Effect of Paper-Bagging on Apple Skin Patterning Associated with MdMYB10 Promoter Methylation
Abstract
1. Introduction
2. Results
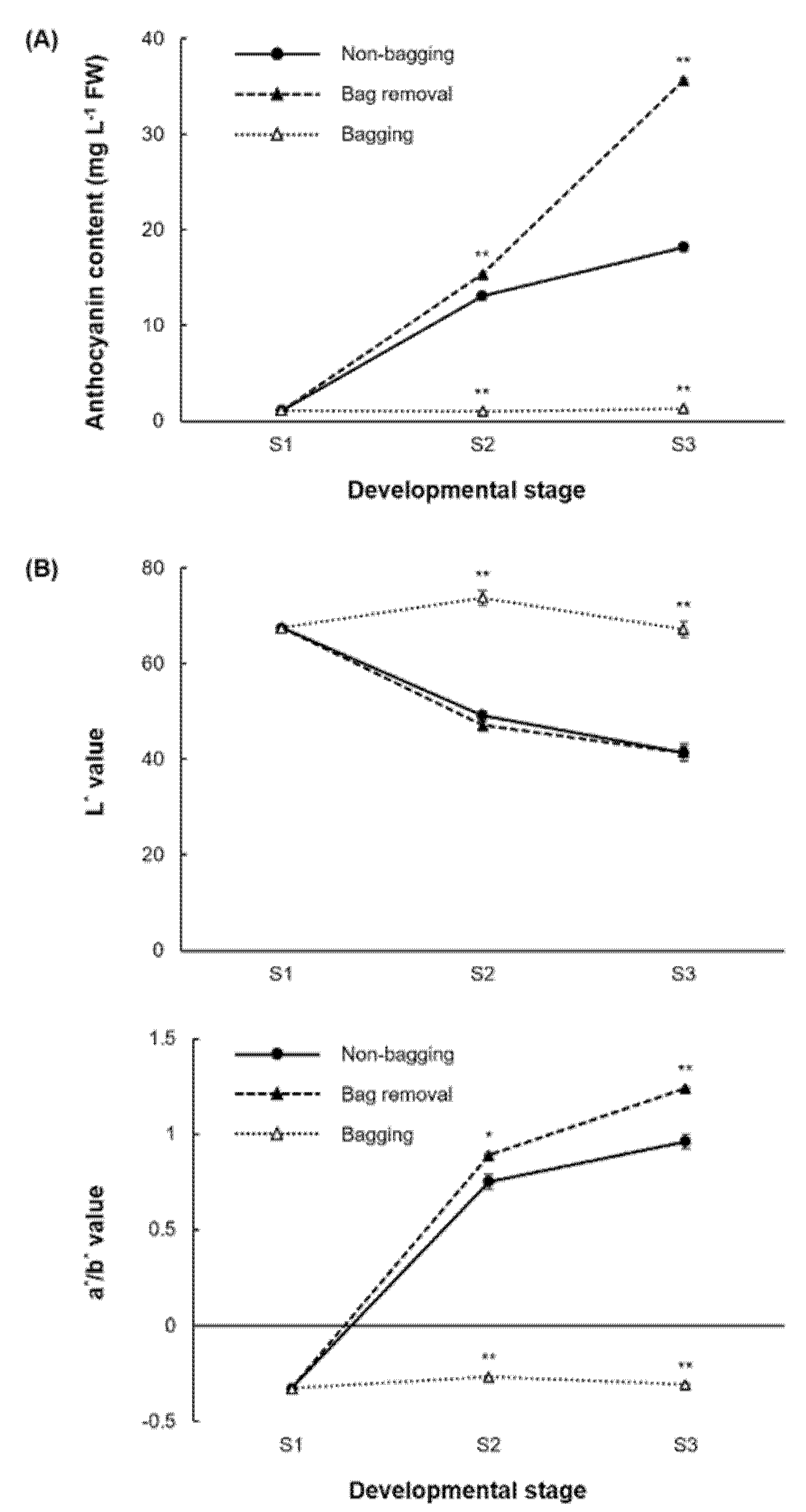
2.1. Anthocyanin Analysis
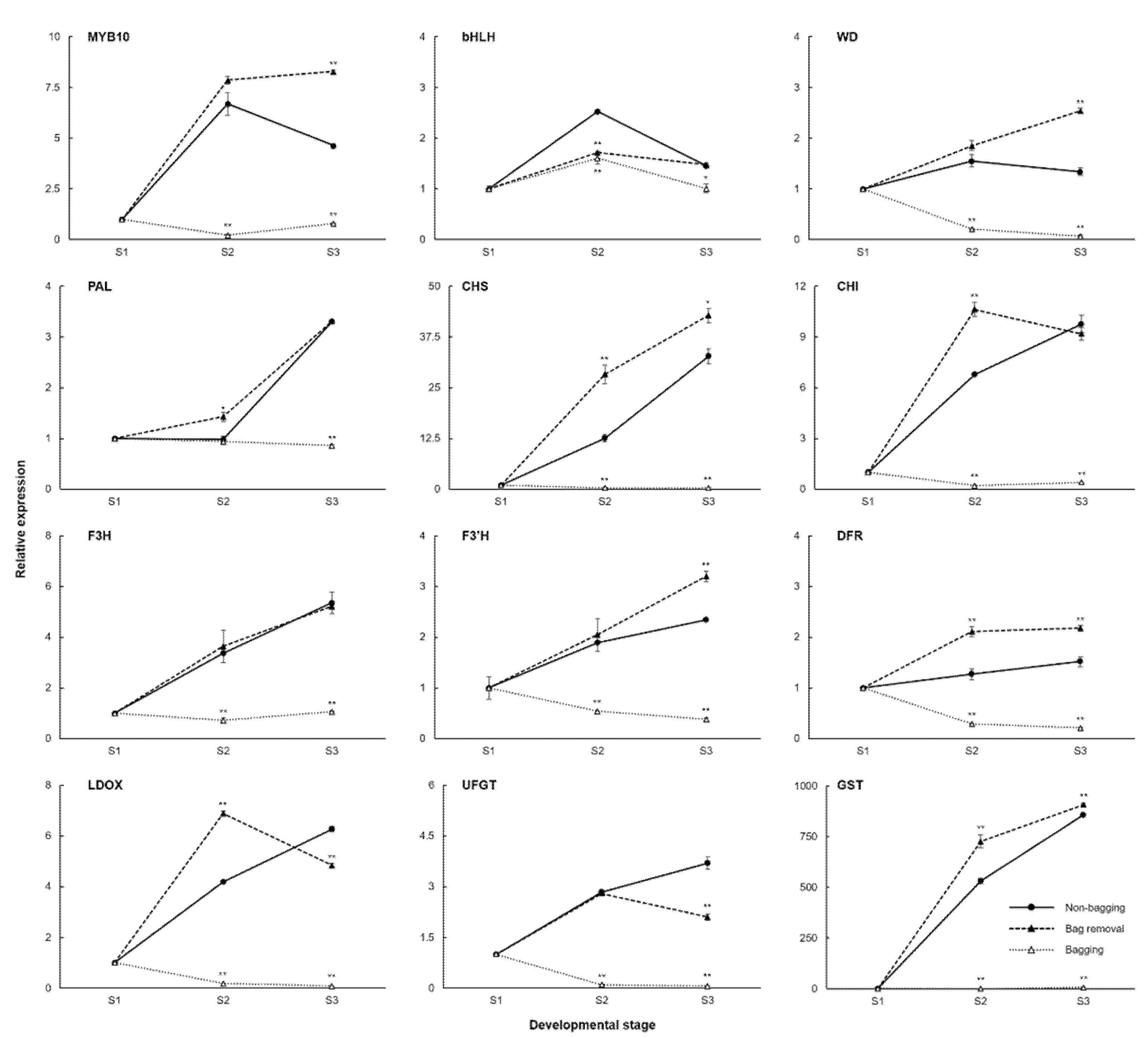
2.2. Expression of the Anthocyanin-Related Genes
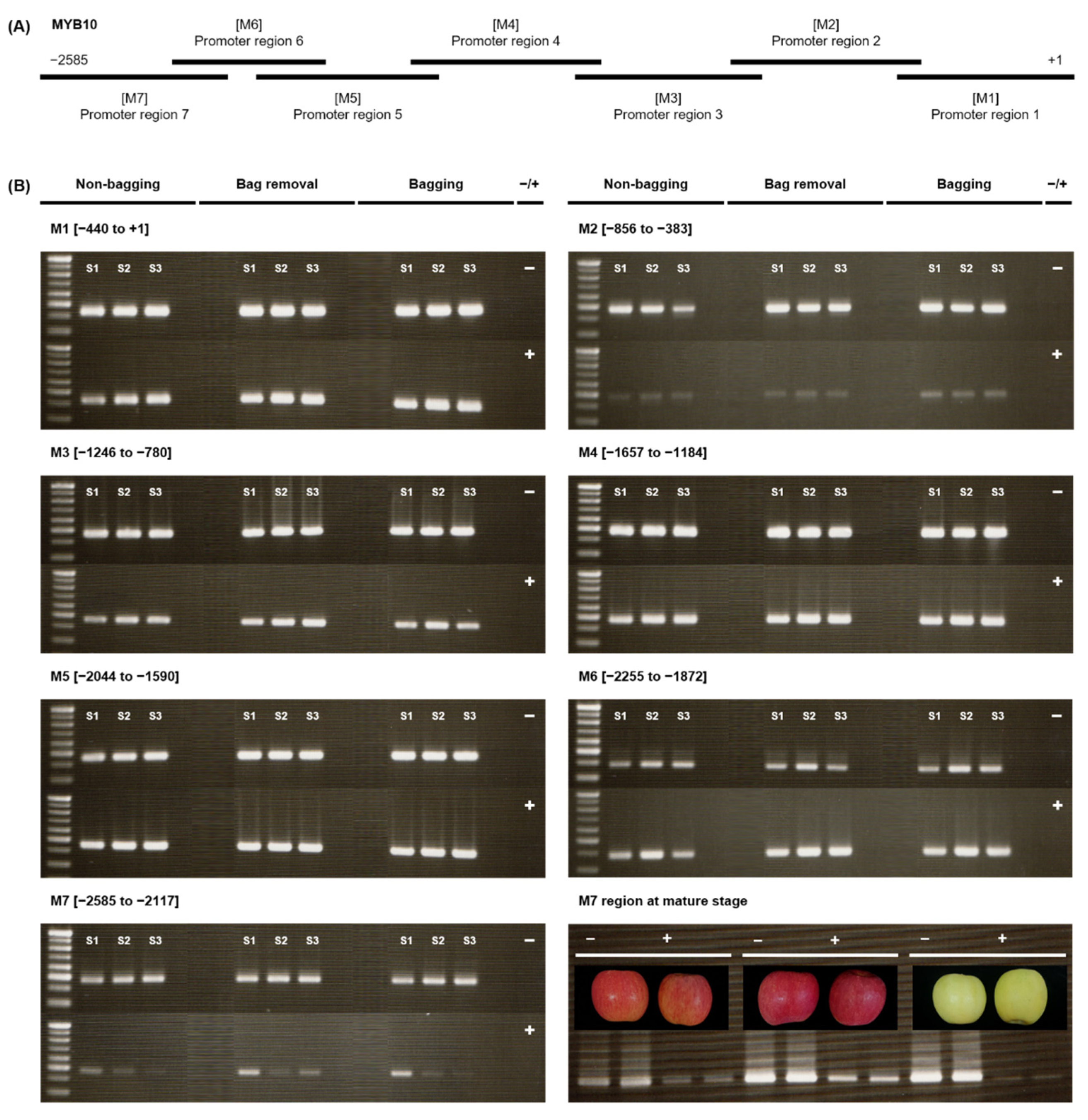
2.3. Effects of Bagging Treatment on the Methylation Level of MdMYB10 Promoter in ‘Fuji’ Apples
2.4. Methyltransferase Expression
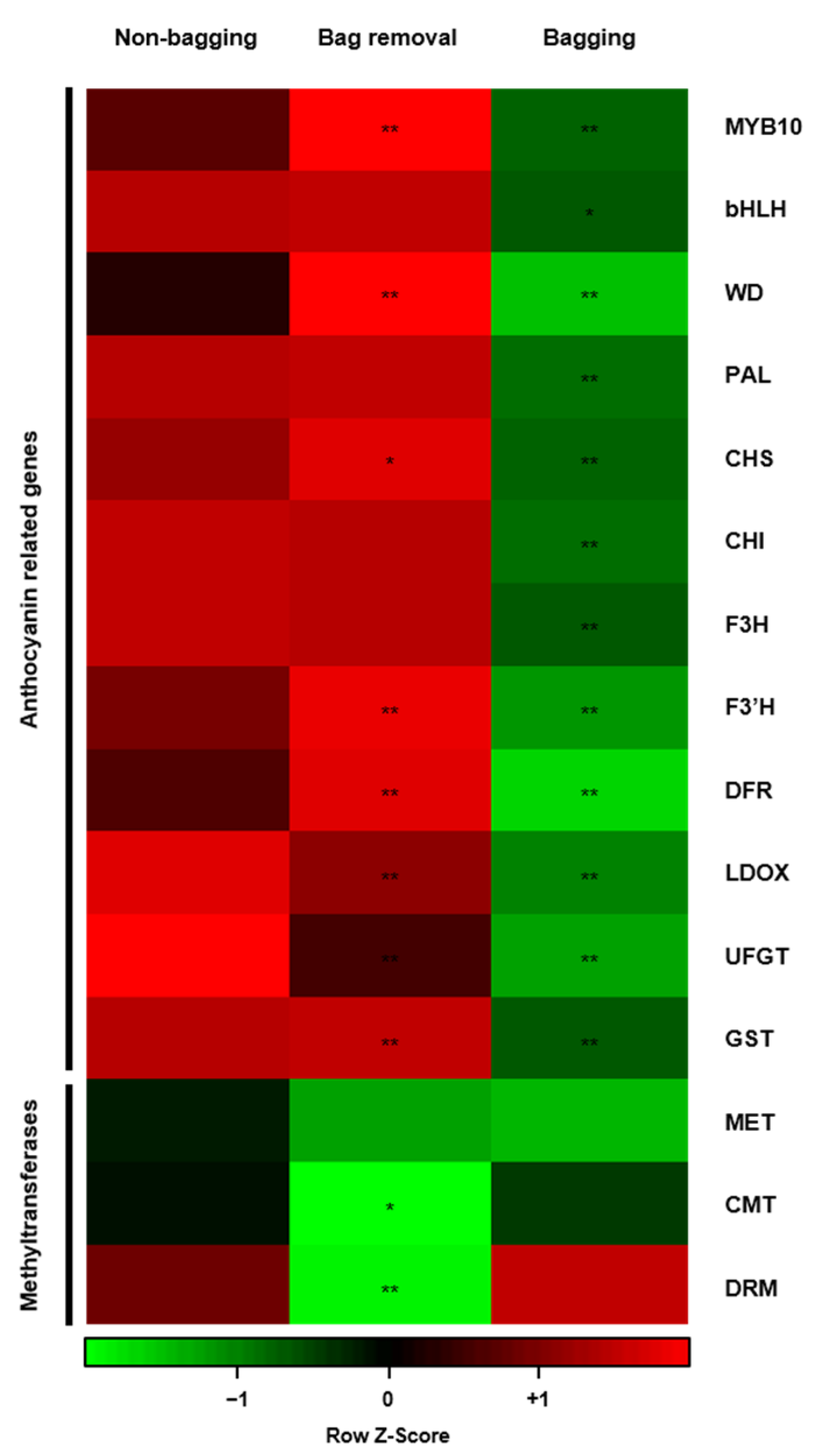
2.5. Visualization of Whole Gene Expression Using Heat Map Software
3. Discussion
3.1. Bag Treatment Induces Changes in Skin Color and Pattern
3.2. Bag Removal Promotes Anthocyanin Biosynthesis and Accumulation
3.3. Demethylation in the M7 Region of MdMYB10 Promoter Causes the Blushed Pattern in Bag Removal Fruit
4. Materials and Methods
4.1. Plant Materials and Paper-Bagging Protocols
4.2. Measurement of Anthocyanin Content and Skin Color Parameters
4.3. RNA Extraction and Quantitative Real-Time PCR Analysis
4.4. DNA Extraction and Methylation Analyses
4.5. Heat Map Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Veberic, R.; Zadravec, P.; Stampar, F. Fruit quality of ‘Fuji’ apple (Malus domestica Borkh.) strains. J. Sci. Food Agric. 2007, 87, 593–599. [Google Scholar] [CrossRef]
- Rojas-Grau, M.A.; Sobrino-Lopez, A.; Soledad Tapia, M.; Martin-Belloso, O. Browning inhibition in fresh-cut ‘Fuji’ apple slices by natural antibrowning agents. J. Food Sci. 2006, 71, 59–65. [Google Scholar] [CrossRef]
- Iglesias, I.; Echeverria, G.; Lopez, M.L. Fruit color development, anthocyanin content, standard quality, volatile compound emissions and consumer acceptability of several ‘Fuji’ apple strains. Sci. Hortic. 2012, 137, 138–147. [Google Scholar] [CrossRef]
- Jakopic, J.; Veberic, R.; Stampar, F. The effect of reflective foil and hail nets on the lighting, color and anthocyanins of ‘Fuji’ apple. Sci. Hortic. 2007, 115, 40–46. [Google Scholar] [CrossRef]
- Takos, A.M.; Jaffe, F.W.; Jacob, S.R.; Bogs, J.; Robinson, S.P.; Walker, A.R. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 2006, 142, 1216–1232. [Google Scholar] [CrossRef]
- Lin-Wang, K.; Micheletti, D.; Palmer, J.; Volz, R.; Lozano, L.; Espley, R.; Hellens, R.P.; Chagne, D.; Rowan, D.D.; Troggio, M.; et al. High temperature reduces apple fruit colour via modulation of the anthocyanin regulatory complex. Plant Cell Environ. 2011, 34, 1176–1190. [Google Scholar] [CrossRef] [PubMed]
- Saure, M.C. External control of anthocyanin formation in apple. Sci. Hortic. 1990, 42, 181–218. [Google Scholar] [CrossRef]
- Marais, E.; Jacobs, G.; Holcroft, D.M. Colour response of ‘Cripps’ Pink’ apples to postharvest irradiation is influenced by maturity and temperature. Sci. Hortic. 2001, 90, 31–41. [Google Scholar] [CrossRef]
- Reay, P.F.; Lancaster, J.E. Accumulation of anthocyanins and quercetin glycosides in ‘Gala’ and ‘Royal Gala’ apple fruit skin with UV-B-visible irradiation: Modifying effects of fruit maturity, fruit side, and temperature. Sci. Hortic. 2001, 90, 57–68. [Google Scholar] [CrossRef]
- Bai, S.; Sun, Y.; Qian, M.; Yang, F.; Ni, J.; Tao, R.; Li, L.; Shu, Q.; Zhang, D.; Teng, Y. Transcriptome analysis of bagging-treated red Chinese sand pear peels reveals light-responsive pathway functions in anthocyanin accumulation. Sci. Rep. 2017, 7, 63–77. [Google Scholar] [CrossRef]
- Ju, Z. Fruit bagging, a useful method for studying anthocyanin synthesis and gene expression in apples. Sci. Hortic. 1998, 77, 155–164. [Google Scholar] [CrossRef]
- Xiao-Ying, D.; Zhong-Fang, D.; Cheng-Lian, L.I.; Geng-Sen, L.I.; Lei, W.; Yong-Zhang, W.; Yong-Bing, Y. Effects of different bag treatments on the absorption of calcium in ‘Red Fuji’ apple fruit. Acta Hortic. Sin. 2007, 34, 835–840. [Google Scholar]
- Teixeira, R.; Boff, M.I.C.; do Amarante, C.V.T.; Steffens, C.A.; Boff, P. Effects of fruit bagging on pests and diseases control and on quality and maturity of ‘Fuji Suprema’ apples. Bragantia 2011, 70, 688–695. [Google Scholar] [CrossRef]
- Yuri, J.A.; Neira, A.; Fuentes, M.; Razmilic, I.; Lepe, V.; Gonzalez, M.F. Bagging cv. Fuji, Raku Raku apple fruit affects their phenolic profile and antioxidant capacity. Erwerbs-Obstbau 2020, 62, 221–229. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, X.; Liu, Y.; Shi, X.; Wang, Y.; Zhang, C.; Zhao, Z. The effect of fruit bagging on the color, phenolic compounds and expression of the anthocyanin biosynthetic and regulatory genes on the ‘Granny Smith’ apples. Eur. Food Res. Technol. 2013, 237, 875–885. [Google Scholar] [CrossRef]
- Feng, F.; Li, M.; Ma, F.; Cheng, L. The effects of bagging and debagging on external fruit quality, metabolites, and the expression of anthocyanin biosynthetic genes in ‘Jonagold’ apple (Malus domestica Borkh.). Sci. Hortic. 2014, 165, 123–131. [Google Scholar] [CrossRef]
- Pikaard, C.S.; Mittelsten Scheid, O. Epigenetic regulation in plants. Cold Spring Harb. Perspect. Biol. 2014, 6, a019315. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Liang, B.; Chang, B.; Liu, L.; Yan, J.; Yang, Y.; Zhao, Z. Transcriptome profiling reveals transcriptional regulation by DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine enhancing red pigmentation in bagged “Granny Smith” apples (Malus domestica). Int. J. Mol. Sci. 2018, 19, 3133. [Google Scholar] [CrossRef]
- Telias, A.; Lin-Wang, K.; Stevenson, D.E.; Cooney, J.M.; Hellens, R.P.; Allan, A.C.; Hoover, E.E.; Bradeen, J.M. Apple skin patterning is associated with differential expression of MYB10. BMC Plant Biol. 2011, 11, 93. [Google Scholar] [CrossRef]
- El-Sharkawy, I.; Liang, D.; Xu, K. Transcriptome analysis of an apple (Malus × domestica) yellow fruit somatic mutation identifies a gene network module highly associated with anthocyanin and epigenetic regulation. J. Exp. Bot. 2015, 66, 7359–7376. [Google Scholar] [CrossRef]
- Ma, C.; Jing, C.; Chang, B.; Yan, J.; Liang, B.; Liu, L.; Yang, Y.; Zhao, Z. The effect of promoter methylation on MdMYB1 expression determines the level of anthocyanin accumulation in skins of two non-red apple cultivars. BMC Plant Biol. 2018, 18, 108. [Google Scholar] [CrossRef]
- Bai, S.; Tuan, P.A.; Saito, T.; Honda, C.; Hatsuyama, Y.; Ito, A.; Moriguchi, T. Epigenetic regulation of MdMYB1 is associated with paper bagging-induced red pigmentation of apples. Planta 2016, 244, 573–586. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.J.; Kim, G.H.; Choi, C. Differential gene expression and epigenetic analyses between striped and blushed skinned sports of ‘Fuji’ apple. Sci. Hortic. 2020, 261, 108944–108949. [Google Scholar] [CrossRef]
- Jiang, S.; Wang, N.; Chen, M.; Zhang, R.; Sun, Q.; Xu, H.; Zhang, Z.; Wang, Y.; Sui, X.; Wang, S.; et al. Methylation of MdMYB1 locus mediated by RdDM pathway regulates anthocyanin biosynthesis in apple. Plant Biotechnol. J. 2020, 19, 212–225. [Google Scholar] [CrossRef] [PubMed]
- Jia, D.; Li, Z.; Dang, Q.; Shang, L.; Shen, J.; Leng, X.; Wang, Y.; Yuan, Y. Anthocyanin biosynthesis and methylation of the MdMYB10 promoter are associated with the red blushed-skin mutant in the red striped-skin “Changfu 2” apple. J. Agric. Food Chem. 2020, 68, 4292–4304. [Google Scholar] [CrossRef]
- Ubi, B. External stimulation of anthocyanin biosynthesis in apple fruit. Food Agric. Environ. 2004, 2, 65–70. [Google Scholar]
- Honda, C.; Iwanami, H.; Naramoto, K.; Maejima, T.; Kanamaru, K.; Moriya-Tanaka, Y.; Hanada, T.; Wada, M. Thinning and bagging treatments and the growing region influence anthocyanin accumulation in red-fleshed apple fruit. Hortic. J. 2017, 86, 291–299. [Google Scholar] [CrossRef]
- Kim, S.-H.; Lee, J.-R.; Hong, S.-T.; Yoo, Y.-K.; An, G.; Kim, S.-R. Molecular cloning and analysis of anthocyanin biosynthesis genes preferentially expressed in apple skin. Plant Sci. 2003, 165, 403–413. [Google Scholar] [CrossRef]
- Darrigues, A.; Hall, J.; van der Knaap, E.; Francis, D.M.; Dujmovic, N.; Gray, S. Tomato analyzer-color test: A new tool for efficient digital phenotyping. J. Am. Soc. Hortic. Sci. 2008, 133, 579–586. [Google Scholar] [CrossRef]
- Telias, A.; Hoover, E.; Rother, D. Plant and environmental factors influencing the pattern of pigment accumulation in ‘Honeycrisp’ apple peels using a novel color analyzer software tool. HortScience 2008, 43, 1441–1445. [Google Scholar] [CrossRef]
- Wang, Z.; Meng, D.; Wang, A.; Li, T.; Jiang, S.; Cong, P.; Li, T. The methylation of the PcMYB10 promoter is associated with green-skinned sport in Max Red Bartlett pear. Plant Physiol. 2013, 162, 885–896. [Google Scholar] [CrossRef] [PubMed]
- Shimazaki, M.; Fujita, K.; Kobayashi, H.; Suzuki, S. Pink-colored grape berry is the result of short insertion in intron of color regulatory gene. PLoS ONE 2011, 6, e21308. [Google Scholar] [CrossRef] [PubMed]
- Honda, C.; Moriya, S. Anthocyanin biosynthesis in apple fruit. Hortic. J. 2018, 87, 305–314. [Google Scholar] [CrossRef]
- Lancaster, J.E.; Dougall, D.K. Regulation of skin color in apples. Crit. Rev. Plant Sci. 1992, 10, 487–502. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Liu, Z.; Wu, Y.; Zheng, L.; Zhang, G. Regulatory mechanisms of anthocyanin biosynthesis in apple and pear. Int. J. Mol. Sci. 2021, 22, 8441. [Google Scholar] [CrossRef]
- Wang, W.; Celton, J.-M.; Buck-Sorlin, G.; Balzergue, S.; Bucher, E.; Laurens, F. Skin color in apple fruit (Malus × domestica): Genetic and epigenetic insights. Epigenomes 2020, 4, 13. [Google Scholar] [CrossRef]
- Du, X.; Wang, Y.; Liu, M.; Liu, X.; Jiang, Z.; Zhao, L.; Tang, Y.; Sun, Y.; Zhang, X.; Liu, D.; et al. The assessment of epigenetic diversity, differentiation, and structure in the ‘Fuji’ mutation line implicates roles of epigenetic modification in the occurrence of different mutant groups as well as spontaneous mutants. PLoS ONE 2020, 15, e0235073. [Google Scholar]
- Lee, J.; Durst, R.W.; Wrolstad, R.E. Determination of total monomeric anthocyanin pigment content of fruit juices, beverages, natural colorants, and wines by the pH differential method: Collaborative study. J. AOAC Int. 2019, 88, 1269–1278. [Google Scholar] [CrossRef]
- Cliff, M.; Sanford, K.; Wismer, W.; Hampson, C. Use of digital images for evaluation of factors responsible for visual preference of apples by consumers. HortScience 2002, 37, 1127–1131. [Google Scholar] [CrossRef]
- Chang, S.; Puryear, J.; Cairney, J. A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 1993, 11, 113–116. [Google Scholar] [CrossRef]
- Hetzl, J.; Foerster, A.M.; Raidl, G.; Mittelsten Scheid, O. CyMATE: A new tool for methylation analysis of plant genomic DNA after bisulphite sequencing. Plant J. 2007, 51, 526–536. [Google Scholar] [CrossRef] [PubMed]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, 147–153. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cho, H.J.; Han, A.R.; Choi, C. Effect of Paper-Bagging on Apple Skin Patterning Associated with MdMYB10 Promoter Methylation. Int. J. Mol. Sci. 2022, 23, 3319. https://doi.org/10.3390/ijms23063319
Cho HJ, Han AR, Choi C. Effect of Paper-Bagging on Apple Skin Patterning Associated with MdMYB10 Promoter Methylation. International Journal of Molecular Sciences. 2022; 23(6):3319. https://doi.org/10.3390/ijms23063319
Chicago/Turabian StyleCho, Hye Jeong, A Reum Han, and Cheol Choi. 2022. "Effect of Paper-Bagging on Apple Skin Patterning Associated with MdMYB10 Promoter Methylation" International Journal of Molecular Sciences 23, no. 6: 3319. https://doi.org/10.3390/ijms23063319
APA StyleCho, H. J., Han, A. R., & Choi, C. (2022). Effect of Paper-Bagging on Apple Skin Patterning Associated with MdMYB10 Promoter Methylation. International Journal of Molecular Sciences, 23(6), 3319. https://doi.org/10.3390/ijms23063319





