthy patients, and a more heterogeneous profile in patients with UC [168]. Antibiotic resistance gene levels were increased in IBD, and their abundance was positively correlated with Escherichia and Bacteroides bacteria [206]. Furthermore, higher than normal levels of hydrogen sulfide generated by gut microbiota have been strongly associated with IBD pathogenesis and indicate increased prevalence of sulfate-reducing bacteria, such as Deltaproteobacteria, Desulfotomaculum, Desulfosporosinus, Thermodesulfobacterium, and Thermodesulfovibrio genera [207].
In CD, metagenomic and metaproteomic studies have characterized a decrease in levels of butanoate and propanoate metabolism genes, butyrate, and other SCFAs, in agreement with the decrease in abundance of SCFA-producing Firmicutes bacteria seen in taxonomic profiling studies [208,209]. Several functional changes in the microbiome of IBD have been identified, including an increase in the activities of pathobionts, alterations in the synthesis of amino acids, neurotransmitters, and vitamins, regulation of mineral absorption, degradation of complex carbohydrates, and effects on pathways related to SCFAs, cysteine, and L-arginine synthesis [168,209,210]. In a metagenomic study with a pediatric CD cohort undergoing anti-TNF-α therapy, greater microbiota changes were correlated with higher levels of fungal and human DNA and variations in microbial genes. Examples of these variations include a decrease in selenocompound metabolic pathway activity and an increase in levels of microbial genes encoding glycerophospholipid metabolism, aminobenzoate degradation, sulfur relay systems, and glutathione metabolism [211]. Together, these data indicate that further metabolomic studies could help differentiate, diagnose, and better characterize disease activity [212].
The measurement of RNA transcripts in tissues from patients with IBD may predict the pathways that are activated and involved in the disease. Using deep RNA sequencing, studies of the transcripts have identified molecular subtypes of CD [213,214,215]. A remarkable report on this topic is the Pediatric RISK Stratification Study, which showed that these molecular signatures may predict disease behavior [216]. However, as RNA does not necessarily represent the proteins produced in cells, there are limitations to this approach. For example, a study on the feasibility of metatranscriptomics for fecal samples observed that transcriptional profiles differed more between individuals than metagenomic functional profiles [168]. Metatranscriptomic data also revealed some species-specific biases in the transcriptional activity of gut bacteria, especially with IBD-specific microbial populations, such as F. prausnitzii [210].
Metabolomics research, including analysis of plasma, serum, urine, stool, and intestinal biopsies, has provided data allowing for differentiation between healthy controls and patients with IBD [217]. In the stool of patients with IBD, a loss of metabolites has been observed in concordance with the loss of microbial diversity [168,218]. There were lower levels of secondary bile acids, sphingolipids, short- and medium-chain fatty acids, and vitamins, whereas primary bile acids, amino acids, polyamines, arachidonate, and acylcarnitines were present in higher levels compared to the controls [219]. In IBD, farnesoid X receptor (FXR) activation, triggered by bile salts, led to the downregulation of proinflammatory cytokines, and in CD, intestinal biopsies showed lower expression levels of FXR [220,221]. Another compound commonly associated with IBD is tryptophan, an essential aromatic amino acid obtained from the diet and a precursor of numerous molecules, such as serotonin, melatonin, nicotinamide, and vitamin B3, as well as other intermediates. Common sources of tryptophan are dairy foods, poultry, fish, and oats, and tryptophan is metabolized by both the host and gut microbiota. Microbiota metabolism leads to indole metabolites that can activate aryl hydrocarbon receptors (AhR) and participate in the onset of IBD [222,223]. Few bacteria produce AhR agonists, such as Peptostreptococcus russellii and members of Lactobacillus, whereas indole-propionic acid (IPA) production has been best characterized in Clostridum sporogenes [224]. Indole induces the release of glucagon-like peptide-1 and its derivatives, indoleacetic acid, indole-3-acetaldehyde, indole-3-aldehyde, indoleacrylic acid, and indole-3-propionic acid. IPA via AhR affects T-cell immunity and exerts anti-inflammatory effects in the gut [225]. AhR expression levels are reduced in patients with CD, whereas tryptophan deficiency promotes more severe colitis in mice [226,227].
Proteomic studies have also been conducted to explore innate and adaptive immune mechanisms in IBD. For example, compared to those in the controls, in patients with UC, 46 proteins, excluding neutrophils and their extracellular trap proteins, were more abundant in the colon tissue [228,229]. In intestinal biopsies of patients with CD, the proteomes of human Th1 and Th1/Th17 clones were studied, and 334 proteins were found to be differentially expressed. Cytotoxic proteins, such as granzyme B and perforin, were more abundant in Th1 cells than in Th17 cells, but only in a subgroup of Th1 cell clones from patients with CD [230]. Regarding regulatory T cells (CD4+ Foxp3+), a proteomics study identified a novel protein, THEMIS, which is important for the suppressive function of Treg cells [231]. In agreement with proteomic studies, the lipidome and immune responses have also been investigated in IBD. For instance, the inflamed mucosa of patients with UC showed increased levels of seven eicosanoids (prostaglandin (PG) E2, PGD2, thromboxane B2, 5-hydroxyeicosatetraenoic acid (HETE), 11-HETE, 12-HETE, and 15-HETE) [232]. Macrophages from patients with CD challenged with heat-inactivated E. coli presented lower levels of newly synthesized phosphatidylinositol [233]. Lipidomic analysis of the phosphatidylcholine lipidome profile of rectal mucus obtained from patients with UC showed lower levels of phosphatidylcholine compared to patients with CD and controls. Interestingly, supplementation with delayed-release phosphatidylcholine was clinically effective [234].
Although knowledge of the mechanisms underlying IBD continues to expand, novel data stemming from individual pathogenic constituents are usually not integrated, leading to only limited data being utilized for achieving relevant progress in the field [64]. Regarding the microbiome, complexity becomes even more evident as modern evolving technologies provide an exponential increase in novel information with an overwhelming accumulation of data. Hence, it is currently believed that a better understanding of the pathogenesis of complex diseases such as IBD will depend on the comprehensive integration of knowledge from different “–omes,” including the microbiome, exposome, and genome.
9. Conclusions
In the last two decades, it has become increasingly evident that the microbiome, immune system, genome, and exposome are comprised of highly complex, dynamic, and mutually interactive systems. Nevertheless, the traditional approach for evaluating the individual components that presumably participate in the pathogenesis of IBD, including the microbiota, has not been sufficient to determine the interconnecting pathways underlying the multiple biological systems involved in the disease development. Even using the best of the currently available methods, including clinical, laboratory, endoscopic, histological, and imaging parameters, we still only have a narrow understanding of the intricate mechanisms responsible for chronic inflammation and the peculiar dynamics and specificities affecting each patient with IBD. Consequently, current treatments continue to be mostly empirical and have limited efficacy.
Regarding the microbial component, although causality remains to be clearly established, evidence indicating an association with IBD pathogenesis is rapidly accumulating. However, a better understanding of the probable microbial basis of IBD depends on more complete, deep, and unbiased investigations at multiple and simultaneous levels, including microbial strains and genomic and functional features, ideally allowing the construction of full transcriptomic and metabolomic profiles. High-throughput technologies capable of analyzing innumerable parameters of the microbiome in conjunction with other system variables have been developed in recent years. Hopefully, more integrative analysis will enable data assembly in a comprehensive fashion to build an IBD network and translate information into useful biological insights with direct influence on specific therapeutic targets, clinical decisions, and disease outcomes, which will preferably be individualized.
Author Contributions
Conceptualization, H.S.P.d.S.; writing—original draft preparation, P.T.S., S.L.B.R., B.E.R., Y.M. and H.S.P.d.S.; writing—review and editing, P.T.S. and S.L.B.R.; visualization, B.E.R. and Y.M.; supervision, H.S.P.d.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brazil (CAPES)—Finance Code 001, National Council for Scientific and Technological Development (CNPq) (306634/2019-8), and the Fundação Carlos Chagas Filho de Amparo a Pesquisa do Estado do Rio de Janeiro (FAPERJ) (E26/202.781/2017 and E26/211.740/2015).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors wish to thank the Brazilian research foundations CAPES, CNPq, and FAPERJ for their financial support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Caradonna, L.; Amati, L.; Magrone, T.; Pellegrino, N.M.; Jirillo, E.; Caccavo, D. Enteric bacteria, lipopolysaccharides and related cytokines in inflammatory bowel disease: Biological and clinical significance. J. Endotoxin Res. 2000, 6, 205–214. [Google Scholar] [PubMed]
- Mahida, Y.R.; Rolfe, V.E. Host-bacterial interactions in inflammatory bowel disease. Clin. Sci. 2004, 107, 331–341. [Google Scholar] [CrossRef] [Green Version]
- Parkes, M. Evidence from genetics for a role of autophagy and innate immunity in IBD pathogenesis. Dig. Dis. 2012, 30, 330–333. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.M.; Abreu, M.T. The innate immune system and inflammatory bowel disease. Scand. J. Gastroenterol. 2015, 50, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Shouval, D.S.; Biswas, A.; Goettel, J.A.; McCann, K.; Conaway, E.; Redhu, N.S.; Mascanfroni, I.D.; Al Adham, Z.; Lavoie, S.; Ibourk, M.; et al. Interleukin-10 receptor signaling in innate immune cells regulates mucosal immune tolerance and anti-inflammatory macrophage function. Immunity 2014, 40, 706–719. [Google Scholar] [CrossRef] [Green Version]
- Taurog, J.D.; Richardson, J.A.; Croft, J.T.; Simmons, W.A.; Zhou, M.; Fernandez-Sueiro, J.L.; Balish, E.; Hammer, R.E. The germfree state prevents development of gut and joint inflammatory disease in HLA-B27 transgenic rats. J. Exp. Med. 1994, 180, 2359–2364. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, J.; Bobe, A.M.; Miyoshi, S.; Huang, Y.; Hubert, N.; Delmont, T.O.; Eren, A.M.; Leone, V.; Chang, E.B. Peripartum Antibiotics Promote Gut Dysbiosis, Loss of Immune Tolerance, and Inflammatory Bowel Disease in Genetically Prone Offspring. Cell Rep. 2017, 20, 491–504. [Google Scholar] [CrossRef] [Green Version]
- Packey, C.D.; Sartor, R.B. Interplay of commensal and pathogenic bacteria, genetic mutations, and immunoregulatory defects in the pathogenesis of inflammatory bowel diseases. J. Intern. Med. 2008, 263, 597–606. [Google Scholar] [CrossRef]
- Cohen, L.J.; Cho, J.H.; Gevers, D.; Chu, H. Genetic Factors and the Intestinal Microbiome Guide Development of Microbe-Based Therapies for Inflammatory Bowel Diseases. Gastroenterology 2019, 156, 2174–2189. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, I.; Roy, B.C.; Khan, S.A.; Septer, S.; Umar, S. Microbiome, Metabolome and Inflammatory Bowel Disease. Microorganisms 2016, 4, 20. [Google Scholar] [CrossRef] [Green Version]
- Petersen, C.; Round, J.L. Defining dysbiosis and its influence on host immunity and disease. Cell. Microbiol. 2014, 16, 1024–1033. [Google Scholar] [CrossRef] [PubMed]
- Mishima, Y.; Sartor, R.B. Manipulating resident microbiota to enhance regulatory immune function to treat inflammatory bowel diseases. J. Gastroenterol. 2020, 55, 4–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turpin, W.; Goethel, A.; Bedrani, L.; Croitoru Mdcm, K. Determinants of IBD Heritability: Genes, Bugs, and More. Inflamm. Bowel. Dis. 2018, 24, 1133–1148. [Google Scholar] [CrossRef] [Green Version]
- Kaser, A.; Lee, A.H.; Franke, A.; Glickman, J.N.; Zeissig, S.; Tilg, H.; Nieuwenhuis, E.E.; Higgins, D.E.; Schreiber, S.; Glimcher, L.H.; et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell 2008, 134, 743–756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khor, B.; Gardet, A.; Xavier, R.J. Genetics and pathogenesis of inflammatory bowel disease. Nature 2011, 474, 307–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lucas Lopez, R.; Grande Burgos, M.J.; Galvez, A.; Perez Pulido, R. The human gastrointestinal tract and oral microbiota in inflammatory bowel disease: A state of the science review. APMIS 2017, 125, 3–10. [Google Scholar] [CrossRef]
- Halfvarson, J.; Brislawn, C.J.; Lamendella, R.; Vazquez-Baeza, Y.; Walters, W.A.; Bramer, L.M.; D’Amato, M.; Bonfiglio, F.; McDonald, D.; Gonzalez, A.; et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat. Microbiol. 2017, 2, 17004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, M.; Chang, E.B. Inflammatory Bowel Diseases (IBD) and the Microbiome-Searching the Crime Scene for Clues. Gastroenterology 2021, 160, 524–537. [Google Scholar] [CrossRef]
- Manichanh, C.; Borruel, N.; Casellas, F.; Guarner, F. The gut microbiota in IBD. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 599–608. [Google Scholar] [CrossRef]
- Tlaskalova-Hogenova, H.; Tuckova, L.; Stepankova, R.; Hudcovic, T.; Palova-Jelinkova, L.; Kozakova, H.; Rossmann, P.; Sanchez, D.; Cinova, J.; Hrncir, T.; et al. Involvement of innate immunity in the development of inflammatory and autoimmune diseases. Ann. N. Y. Acad. Sci. 2005, 1051, 787–798. [Google Scholar] [CrossRef]
- Sartor, R.B. Therapeutic manipulation of the enteric microflora in inflammatory bowel diseases: Antibiotics, probiotics, and prebiotics. Gastroenterology 2004, 126, 1620–1633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eun, C.S.; Mishima, Y.; Wohlgemuth, S.; Liu, B.; Bower, M.; Carroll, I.M.; Sartor, R.B. Induction of bacterial antigen-specific colitis by a simplified human microbiota consortium in gnotobiotic interleukin-10-/- mice. Infect. Immun. 2014, 82, 2239–2246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Britton, G.J.; Contijoch, E.J.; Mogno, I.; Vennaro, O.H.; Llewellyn, S.R.; Ng, R.; Li, Z.; Mortha, A.; Merad, M.; Das, A.; et al. Microbiotas from Humans with Inflammatory Bowel Disease Alter the Balance of Gut Th17 and RORgammat(+) Regulatory T Cells and Exacerbate Colitis in Mice. Immunity 2019, 50, 212–224.e4. [Google Scholar] [CrossRef] [Green Version]
- Clooney, A.G.; Eckenberger, J.; Laserna-Mendieta, E.; Sexton, K.A.; Bernstein, M.T.; Vagianos, K.; Sargent, M.; Ryan, F.J.; Moran, C.; Sheehan, D.; et al. Ranking microbiome variance in inflammatory bowel disease: A large longitudinal intercontinental study. Gut 2021, 70, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Miquel, S.; Martin, R.; Rossi, O.; Bermudez-Humaran, L.G.; Chatel, J.M.; Sokol, H.; Thomas, M.; Wells, J.M.; Langella, P. Faecalibacterium prausnitzii and human intestinal health. Curr. Opin. Microbiol. 2013, 16, 255–261. [Google Scholar] [CrossRef]
- Oyri, S.F.; Muzes, G.; Sipos, F. Dysbiotic gut microbiome: A key element of Crohn’s disease. Comp. Immunol. Microbiol. Infect. Dis. 2015, 43, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermudez-Humaran, L.G.; Gratadoux, J.J.; Blugeon, S.; Bridonneau, C.; Furet, J.P.; Corthier, G.; et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, A.; Lordan, C.; Ross, R.P.; Cotter, P.D. Gut microbes from the phylogenetically diverse genus Eubacterium and their various contributions to gut health. Gut Microbes 2020, 12, 1802866. [Google Scholar] [CrossRef]
- Kang, S.; Denman, S.E.; Morrison, M.; Yu, Z.; Dore, J.; Leclerc, M.; McSweeney, C.S. Dysbiosis of fecal microbiota in Crohn’s disease patients as revealed by a custom phylogenetic microarray. Inflamm. Bowel. Dis. 2010, 16, 2034–2042. [Google Scholar] [CrossRef]
- Gevers, D.; Kugathasan, S.; Denson, L.A.; Vazquez-Baeza, Y.; Van Treuren, W.; Ren, B.; Schwager, E.; Knights, D.; Song, S.J.; Yassour, M.; et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014, 15, 382–392. [Google Scholar] [CrossRef] [Green Version]
- Azimi, T.; Nasiri, M.J.; Chirani, A.S.; Pouriran, R.; Dabiri, H. The role of bacteria in the inflammatory bowel disease development: A narrative review. APMIS 2018, 126, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, W.; Marya, N.; Fahed, J.; Leslie, G.; Patel, K.; Cave, D.R. Clostridium difficile in Inflammatory Bowel Disease: A Retrospective Study. Gastroenterol. Res. Pract. 2017, 2017, 4803262. [Google Scholar] [CrossRef] [Green Version]
- Miranda-Bautista, J.; Padilla-Suarez, C.; Bouza, E.; Munoz, P.; Menchen, L.; Marin-Jimenez, I. Listeria monocytogenes infection in inflammatory bowel disease patients: Case series and review of the literature. Eur. J. Gastroenterol. Hepatol. 2014, 26, 1247–1252. [Google Scholar] [CrossRef] [PubMed]
- Lo Presti, A.; Del Chierico, F.; Altomare, A.; Zorzi, F.; Cella, E.; Putignani, L.; Guarino, M.P.L.; Monteleone, G.; Cicala, M.; Angeletti, S.; et al. Exploring the genetic diversity of the 16S rRNA gene of Akkermansia muciniphila in IBD and IBS. Future Microbiol. 2019, 14, 1497–1509. [Google Scholar] [CrossRef] [PubMed]
- Zhai, R.; Xue, X.; Zhang, L.; Yang, X.; Zhao, L.; Zhang, C. Strain-Specific Anti-inflammatory Properties of Two Akkermansia muciniphila Strains on Chronic Colitis in Mice. Front. Cell. Infect. Microbiol. 2019, 9, 239. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Ji, X.; Lu, G.; Zhang, F. The potential of Akkermansia muciniphila in inflammatory bowel disease. Appl. Microbiol. Biotechnol. 2021, 105, 5785–5794. [Google Scholar] [CrossRef]
- Zocco, M.A.; dal Verme, L.Z.; Cremonini, F.; Piscaglia, A.C.; Nista, E.C.; Candelli, M.; Novi, M.; Rigante, D.; Cazzato, I.A.; Ojetti, V.; et al. Efficacy of Lactobacillus GG in maintaining remission of ulcerative colitis. Aliment. Pharm. 2006, 23, 1567–1574. [Google Scholar] [CrossRef]
- Turroni, F.; Duranti, S.; Milani, C.; Lugli, G.A.; van Sinderen, D.; Ventura, M. Bifidobacterium bifidum: A Key Member of the Early Human Gut Microbiota. Microorganisms 2019, 7, 544. [Google Scholar] [CrossRef] [Green Version]
- Elguezabal, N.; Chamorro, S.; Molina, E.; Garrido, J.M.; Izeta, A.; Rodrigo, L.; Juste, R.A. Lactase persistence, NOD2 status and Mycobacterium avium subsp. paratuberculosis infection associations to Inflammatory Bowel Disease. Gut Pathog. 2012, 4, 6. [Google Scholar] [CrossRef] [Green Version]
- Sibartie, S.; Scully, P.; Keohane, J.; O’Neill, S.; O’Mahony, J.; O’Hanlon, D.; Kirwan, W.O.; O’Mahony, L.; Shanahan, F. Mycobacterium avium subsp. Paratuberculosis (MAP) as a modifying factor in Crohn’s disease. Inflamm. Bowel. Dis. 2010, 16, 296–304. [Google Scholar] [CrossRef]
- Palmela, C.; Chevarin, C.; Xu, Z.; Torres, J.; Sevrin, G.; Hirten, R.; Barnich, N.; Ng, S.C.; Colombel, J.F. Adherent-invasive Escherichia coli in inflammatory bowel disease. Gut 2018, 67, 574–587. [Google Scholar] [CrossRef] [PubMed]
- Mirsepasi-Lauridsen, H.C.; Vallance, B.A.; Krogfelt, K.A.; Petersen, A.M. Escherichia coli Pathobionts Associated with Inflammatory Bowel Disease. Clin. Microbiol. Rev. 2019, 32, e00060-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brennan, C.A.; Garrett, W.S. Gut Microbiota, Inflammation, and Colorectal Cancer. Annu. Rev. Microbiol. 2016, 70, 395–411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neut, C.; Bulois, P.; Desreumaux, P.; Membre, J.M.; Lederman, E.; Gambiez, L.; Cortot, A.; Quandalle, P.; van Kruiningen, H.; Colombel, J.F. Changes in the bacterial flora of the neoterminal ileum after ileocolonic resection for Crohn’s disease. Am. J. Gastroenterol. 2002, 97, 939–946. [Google Scholar] [CrossRef]
- Wexler, H.M. Bacteroides: The good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 2007, 20, 593–621. [Google Scholar] [CrossRef] [Green Version]
- Zafar, H.; Saier, M.H., Jr. Gut Bacteroides species in health and disease. Gut Microbes 2021, 13, 1848158. [Google Scholar] [CrossRef]
- Goethel, A.; Croitoru, K.; Philpott, D.J. The interplay between microbes and the immune response in inflammatory bowel disease. J. Physiol. 2018, 596, 3869–3882. [Google Scholar] [CrossRef] [Green Version]
- Kolho, K.L.; Korpela, K.; Jaakkola, T.; Pichai, M.V.; Zoetendal, E.G.; Salonen, A.; de Vos, W.M. Fecal Microbiota in Pediatric Inflammatory Bowel Disease and Its Relation to Inflammation. Am. J. Gastroenterol. 2015, 110, 921–930. [Google Scholar] [CrossRef]
- Zhuang, X.; Liu, C.; Zhan, S.; Tian, Z.; Li, N.; Mao, R.; Zeng, Z.; Chen, M. Gut Microbiota Profile in Pediatric Patients With Inflammatory Bowel Disease: A Systematic Review. Front. Pediatr. 2021, 9, 626232. [Google Scholar] [CrossRef]
- Lloyd-Price, J.; Arze, C.; Ananthakrishnan, A.N.; Schirmer, M.; Avila-Pacheco, J.; Poon, T.W.; Andrews, E.; Ajami, N.J.; Bonham, K.S.; Brislawn, C.J.; et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019, 569, 655–662. [Google Scholar] [CrossRef]
- Ribaldone, D.G.; Caviglia, G.P.; Abdulle, A.; Pellicano, R.; Ditto, M.C.; Morino, M.; Fusaro, E.; Saracco, G.M.; Bugianesi, E.; Astegiano, M. Adalimumab Therapy Improves Intestinal Dysbiosis in Crohn’s Disease. J. Clin. Med. 2019, 8, 1646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Powell, N.; MacDonald, T.T. Recent advances in gut immunology. Parasite Immunol. 2017, 39, e12430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nkamga, V.D.; Lotte, R.; Chirio, D.; Lonjon, M.; Roger, P.M.; Drancourt, M.; Ruimy, R. Methanobrevibacter oralis detected along with Aggregatibacter actinomycetemcomitans in a series of community-acquired brain abscesses. Clin. Microbiol. Infect. 2018, 24, 207–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matijasic, M.; Mestrovic, T.; Paljetak, H.C.; Peric, M.; Baresic, A.; Verbanac, D. Gut Microbiota beyond Bacteria-Mycobiome, Virome, Archaeome, and Eukaryotic Parasites in IBD. Int. J. Mol. Sci. 2020, 21, 2668. [Google Scholar] [CrossRef] [Green Version]
- Ghavami, S.B.; Rostami, E.; Sephay, A.A.; Shahrokh, S.; Balaii, H.; Aghdaei, H.A.; Zali, M.R. Alterations of the human gut Methanobrevibacter smithii as a biomarker for inflammatory bowel diseases. Microb. Pathog. 2018, 117, 285–289. [Google Scholar] [CrossRef]
- Blais Lecours, P.; Marsolais, D.; Cormier, Y.; Berberi, M.; Hache, C.; Bourdages, R.; Duchaine, C. Increased prevalence of Methanosphaera stadtmanae in inflammatory bowel diseases. PLoS ONE 2014, 9, e87734. [Google Scholar] [CrossRef]
- Norman, J.M.; Handley, S.A.; Baldridge, M.T.; Droit, L.; Liu, C.Y.; Keller, B.C.; Kambal, A.; Monaco, C.L.; Zhao, G.; Fleshner, P.; et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 2015, 160, 447–460. [Google Scholar] [CrossRef] [Green Version]
- Cadwell, K.; Patel, K.K.; Maloney, N.S.; Liu, T.C.; Ng, A.C.; Storer, C.E.; Head, R.D.; Xavier, R.; Stappenbeck, T.S.; Virgin, H.W. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell 2010, 141, 1135–1145. [Google Scholar] [CrossRef] [Green Version]
- Perez-Brocal, V.; Garcia-Lopez, R.; Nos, P.; Beltran, B.; Moret, I.; Moya, A. Metagenomic Analysis of Crohn’s Disease Patients Identifies Changes in the Virome and Microbiome Related to Disease Status and Therapy, and Detects Potential Interactions and Biomarkers. Inflamm. Bowel. Dis. 2015, 21, 2515–2532. [Google Scholar] [CrossRef]
- Santiago-Rodriguez, T.M.; Hollister, E.B. Human Virome and Disease: High-Throughput Sequencing for Virus Discovery, Identification of Phage-Bacteria Dysbiosis and Development of Therapeutic Approaches with Emphasis on the Human Gut. Viruses 2019, 11, 656. [Google Scholar] [CrossRef] [Green Version]
- Beheshti-Maal, A.; Shahrokh, S.; Ansari, S.; Mirsamadi, E.S.; Yadegar, A.; Mirjalali, H.; Zali, M.R. Gut mycobiome: The probable determinative role of fungi in IBD patients. Mycoses 2021, 64, 468–476. [Google Scholar] [CrossRef] [PubMed]
- Mason, K.L.; Erb Downward, J.R.; Mason, K.D.; Falkowski, N.R.; Eaton, K.A.; Kao, J.Y.; Young, V.B.; Huffnagle, G.B. Candida albicans and bacterial microbiota interactions in the cecum during recolonization following broad-spectrum antibiotic therapy. Infect. Immun. 2012, 80, 3371–3380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Tilburg Bernardes, E.; Pettersen, V.K.; Gutierrez, M.W.; Laforest-Lapointe, I.; Jendzjowsky, N.G.; Cavin, J.B.; Vicentini, F.A.; Keenan, C.M.; Ramay, H.R.; Samara, J.; et al. Intestinal fungi are causally implicated in microbiome assembly and immune development in mice. Nat. Commun. 2020, 11, 2577. [Google Scholar] [CrossRef] [PubMed]
- de Souza, H.S.P.; Fiocchi, C.; Iliopoulos, D. The IBD interactome: An integrated view of aetiology, pathogenesis and therapy. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 739–749. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 2012, 489, 220–230. [Google Scholar] [CrossRef] [Green Version]
- Forbes, J.D.; Van Domselaar, G.; Bernstein, C.N. The Gut Microbiota in Immune-Mediated Inflammatory Diseases. Front. Microbiol. 2016, 7, 1081. [Google Scholar] [CrossRef] [Green Version]
- Kaplan, G.G. The global burden of IBD: From 2015 to 2025. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 720–727. [Google Scholar] [CrossRef]
- Ng, S.C.; Tang, W.; Leong, R.W.; Chen, M.; Ko, Y.; Studd, C.; Niewiadomski, O.; Bell, S.; Kamm, M.A.; de Silva, H.J.; et al. Environmental risk factors in inflammatory bowel disease: A population-based case-control study in Asia-Pacific. Gut 2015, 64, 1063–1071. [Google Scholar] [CrossRef] [Green Version]
- Cui, G.; Liu, H.; Xu, G.; Laugsand, J.B.; Pang, Z. Exploring Links Between Industrialization, Urbanization, and Chinese Inflammatory Bowel Disease. Front. Med. 2021, 8, 757025. [Google Scholar] [CrossRef]
- Zuo, T.; Kamm, M.A.; Colombel, J.F.; Ng, S.C. Urbanization and the gut microbiota in health and inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 440–452. [Google Scholar] [CrossRef]
- Nielsen, C.C.; Gascon, M.; Osornio-Vargas, A.R.; Shier, C.; Guttman, D.S.; Becker, A.B.; Azad, M.B.; Sears, M.R.; Lefebvre, D.L.; Moraes, T.J.; et al. Natural environments in the urban context and gut microbiota in infants. Environ. Int. 2020, 142, 105881. [Google Scholar] [CrossRef] [PubMed]
- Rogler, G.; Vavricka, S. Exposome in IBD: Recent insights in environmental factors that influence the onset and course of IBD. Inflamm. Bowel. Dis. 2015, 21, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Saidel-Odes, L.; Odes, S. Hygiene hypothesis in inflammatory bowel disease. Ann. Gastroenterol. 2014, 27, 189–190. [Google Scholar] [PubMed]
- Probert, C.S.; Jayanthi, V.; Pinder, D.; Wicks, A.C.; Mayberry, J.F. Epidemiological study of ulcerative proctocolitis in Indian migrants and the indigenous population of Leicestershire. Gut 1992, 33, 687–693. [Google Scholar] [CrossRef] [Green Version]
- Carr, I.; Mayberry, J.F. The effects of migration on ulcerative colitis: A three-year prospective study among Europeans and first- and second- generation South Asians in Leicester (1991–1994). Am. J. Gastroenterol. 1999, 94, 2918–2922. [Google Scholar] [CrossRef]
- Benchimol, E.I.; Manuel, D.G.; To, T.; Mack, D.R.; Nguyen, G.C.; Gommerman, J.L.; Croitoru, K.; Mojaverian, N.; Wang, X.; Quach, P.; et al. Asthma, type 1 and type 2 diabetes mellitus, and inflammatory bowel disease amongst South Asian immigrants to Canada and their children: A population-based cohort study. PLoS ONE 2015, 10, e0123599. [Google Scholar] [CrossRef] [Green Version]
- Ko, Y.; Kariyawasam, V.; Karnib, M.; Butcher, R.; Samuel, D.; Alrubaie, A.; Rahme, N.; McDonald, C.; Cowlishaw, J.; Katelaris, P.; et al. Inflammatory Bowel Disease Environmental Risk Factors: A Population-Based Case-Control Study of Middle Eastern Migration to Australia. Clin. Gastroenterol. Hepatol. 2015, 13, 1453–1463.e1. [Google Scholar] [CrossRef]
- Carr, E.J.; Dooley, J.; Garcia-Perez, J.E.; Lagou, V.; Lee, J.C.; Wouters, C.; Meyts, I.; Goris, A.; Boeckxstaens, G.; Linterman, M.A.; et al. The cellular composition of the human immune system is shaped by age and cohabitation. Nat. Immunol. 2016, 17, 461–468. [Google Scholar] [CrossRef]
- Valles-Colomer, M.; Bacigalupe, R.; Vieira-Silva, S.; Suzuki, S.; Darzi, Y.; Tito, R.Y.; Yamada, T.; Segata, N.; Raes, J.; Falony, G. Variation and transmission of the human gut microbiota across multiple familial generations. Nat. Microbiol. 2022, 7, 87–96. [Google Scholar] [CrossRef]
- Costello, E.K.; Stagaman, K.; Dethlefsen, L.; Bohannan, B.J.; Relman, D.A. The application of ecological theory toward an understanding of the human microbiome. Science 2012, 336, 1255–1262. [Google Scholar] [CrossRef] [Green Version]
- Falony, G.; Joossens, M.; Vieira-Silva, S.; Wang, J.; Darzi, Y.; Faust, K.; Kurilshikov, A.; Bonder, M.J.; Valles-Colomer, M.; Vandeputte, D.; et al. Population-level analysis of gut microbiome variation. Science 2016, 352, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Zhernakova, A.; Kurilshikov, A.; Bonder, M.J.; Tigchelaar, E.F.; Schirmer, M.; Vatanen, T.; Mujagic, Z.; Vila, A.V.; Falony, G.; Vieira-Silva, S.; et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 2016, 352, 565–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Ning, D. Stochastic Community Assembly: Does It Matter in Microbial Ecology? Microbiol. Mol. Biol. Rev. 2017, 81, e00002-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef] [Green Version]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef]
- Pasolli, E.; Asnicar, F.; Manara, S.; Zolfo, M.; Karcher, N.; Armanini, F.; Beghini, F.; Manghi, P.; Tett, A.; Ghensi, P.; et al. Extensive Unexplored Human Microbiome Diversity Revealed by over 150,000 Genomes from Metagenomes Spanning Age, Geography, and Lifestyle. Cell 2019, 176, 649–662.e620. [Google Scholar] [CrossRef] [Green Version]
- Hanski, I.; von Hertzen, L.; Fyhrquist, N.; Koskinen, K.; Torppa, K.; Laatikainen, T.; Karisola, P.; Auvinen, P.; Paulin, L.; Makela, M.J.; et al. Environmental biodiversity, human microbiota, and allergy are interrelated. Proc. Natl. Acad. Sci. USA 2012, 109, 8334–8339. [Google Scholar] [CrossRef] [Green Version]
- Gascon, M.; Vrijheid, M.; Nieuwenhuijsen, M.J. The Built Environment and Child Health: An Overview of Current Evidence. Curr. Environ. Health Rep. 2016, 3, 250–257. [Google Scholar] [CrossRef]
- Genuneit, J.; Standl, M. Epidemiology of Allergy: Natural Course and Risk Factors of Allergic Diseases. Handb. Exp. Pharm. 2022, 268, 21–27. [Google Scholar] [CrossRef]
- Kuenzig, M.E.; Bishay, K.; Leigh, R.; Kaplan, G.G.; Benchimol, E.I.; Crowdscreen, S.R.R.T. Co-occurrence of Asthma and the Inflammatory Bowel Diseases: A Systematic Review and Meta-analysis. Clin. Transl. Gastroenterol. 2018, 9, 188. [Google Scholar] [CrossRef]
- da Luz Moreira, A.; de Campos Lobato, L.F.; de Lima Moreira, J.P.; Luiz, R.R.; Elia, C.; Fiocchi, C.; de Souza, H.S.P. Geosocial Features and Loss of Biodiversity Underlie Variable Rates of Inflammatory Bowel Disease in a Large Developing Country: A Population-Based Study. Inflamm. Bowel. Dis. 2022. [Google Scholar] [CrossRef] [PubMed]
- Ungaro, R.; Bernstein, C.N.; Gearry, R.; Hviid, A.; Kolho, K.L.; Kronman, M.P.; Shaw, S.; Van Kruiningen, H.; Colombel, J.F.; Atreja, A. Antibiotics associated with increased risk of new-onset Crohn’s disease but not ulcerative colitis: A meta-analysis. Am. J. Gastroenterol. 2014, 109, 1728–1738. [Google Scholar] [CrossRef] [PubMed]
- Aniwan, S.; Tremaine, W.J.; Raffals, L.E.; Kane, S.V.; Loftus, E.V., Jr. Antibiotic Use and New-Onset Inflammatory Bowel Disease in Olmsted County, Minnesota: A Population-Based Case-Control Study. J. Crohns Colitis 2018, 12, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Shaw, S.Y.; Blanchard, J.F.; Bernstein, C.N. Association between the use of antibiotics in the first year of life and pediatric inflammatory bowel disease. Am. J. Gastroenterol. 2010, 105, 2687–2692. [Google Scholar] [CrossRef]
- Hviid, A.; Svanstrom, H.; Frisch, M. Antibiotic use and inflammatory bowel diseases in childhood. Gut 2011, 60, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Ortqvist, A.K.; Lundholm, C.; Halfvarson, J.; Ludvigsson, J.F.; Almqvist, C. Fetal and early life antibiotics exposure and very early onset inflammatory bowel disease: A population-based study. Gut 2019, 68, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Hildebrand, H.; Malmborg, P.; Askling, J.; Ekbom, A.; Montgomery, S.M. Early-life exposures associated with antibiotic use and risk of subsequent Crohn’s disease. Scand. J. Gastroenterol. 2008, 43, 961–966. [Google Scholar] [CrossRef]
- Vu Thi Ngoc, B.; Ho Bich, H.; Galazzo, G.; Vu Tien Viet, D.; Oomen, M.; Nghiem Nguyen Minh, T.; Tran Huy, H.; van Doorn, H.R.; Wertheim, H.F.L.; Penders, J. Cross-Sectional Analysis of the Microbiota of Human Gut and Its Direct Environment in a Household Cohort with High Background of Antibiotic Use. Microorganisms 2021, 9, 2115. [Google Scholar] [CrossRef]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [Green Version]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [Green Version]
- Hou, J.K.; Abraham, B.; El-Serag, H. Dietary intake and risk of developing inflammatory bowel disease: A systematic review of the literature. Am. J. Gastroenterol. 2011, 106, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Ananthakrishnan, A.N. Epidemiology and risk factors for IBD. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Sartorio, M.U.A.; Pendezza, E.; Coppola, S.; Paparo, L.; D’Auria, E.; Zuccotti, G.V.; Berni Canani, R. Potential Role of Omega-3 Polyunsaturated Fatty Acids in Pediatric Food Allergy. Nutrients 2021, 14, 152. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Wang, Y.; Gao, H.; Li, D.; Jiang, R.; Ge, L.; Tong, C.; Xu, K. Associations among Dietary Omega-3 Polyunsaturated Fatty Acids, the Gut Microbiota, and Intestinal Immunity. Mediat. Inflamm. 2021, 2021, 8879227. [Google Scholar] [CrossRef]
- Mayneris-Perxachs, J.; Bolick, D.T.; Leng, J.; Medlock, G.L.; Kolling, G.L.; Papin, J.A.; Swann, J.R.; Guerrant, R.L. Protein- and zinc-deficient diets modulate the murine microbiome and metabolic phenotype. Am. J. Clin. Nutr. 2016, 104, 1253–1262. [Google Scholar] [CrossRef] [PubMed]
- Virtanen, S.M. Dietary factors in the development of type 1 diabetes. Pediatr. Diabetes 2016, 17 (Suppl. S22), 49–55. [Google Scholar] [CrossRef] [PubMed]
- Del Pinto, R.; Pietropaoli, D.; Chandar, A.K.; Ferri, C.; Cominelli, F. Association Between Inflammatory Bowel Disease and Vitamin D Deficiency: A Systematic Review and Meta-analysis. Inflamm. Bowel. Dis. 2015, 21, 2708–2717. [Google Scholar] [CrossRef] [Green Version]
- de Bruyn, J.R.; van Heeckeren, R.; Ponsioen, C.Y.; van den Brink, G.R.; Lowenberg, M.; Bredenoord, A.J.; Frijstein, G.; D’Haens, G.R. Vitamin D deficiency in Crohn’s disease and healthy controls: A prospective case-control study in the Netherlands. J. Crohns Colitis 2014, 8, 1267–1273. [Google Scholar] [CrossRef] [Green Version]
- Kearney, J. Food consumption trends and drivers. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 2793–2807. [Google Scholar] [CrossRef]
- Cho, J.H.; Brant, S.R. Recent insights into the genetics of inflammatory bowel disease. Gastroenterology 2011, 140, 1704–1712. [Google Scholar] [CrossRef] [Green Version]
- Ramos, G.P.; Papadakis, K.A. Mechanisms of Disease: Inflammatory Bowel Diseases. Mayo. Clin. Proc. 2019, 94, 155–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Lange, K.M.; Barrett, J.C. Understanding inflammatory bowel disease via immunogenetics. J. Autoimmun. 2015, 64, 91–100. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Oshima, T.; Tomita, T.; Fukui, H.; Miwa, H. Butyrate Alleviates Cytokine-Induced Barrier Dysfunction by Modifying Claudin-2 Levels. Biology 2021, 10, 205. [Google Scholar] [CrossRef]
- Imhann, F.; Vich Vila, A.; Bonder, M.J.; Fu, J.; Gevers, D.; Visschedijk, M.C.; Spekhorst, L.M.; Alberts, R.; Franke, L.; van Dullemen, H.M.; et al. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut 2018, 67, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Glick, D.; Barth, S.; Macleod, K.F. Autophagy: Cellular and molecular mechanisms. J. Pathol. 2010, 221, 3–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campoy, E.; Colombo, M.I. Autophagy in intracellular bacterial infection. Biochim. Biophys. Acta 2009, 1793, 1465–1477. [Google Scholar] [CrossRef] [Green Version]
- Elliott, T.R.; Hudspith, B.N.; Rayment, N.B.; Prescott, N.J.; Petrovska, L.; Hermon-Taylor, J.; Brostoff, J.; Boussioutas, A.; Mathew, C.G.; Sanderson, J.D. Defective macrophage handling of Escherichia coli in Crohn’s disease. J. Gastroenterol. Hepatol. 2015, 30, 1265–1274. [Google Scholar] [CrossRef]
- Hampe, J.; Franke, A.; Rosenstiel, P.; Till, A.; Teuber, M.; Huse, K.; Albrecht, M.; Mayr, G.; De La Vega, F.M.; Briggs, J.; et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet. 2007, 39, 207–211. [Google Scholar] [CrossRef]
- Rioux, J.D.; Xavier, R.J.; Taylor, K.D.; Silverberg, M.S.; Goyette, P.; Huett, A.; Green, T.; Kuballa, P.; Barmada, M.M.; Datta, L.W.; et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 2007, 39, 596–604. [Google Scholar] [CrossRef]
- Messer, J.S.; Murphy, S.F.; Logsdon, M.F.; Lodolce, J.P.; Grimm, W.A.; Bartulis, S.J.; Vogel, T.P.; Burn, M.; Boone, D.L. The Crohn’s disease: Associated ATG16L1 variant and Salmonella invasion. BMJ Open 2013, 3, e002790. [Google Scholar] [CrossRef] [Green Version]
- Conway, K.L.; Kuballa, P.; Song, J.H.; Patel, K.K.; Castoreno, A.B.; Yilmaz, O.H.; Jijon, H.B.; Zhang, M.; Aldrich, L.N.; Villablanca, E.J.; et al. Atg16l1 is required for autophagy in intestinal epithelial cells and protection of mice from Salmonella infection. Gastroenterology 2013, 145, 1347–1357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scharl, M.; Rogler, G. Inflammatory bowel disease: Dysfunction of autophagy? Dig. Dis. 2012, 30 (Suppl. S3), 12–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cadwell, K.; Liu, J.Y.; Brown, S.L.; Miyoshi, H.; Loh, J.; Lennerz, J.K.; Kishi, C.; Kc, W.; Carrero, J.A.; Hunt, S.; et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 2008, 456, 259–263. [Google Scholar] [CrossRef] [Green Version]
- Cooney, R.; Baker, J.; Brain, O.; Danis, B.; Pichulik, T.; Allan, P.; Ferguson, D.J.; Campbell, B.J.; Jewell, D.; Simmons, A. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat. Med. 2010, 16, 90–97. [Google Scholar] [CrossRef]
- Sorbara, M.T.; Ellison, L.K.; Ramjeet, M.; Travassos, L.H.; Jones, N.L.; Girardin, S.E.; Philpott, D.J. The protein ATG16L1 suppresses inflammatory cytokines induced by the intracellular sensors Nod1 and Nod2 in an autophagy-independent manner. Immunity 2013, 39, 858–873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rocha, J.D.; Schlossmacher, M.G.; Philpott, D.J. LRRK2 and Nod2 promote lysozyme sorting in Paneth cells. Nat. Immunol. 2015, 16, 898–900. [Google Scholar] [CrossRef]
- Cadwell, K.; Patel, K.K.; Komatsu, M.; Virgin, H.W.t.; Stappenbeck, T.S. A common role for Atg16L1, Atg5 and Atg7 in small intestinal Paneth cells and Crohn disease. Autophagy 2009, 5, 250–252. [Google Scholar] [CrossRef] [Green Version]
- Adolph, T.E.; Tomczak, M.F.; Niederreiter, L.; Ko, H.J.; Bock, J.; Martinez-Naves, E.; Glickman, J.N.; Tschurtschenthaler, M.; Hartwig, J.; Hosomi, S.; et al. Paneth cells as a site of origin for intestinal inflammation. Nature 2013, 503, 272–276. [Google Scholar] [CrossRef] [Green Version]
- Baxt, L.A.; Xavier, R.J. Role of Autophagy in the Maintenance of Intestinal Homeostasis. Gastroenterology 2015, 149, 553–562. [Google Scholar] [CrossRef] [Green Version]
- Caruso, R.; Warner, N.; Inohara, N.; Nunez, G. NOD1 and NOD2: Signaling, host defense, and inflammatory disease. Immunity 2014, 41, 898–908. [Google Scholar] [CrossRef] [Green Version]
- Negroni, A.; Pierdomenico, M.; Cucchiara, S.; Stronati, L. NOD2 and inflammation: Current insights. J. Inflamm. Res. 2018, 11, 49–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naser, S.A.; Arce, M.; Khaja, A.; Fernandez, M.; Naser, N.; Elwasila, S.; Thanigachalam, S. Role of ATG16L, NOD2 and IL23R in Crohn’s disease pathogenesis. World J. Gastroenterol. 2012, 18, 412–424. [Google Scholar] [CrossRef] [PubMed]
- Knights, D.; Lassen, K.G.; Xavier, R.J. Advances in inflammatory bowel disease pathogenesis: Linking host genetics and the microbiome. Gut 2013, 62, 1505–1510. [Google Scholar] [CrossRef]
- Wehkamp, J.; Salzman, N.H.; Porter, E.; Nuding, S.; Weichenthal, M.; Petras, R.E.; Shen, B.; Schaeffeler, E.; Schwab, M.; Linzmeier, R.; et al. Reduced Paneth cell alpha-defensins in ileal Crohn’s disease. Proc. Natl. Acad. Sci. USA 2005, 102, 18129–18134. [Google Scholar] [CrossRef] [Green Version]
- Uhlig, H.H.; McKenzie, B.S.; Hue, S.; Thompson, C.; Joyce-Shaikh, B.; Stepankova, R.; Robinson, N.; Buonocore, S.; Tlaskalova-Hogenova, H.; Cua, D.J.; et al. Differential activity of IL-12 and IL-23 in mucosal and systemic innate immune pathology. Immunity 2006, 25, 309–318. [Google Scholar] [CrossRef] [Green Version]
- Yen, D.; Cheung, J.; Scheerens, H.; Poulet, F.; McClanahan, T.; McKenzie, B.; Kleinschek, M.A.; Owyang, A.; Mattson, J.; Blumenschein, W.; et al. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J. Clin. Investig. 2006, 116, 1310–1316. [Google Scholar] [CrossRef] [PubMed]
- Elson, C.O.; Cong, Y.; Weaver, C.T.; Schoeb, T.R.; McClanahan, T.K.; Fick, R.B.; Kastelein, R.A. Monoclonal anti-interleukin 23 reverses active colitis in a T cell-mediated model in mice. Gastroenterology 2007, 132, 2359–2370. [Google Scholar] [CrossRef]
- Yu, R.Y.; Gallagher, G. A naturally occurring, soluble antagonist of human IL-23 inhibits the development and in vitro function of human Th17 cells. J. Immunol. 2010, 185, 7302–7308. [Google Scholar] [CrossRef]
- Liu, B.; Gulati, A.S.; Cantillana, V.; Henry, S.C.; Schmidt, E.A.; Daniell, X.; Grossniklaus, E.; Schoenborn, A.A.; Sartor, R.B.; Taylor, G.A. Irgm1-deficient mice exhibit Paneth cell abnormalities and increased susceptibility to acute intestinal inflammation. Am. J. Physiol. Liver Physiol. 2013, 305, G573–G584. [Google Scholar] [CrossRef] [Green Version]
- Spalinger, M.R.; McCole, D.F.; Rogler, G.; Scharl, M. Protein tyrosine phosphatase non-receptor type 2 and inflammatory bowel disease. World J. Gastroenterol. 2016, 22, 1034–1044. [Google Scholar] [CrossRef] [Green Version]
- Spalinger, M.R.; Manzini, R.; Hering, L.; Riggs, J.B.; Gottier, C.; Lang, S.; Atrott, K.; Fettelschoss, A.; Olomski, F.; Kundig, T.M.; et al. PTPN2 Regulates Inflammasome Activation and Controls Onset of Intestinal Inflammation and Colon Cancer. Cell Rep. 2018, 22, 1835–1848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scharl, M.; Mwinyi, J.; Fischbeck, A.; Leucht, K.; Eloranta, J.J.; Arikkat, J.; Pesch, T.; Kellermeier, S.; Mair, A.; Kullak-Ublick, G.A.; et al. Crohn’s disease-associated polymorphism within the PTPN2 gene affects muramyl-dipeptide-induced cytokine secretion and autophagy. Inflamm. Bowel. Dis. 2012, 18, 900–912. [Google Scholar] [CrossRef] [PubMed]
- Spalinger, M.R.; Kasper, S.; Chassard, C.; Raselli, T.; Frey-Wagner, I.; Gottier, C.; Lang, S.; Atrott, K.; Vavricka, S.R.; Mair, F.; et al. PTPN2 controls differentiation of CD4(+) T cells and limits intestinal inflammation and intestinal dysbiosis. Mucosal. Immunol. 2015, 8, 918–929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takagawa, T.; Kitani, A.; Fuss, I.; Levine, B.; Brant, S.R.; Peter, I.; Tajima, M.; Nakamura, S.; Strober, W. An increase in LRRK2 suppresses autophagy and enhances Dectin-1-induced immunity in a mouse model of colitis. Sci. Transl. Med. 2018, 10, eaan8162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kellermayer, R. Challenges for epigenetic research in inflammatory bowel diseases. Epigenomics 2017, 9, 527–538. [Google Scholar] [CrossRef]
- Benchimol, E.I.; Mack, D.R.; Guttmann, A.; Nguyen, G.C.; To, T.; Mojaverian, N.; Quach, P.; Manuel, D.G. Inflammatory bowel disease in immigrants to Canada and their children: A population-based cohort study. Am. J. Gastroenterol. 2015, 110, 553–563. [Google Scholar] [CrossRef]
- Dabritz, J.; Menheniott, T.R. Linking immunity, epigenetics, and cancer in inflammatory bowel disease. Inflamm. Bowel. Dis. 2014, 20, 1638–1654. [Google Scholar] [CrossRef] [Green Version]
- Amatullah, H.; Jeffrey, K.L. Epigenome-metabolome-microbiome axis in health and IBD. Curr. Opin. Microbiol. 2020, 56, 97–108. [Google Scholar] [CrossRef]
- Woo, V.; Alenghat, T. Epigenetic regulation by gut microbiota. Gut Microbes 2022, 14, 2022407. [Google Scholar] [CrossRef]
- Wawrzyniak, M.; Scharl, M. Genetics and epigenetics of inflammatory bowel disease. Swiss Med. Wkly 2018, 148, w14671. [Google Scholar] [CrossRef]
- Ray, G.; Longworth, M.S. Epigenetics, DNA Organization, and Inflammatory Bowel Disease. Inflamm. Bowel. Dis. 2019, 25, 235–247. [Google Scholar] [CrossRef] [PubMed]
- Kalla, R.; Ventham, N.T.; Kennedy, N.A. MicroRNAs: New players in inflammatory bowel disease. Gut 2015, 64, 1008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, W.; Chen, L.; Xu, L.; Bilotta, A.J.; Yao, S.; Liu, Z.; Cong, Y. MicroRNA-10a Negatively Regulates CD4(+) T Cell IL-10 Production through Suppression of Blimp1. J. Immunol. 2021, 207, 985–995. [Google Scholar] [CrossRef] [PubMed]
- Chapman, C.G.; Pekow, J. The emerging role of miRNAs in inflammatory bowel disease: A review. Ther. Adv. Gastroenterol. 2015, 8, 4–22. [Google Scholar] [CrossRef] [Green Version]
- Taganov, K.D.; Boldin, M.P.; Chang, K.J.; Baltimore, D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar] [CrossRef] [Green Version]
- Ceppi, M.; Pereira, P.M.; Dunand-Sauthier, I.; Barras, E.; Reith, W.; Santos, M.A.; Pierre, P. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc. Natl. Acad. Sci. USA 2009, 106, 2735–2740. [Google Scholar] [CrossRef] [Green Version]
- Aleksandrova, K.; Romero-Mosquera, B.; Hernandez, V. Diet, Gut Microbiome and Epigenetics: Emerging Links with Inflammatory Bowel Diseases and Prospects for Management and Prevention. Nutrients 2017, 9, 962. [Google Scholar] [CrossRef]
- Bourassa, M.W.; Alim, I.; Bultman, S.J.; Ratan, R.R. Butyrate, neuroepigenetics and the gut microbiome: Can a high fiber diet improve brain health? Neurosci. Lett. 2016, 625, 56–63. [Google Scholar] [CrossRef] [Green Version]
- Brown, A.J.; Goldsworthy, S.M.; Barnes, A.A.; Eilert, M.M.; Tcheang, L.; Daniels, D.; Muir, A.I.; Wigglesworth, M.J.; Kinghorn, I.; Fraser, N.J.; et al. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J. Biol. Chem. 2003, 278, 11312–11319. [Google Scholar] [CrossRef] [Green Version]
- Puddu, A.; Sanguineti, R.; Montecucco, F.; Viviani, G.L. Evidence for the gut microbiota short-chain fatty acids as key pathophysiological molecules improving diabetes. Mediat. Inflamm. 2014, 2014, 162021. [Google Scholar] [CrossRef]
- Paul, B.; Barnes, S.; Demark-Wahnefried, W.; Morrow, C.; Salvador, C.; Skibola, C.; Tollefsbol, T.O. Influences of diet and the gut microbiome on epigenetic modulation in cancer and other diseases. Clin. Epigenetics 2015, 7, 112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miro-Blanch, J.; Yanes, O. Epigenetic Regulation at the Interplay Between Gut Microbiota and Host Metabolism. Front. Genet. 2019, 10, 638. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Kumar, A. Microbial pathogenesis in inflammatory bowel diseases. Microb. Pathog. 2022, 163, 105383. [Google Scholar] [CrossRef]
- Lecuyer, E.; Rakotobe, S.; Lengline-Garnier, H.; Lebreton, C.; Picard, M.; Juste, C.; Fritzen, R.; Eberl, G.; McCoy, K.D.; Macpherson, A.J.; et al. Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity 2014, 40, 608–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, B.; Chen, H.; Shu, X.; Yin, Y.; Li, J.; Qin, J.; Chen, L.; Peng, K.; Xu, F.; Gu, W.; et al. Presence of Segmented Filamentous Bacteria in Human Children and Its Potential Role in the Modulation of Human Gut Immunity. Front. Microbiol. 2018, 9, 1403. [Google Scholar] [CrossRef]
- Hansen, J.J.; Sartor, R.B. Therapeutic Manipulation of the Microbiome in IBD: Current Results and Future Approaches. Curr. Treat. Options Gastroenterol. 2015, 13, 105–120. [Google Scholar] [CrossRef] [Green Version]
- Richard, M.L.; Sokol, H. The gut mycobiota: Insights into analysis, environmental interactions and role in gastrointestinal diseases. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 331–345. [Google Scholar] [CrossRef]
- Franzosa, E.A.; Sirota-Madi, A.; Avila-Pacheco, J.; Fornelos, N.; Haiser, H.J.; Reinker, S.; Vatanen, T.; Hall, A.B.; Mallick, H.; McIver, L.J.; et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat. Microbiol. 2019, 4, 293–305. [Google Scholar] [CrossRef]
- Shin, N.R.; Whon, T.W.; Bae, J.W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef]
- Sanders, D.J.; Inniss, S.; Sebepos-Rogers, G.; Rahman, F.Z.; Smith, A.M. The role of the microbiome in gastrointestinal inflammation. Biosci. Rep. 2021, 41, BSR20203850. [Google Scholar] [CrossRef]
- Nanini, H.F.; Bernardazzi, C.; Castro, F.; de Souza, H.S.P. Damage-associated molecular patterns in inflammatory bowel disease: From biomarkers to therapeutic targets. World J. Gastroenterol. 2018, 24, 4622–4634. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Akira, S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 2011, 34, 637–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.H.; Liu, L.; Hou, Y.Y.; Shen, S.N.; Wang, T.T. C-type lectin receptor-mediated immune recognition and response of the microbiota in the gut. Gastroenterol. Rep. 2019, 7, 312–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Y.; Li, X.; Liu, S.; Zhang, Y.; Zhang, D. Toll-like Receptors and Inflammatory Bowel Disease. Front. Immunol. 2018, 9, 72. [Google Scholar] [CrossRef] [Green Version]
- Lavin, Y.; Winter, D.; Blecher-Gonen, R.; David, E.; Keren-Shaul, H.; Merad, M.; Jung, S.; Amit, I. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 2014, 159, 1312–1326. [Google Scholar] [CrossRef] [Green Version]
- Okabe, Y.; Medzhitov, R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell 2014, 157, 832–844. [Google Scholar] [CrossRef] [Green Version]
- Bernink, J.H.; Peters, C.P.; Munneke, M.; te Velde, A.A.; Meijer, S.L.; Weijer, K.; Hreggvidsdottir, H.S.; Heinsbroek, S.E.; Legrand, N.; Buskens, C.J.; et al. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat. Immunol. 2013, 14, 221–229. [Google Scholar] [CrossRef]
- Ebihara, T. Dichotomous Regulation of Acquired Immunity by Innate Lymphoid Cells. Cells 2020, 9, 1193. [Google Scholar] [CrossRef]
- Martinon, F.; Mayor, A.; Tschopp, J. The inflammasomes: Guardians of the body. Annu. Rev. Immunol. 2009, 27, 229–265. [Google Scholar] [CrossRef] [Green Version]
- Lamkanfi, M.; Dixit, V.M. Modulation of inflammasome pathways by bacterial and viral pathogens. J. Immunol. 2011, 187, 597–602. [Google Scholar] [CrossRef] [Green Version]
- Vanhove, W.; Peeters, P.M.; Staelens, D.; Schraenen, A.; Van der Goten, J.; Cleynen, I.; De Schepper, S.; Van Lommel, L.; Reynaert, N.L.; Schuit, F.; et al. Strong Upregulation of AIM2 and IFI16 Inflammasomes in the Mucosa of Patients with Active Inflammatory Bowel Disease. Inflamm. Bowel. Dis. 2015, 21, 2673–2682. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Dong, Y.; Ye, M.; Jin, S.; Yang, J.; Joosse, M.E.; Sun, Y.; Zhang, J.; Lazarev, M.; Brant, S.R.; et al. The Pathogenic Role of NLRP3 Inflammasome Activation in Inflammatory Bowel Diseases of Both Mice and Humans. J. Crohns Colitis 2017, 11, 737–750. [Google Scholar] [CrossRef] [PubMed]
- Zaki, M.H.; Boyd, K.L.; Vogel, P.; Kastan, M.B.; Lamkanfi, M.; Kanneganti, T.D. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity 2010, 32, 379–391. [Google Scholar] [CrossRef] [Green Version]
- Geremia, A.; Arancibia-Carcamo, C.V. Innate Lymphoid Cells in Intestinal Inflammation. Front. Immunol. 2017, 8, 1296. [Google Scholar] [CrossRef] [PubMed]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450. [Google Scholar] [CrossRef]
- Tanoue, T.; Atarashi, K.; Honda, K. Development and maintenance of intestinal regulatory T cells. Nat. Rev. Immunol. 2016, 16, 295–309. [Google Scholar] [CrossRef]
- Shepherd, F.R.; McLaren, J.E. T Cell Immunity to Bacterial Pathogens: Mechanisms of Immune Control and Bacterial Evasion. Int. J. Mol. Sci. 2020, 21, 6144. [Google Scholar] [CrossRef]
- Silberger, D.J.; Zindl, C.L.; Weaver, C.T. Citrobacter rodentium: A model enteropathogen for understanding the interplay of innate and adaptive components of type 3 immunity. Mucosal. Immunol. 2017, 10, 1108–1117. [Google Scholar] [CrossRef] [Green Version]
- Gagliani, N.; Amezcua Vesely, M.C.; Iseppon, A.; Brockmann, L.; Xu, H.; Palm, N.W.; de Zoete, M.R.; Licona-Limon, P.; Paiva, R.S.; Ching, T.; et al. Th17 cells transdifferentiate into regulatory T cells during resolution of inflammation. Nature 2015, 523, 221–225. [Google Scholar] [CrossRef]
- Ledder, O.; Turner, D. Antibiotics in IBD: Still a Role in the Biological Era? Inflamm. Bowel. Dis. 2018, 24, 1676–1688. [Google Scholar] [CrossRef]
- Selby, W.; Pavli, P.; Crotty, B.; Florin, T.; Radford-Smith, G.; Gibson, P.; Mitchell, B.; Connell, W.; Read, R.; Merrett, M.; et al. Two-year combination antibiotic therapy with clarithromycin, rifabutin, and clofazimine for Crohn’s disease. Gastroenterology 2007, 132, 2313–2319. [Google Scholar] [CrossRef]
- Thia, K.T.; Mahadevan, U.; Feagan, B.G.; Wong, C.; Cockeram, A.; Bitton, A.; Bernstein, C.N.; Sandborn, W.J. Ciprofloxacin or metronidazole for the treatment of perianal fistulas in patients with Crohn’s disease: A randomized, double-blind, placebo-controlled pilot study. Inflamm. Bowel. Dis. 2009, 15, 17–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matthes, H.; Krummenerl, T.; Giensch, M.; Wolff, C.; Schulze, J. Clinical trial: Probiotic treatment of acute distal ulcerative colitis with rectally administered Escherichia coli Nissle 1917 (EcN). BMC Complement. Altern. Med. 2010, 10, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fedorak, R.N. Probiotics in the management of ulcerative colitis. Gastroenterol. Hepatol. 2010, 6, 688–690. [Google Scholar]
- Bibiloni, R.; Fedorak, R.N.; Tannock, G.W.; Madsen, K.L.; Gionchetti, P.; Campieri, M.; De Simone, C.; Sartor, R.B. VSL#3 probiotic-mixture induces remission in patients with active ulcerative colitis. Am. J. Gastroenterol. 2005, 100, 1539–1546. [Google Scholar] [CrossRef] [PubMed]
- Weingarden, A.R.; Vaughn, B.P. Intestinal microbiota, fecal microbiota transplantation, and inflammatory bowel disease. Gut Microbes 2017, 8, 238–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wasilewski, A.; Zielinska, M.; Storr, M.; Fichna, J. Beneficial Effects of Probiotics, Prebiotics, Synbiotics, and Psychobiotics in Inflammatory Bowel Disease. Inflamm. Bowel. Dis. 2015, 21, 1674–1682. [Google Scholar] [CrossRef]
- Motta, J.P.; Bermudez-Humaran, L.G.; Deraison, C.; Martin, L.; Rolland, C.; Rousset, P.; Boue, J.; Dietrich, G.; Chapman, K.; Kharrat, P.; et al. Food-grade bacteria expressing elafin protect against inflammation and restore colon homeostasis. Sci. Transl. Med. 2012, 4, 158ra144. [Google Scholar] [CrossRef]
- Hanson, M.L.; Hixon, J.A.; Li, W.; Felber, B.K.; Anver, M.R.; Stewart, C.A.; Janelsins, B.M.; Datta, S.K.; Shen, W.; McLean, M.H.; et al. Oral delivery of IL-27 recombinant bacteria attenuates immune colitis in mice. Gastroenterology 2014, 146, 210–221.e213. [Google Scholar] [CrossRef] [Green Version]
- Steidler, L.; Hans, W.; Schotte, L.; Neirynck, S.; Obermeier, F.; Falk, W.; Fiers, W.; Remaut, E. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science 2000, 289, 1352–1355. [Google Scholar] [CrossRef] [Green Version]
- LeBlanc, J.G.; del Carmen, S.; Miyoshi, A.; Azevedo, V.; Sesma, F.; Langella, P.; Bermudez-Humaran, L.G.; Watterlot, L.; Perdigon, G.; de Moreno de LeBlanc, A. Use of superoxide dismutase and catalase producing lactic acid bacteria in TNBS induced Crohn’s disease in mice. J. Biotechnol. 2011, 151, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Del Carmen, S.; de Moreno de LeBlanc, A.; Martin, R.; Chain, F.; Langella, P.; Bermudez-Humaran, L.G.; LeBlanc, J.G. Genetically engineered immunomodulatory Streptococcus thermophilus strains producing antioxidant enzymes exhibit enhanced anti-inflammatory activities. Appl. Environ. Microbiol. 2014, 80, 869–877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moayyedi, P.; Surette, M.G.; Kim, P.T.; Libertucci, J.; Wolfe, M.; Onischi, C.; Armstrong, D.; Marshall, J.K.; Kassam, Z.; Reinisch, W.; et al. Fecal Microbiota Transplantation Induces Remission in Patients With Active Ulcerative Colitis in a Randomized Controlled Trial. Gastroenterology 2015, 149, 102–109.e106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sood, A.; Mahajan, R.; Singh, A.; Midha, V.; Mehta, V.; Narang, V.; Singh, T.; Singh Pannu, A. Role of Faecal Microbiota Transplantation for Maintenance of Remission in Patients With Ulcerative Colitis: A Pilot Study. J. Crohns Colitis 2019, 13, 1311–1317. [Google Scholar] [CrossRef]
- Dang, X.; Xu, M.; Liu, D.; Zhou, D.; Yang, W. Assessing the efficacy and safety of fecal microbiota transplantation and probiotic VSL#3 for active ulcerative colitis: A systematic review and meta-analysis. PLoS ONE 2020, 15, e0228846. [Google Scholar] [CrossRef]
- Vich Vila, A.; Imhann, F.; Collij, V.; Jankipersadsing, S.A.; Gurry, T.; Mujagic, Z.; Kurilshikov, A.; Bonder, M.J.; Jiang, X.; Tigchelaar, E.F.; et al. Gut microbiota composition and functional changes in inflammatory bowel disease and irritable bowel syndrome. Sci. Transl. Med. 2018, 10, eaap8914. [Google Scholar] [CrossRef] [Green Version]
- Blachier, F.; Davila, A.M.; Mimoun, S.; Benetti, P.H.; Atanasiu, C.; Andriamihaja, M.; Benamouzig, R.; Bouillaud, F.; Tome, D. Luminal sulfide and large intestine mucosa: Friend or foe? Amino Acids 2010, 39, 335–347. [Google Scholar] [CrossRef]
- Kostic, A.D.; Xavier, R.J.; Gevers, D. The microbiome in inflammatory bowel disease: Current status and the future ahead. Gastroenterology 2014, 146, 1489–1499. [Google Scholar] [CrossRef] [Green Version]
- Morgan, X.C.; Tickle, T.L.; Sokol, H.; Gevers, D.; Devaney, K.L.; Ward, D.V.; Reyes, J.A.; Shah, S.A.; LeLeiko, N.; Snapper, S.B.; et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012, 13, R79. [Google Scholar] [CrossRef]
- Schirmer, M.; Garner, A.; Vlamakis, H.; Xavier, R.J. Microbial genes and pathways in inflammatory bowel disease. Nat. Rev. Microbiol. 2019, 17, 497–511. [Google Scholar] [CrossRef]
- Lewis, J.D.; Chen, E.Z.; Baldassano, R.N.; Otley, A.R.; Griffiths, A.M.; Lee, D.; Bittinger, K.; Bailey, A.; Friedman, E.S.; Hoffmann, C.; et al. Inflammation, Antibiotics, and Diet as Environmental Stressors of the Gut Microbiome in Pediatric Crohn’s Disease. Cell Host Microbe 2015, 18, 489–500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serrano-Gomez, G.; Mayorga, L.; Oyarzun, I.; Roca, J.; Borruel, N.; Casellas, F.; Varela, E.; Pozuelo, M.; Machiels, K.; Guarner, F.; et al. Dysbiosis and relapse-related microbiome in inflammatory bowel disease: A shotgun metagenomic approach. Comput. Struct. Biotechnol. J. 2021, 19, 6481–6489. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.A.; Wang, Z. Next-generation transcriptome assembly. Nat. Rev. Genet. 2011, 12, 671–682. [Google Scholar] [CrossRef] [PubMed]
- Haberman, Y.; Tickle, T.L.; Dexheimer, P.J.; Kim, M.O.; Tang, D.; Karns, R.; Baldassano, R.N.; Noe, J.D.; Rosh, J.; Markowitz, J.; et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J. Clin. Investig. 2014, 124, 3617–3633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiser, M.; Simon, J.M.; Kochar, B.; Tovar, A.; Israel, J.W.; Robinson, A.; Gipson, G.R.; Schaner, M.S.; Herfarth, H.H.; Sartor, R.B.; et al. Molecular classification of Crohn’s disease reveals two clinically relevant subtypes. Gut 2018, 67, 36–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kugathasan, S.; Denson, L.A.; Walters, T.D.; Kim, M.O.; Marigorta, U.M.; Schirmer, M.; Mondal, K.; Liu, C.; Griffiths, A.; Noe, J.D.; et al. Prediction of complicated disease course for children newly diagnosed with Crohn’s disease: A multicentre inception cohort study. Lancet 2017, 389, 1710–1718. [Google Scholar] [CrossRef] [Green Version]
- Gallagher, K.; Catesson, A.; Griffin, J.L.; Holmes, E.; Williams, H.R.T. Metabolomic Analysis in Inflammatory Bowel Disease: A Systematic Review. J. Crohns Colitis 2021, 15, 813–826. [Google Scholar] [CrossRef]
- Santoru, M.L.; Piras, C.; Murgia, A.; Palmas, V.; Camboni, T.; Liggi, S.; Ibba, I.; Lai, M.A.; Orru, S.; Blois, S.; et al. Cross sectional evaluation of the gut-microbiome metabolome axis in an Italian cohort of IBD patients. Sci. Rep. 2017, 7, 9523. [Google Scholar] [CrossRef]
- Duboc, H.; Rajca, S.; Rainteau, D.; Benarous, D.; Maubert, M.A.; Quervain, E.; Thomas, G.; Barbu, V.; Humbert, L.; Despras, G.; et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut 2013, 62, 531–539. [Google Scholar] [CrossRef]
- Vavassori, P.; Mencarelli, A.; Renga, B.; Distrutti, E.; Fiorucci, S. The bile acid receptor FXR is a modulator of intestinal innate immunity. J. Immunol. 2009, 183, 6251–6261. [Google Scholar] [CrossRef] [Green Version]
- Gadaleta, R.M.; van Erpecum, K.J.; Oldenburg, B.; Willemsen, E.C.; Renooij, W.; Murzilli, S.; Klomp, L.W.; Siersema, P.D.; Schipper, M.E.; Danese, S.; et al. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut 2011, 60, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Desbonnet, L.; Clarke, G.; Traplin, A.; O’Sullivan, O.; Crispie, F.; Moloney, R.D.; Cotter, P.D.; Dinan, T.G.; Cryan, J.F. Gut microbiota depletion from early adolescence in mice: Implications for brain and behaviour. Brain Behav. Immun. 2015, 48, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Yano, J.M.; Yu, K.; Donaldson, G.P.; Shastri, G.G.; Ann, P.; Ma, L.; Nagler, C.R.; Ismagilov, R.F.; Mazmanian, S.K.; Hsiao, E.Y. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 2015, 161, 264–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wikoff, W.R.; Anfora, A.T.; Liu, J.; Schultz, P.G.; Lesley, S.A.; Peters, E.C.; Siuzdak, G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. USA 2009, 106, 3698–3703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, J.; Heller, J.J.; Guo, X.; Chen, Z.M.; Fish, K.; Fu, Y.X.; Zhou, L. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity 2012, 36, 92–104. [Google Scholar] [CrossRef] [Green Version]
- Monteleone, I.; Rizzo, A.; Sarra, M.; Sica, G.; Sileri, P.; Biancone, L.; MacDonald, T.T.; Pallone, F.; Monteleone, G. Aryl hydrocarbon receptor-induced signals up-regulate IL-22 production and inhibit inflammation in the gastrointestinal tract. Gastroenterology 2011, 141, 237–248.e1. [Google Scholar] [CrossRef]
- Hashimoto, T.; Perlot, T.; Rehman, A.; Trichereau, J.; Ishiguro, H.; Paolino, M.; Sigl, V.; Hanada, T.; Hanada, R.; Lipinski, S.; et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature 2012, 487, 477–481. [Google Scholar] [CrossRef]
- Bennike, T.B.; Carlsen, T.G.; Ellingsen, T.; Bonderup, O.K.; Glerup, H.; Bogsted, M.; Christiansen, G.; Birkelund, S.; Stensballe, A.; Andersen, V. Neutrophil Extracellular Traps in Ulcerative Colitis: A Proteome Analysis of Intestinal Biopsies. Inflamm. Bowel. Dis. 2015, 21, 2052–2067. [Google Scholar] [CrossRef] [Green Version]
- Brinkmann, V.; Reichard, U.; Goosmann, C.; Fauler, B.; Uhlemann, Y.; Weiss, D.S.; Weinrauch, Y.; Zychlinsky, A. Neutrophil extracellular traps kill bacteria. Science 2004, 303, 1532–1535. [Google Scholar] [CrossRef]
- Riaz, T.; Sollid, L.M.; Olsen, I.; de Souza, G.A. Quantitative Proteomics of Gut-Derived Th1 and Th1/Th17 Clones Reveal the Presence of CD28+ NKG2D- Th1 Cytotoxic CD4+ T cells. Mol. Cell Proteom. 2016, 15, 1007–1016. [Google Scholar] [CrossRef] [Green Version]
- Duguet, F.; Locard-Paulet, M.; Marcellin, M.; Chaoui, K.; Bernard, I.; Andreoletti, O.; Lesourne, R.; Burlet-Schiltz, O.; Gonzalez de Peredo, A.; Saoudi, A. Proteomic Analysis of Regulatory T Cells Reveals the Importance of Themis1 in the Control of Their Suppressive Function. Mol. Cell Proteom. 2017, 16, 1416–1432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masoodi, M.; Pearl, D.S.; Eiden, M.; Shute, J.K.; Brown, J.F.; Calder, P.C.; Trebble, T.M. Altered colonic mucosal Polyunsaturated Fatty Acid (PUFA) derived lipid mediators in ulcerative colitis: New insight into relationship with disease activity and pathophysiology. PLoS ONE 2013, 8, e76532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sewell, G.W.; Hannun, Y.A.; Han, X.; Koster, G.; Bielawski, J.; Goss, V.; Smith, P.J.; Rahman, F.Z.; Vega, R.; Bloom, S.L.; et al. Lipidomic profiling in Crohn’s disease: Abnormalities in phosphatidylinositols, with preservation of ceramide, phosphatidylcholine and phosphatidylserine composition. Int. J. Biochem. Cell Biol. 2012, 44, 1839–1846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karner, M.; Kocjan, A.; Stein, J.; Schreiber, S.; von Boyen, G.; Uebel, P.; Schmidt, C.; Kupcinskas, L.; Dina, I.; Zuelch, F.; et al. First multicenter study of modified release phosphatidylcholine “LT-02” in ulcerative colitis: A randomized, placebo-controlled trial in mesalazine-refractory courses. Am. J. Gastroenterol. 2014, 109, 1041–1051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).