Non-Coding RNAs: New Biomarkers and Therapeutic Targets for Temporal Lobe Epilepsy
Abstract
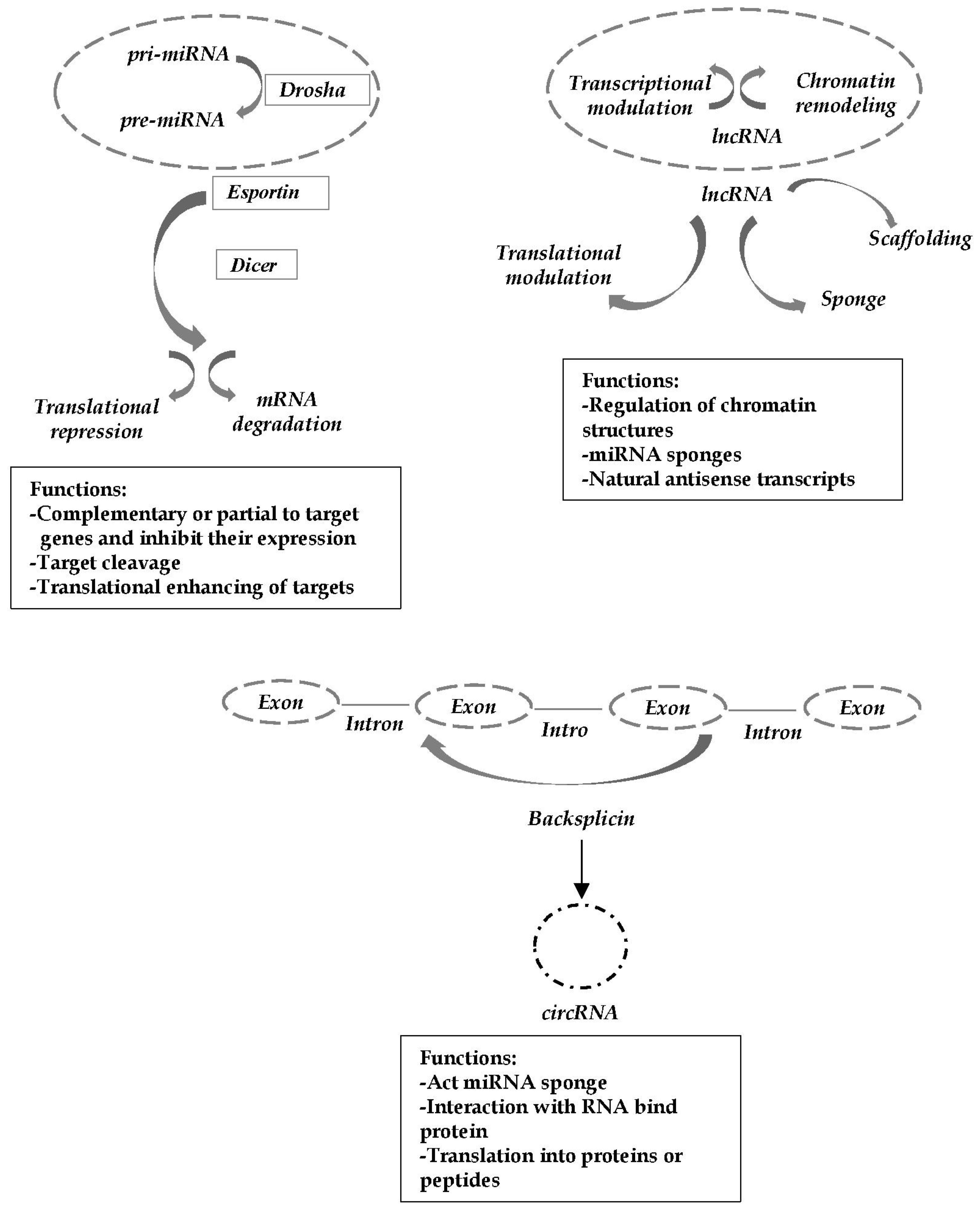
:1. Introduction
2. Role of miRNAs in TLE Pathogenesis
2.1. Studies Suggested the Role of miRNA in TLE
2.2. Principal Pathogenic Mechanisms and Related miRNAs Involved in TLE
3. Impact on Diagnosis and Prognosis
3.1. miRNAs as Putative Biomarkers in TLE
3.2. Prospects for miRNAs Therapeutics in TLE
4. Role of lncRNAs in TLE Pathogenesis
4.1. Studies Suggested the Role of lncRNA in TLE
4.2. Principal Pathogenic Mechanisms and Related lncRNAs Involved in TLE
5. Impact on Diagnosis and Prognosis
5.1. lncRNA as Putative Biomarkers in TLE
5.2. Prospects for lncRNAs Therapeutics in TLE
6. Role of circRNAs in TLE Pathogenesis
6.1. Studies Suggested the Role of circRNA in TLE
6.2. Principal Pathogenic Mechanisms and Related circRNAs Involved in TLE
7. Impact on Diagnosis and Prognosis
circRNAs as Therapeutic Targets and Diagnostic Biomarkers in TLE
8. Summary: Outlooks and Next Steps
Author Contributions
Funding
Conflicts of Interest
References
- Manford, M.; Hart, Y.M.; Sander, J.W.; Shorvon, S.D. National General Practice Study of Epilepsy (NGPSE): Partial seizure patterns in a general population. Neurology 1992, 42, 1911–1917. [Google Scholar]
- Labate, A.; Gambardella, A.; Andermann, E.; Aguglia, U.; Cendes, F.; Berkovic, S.F.; Andermann, F. Benign mesial temporal lobe epilepsy. Nat. Rev. Neurol. 2011, 7, 237–240. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Hartz, A.M.S.; Bauer, B. Drug-Resistant Epilepsy: Multiple Hypotheses, Few Answers. Front. Neurol. 2017, 8, 301. [Google Scholar] [CrossRef]
- Labate, A.; Aguglia, U.; Tripepi, G.; Mumoli, L.; Ferlazzo, E.; Baggetta, R.; Quattrone, A.; Gambardella, A. Long-term outcome of mild mesial temporal lobe epilepsy: A prospective longitudinal cohort study. Neurology 2016, 86, 1904–1910. [Google Scholar] [CrossRef] [PubMed]
- Blümcke, I.; Thom, M.; Aronica, E.; Armstrong, D.D.; Bartolomei, F.; Bernasconi, A.; Bernasconi, N.; Bien, C.G.; Cendes, F.; Coras, R.; et al. International consensus classification of hippocampal sclerosis in temporal lobe epilepsy: A task force report from the ILAE commission on diagnostic methods. Epilepsia 2013, 54, 1315–1329. [Google Scholar] [CrossRef] [PubMed]
- Gambardella, A.; Messina, D.; Le Piane, E.; Oliveri, R.L.; Annesi, G.; Zappia, M.; Andermann, M.; Quattrone, A.; Aguglia, U. Familial temporal lobe epilepsy autosomal dominant inheritance in a large pedigree from southern Italy. Epilepsy Res. 2000, 38, 127–132. [Google Scholar] [CrossRef]
- Patel, D.C.; Tewari, B.P.; Chaunsali, L.; Sontheimer, H. Neuron-glia interactions in the pathophysiology of epilepsy. Nat. Rev. Neurosci. 2019, 20, 282–297. [Google Scholar] [CrossRef] [PubMed]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef] [Green Version]
- Peschansky, V.J.; Wahlestedt, C. Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics 2014, 9, 3–12. [Google Scholar] [CrossRef] [Green Version]
- Mills, J.D.; van Vliet, E.A.; Chen, B.J.; Janitz, M.; Anink, J.J.; Baayen, J.; Idema, S.; Devore, S.; Friedman, D.; Diehl, B.; et al. Coding and non-coding transcriptome of mesial temporal lobe epilepsy: Critical role of small non-coding RNAs. Neurobiol. Dis. 2020, 134, 104612. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Chen, Y. Pathophysiology and clinical utility of non-coding RNAs in epilepsy. Front. Mol. Neurosci. 2017, 10, 249. [Google Scholar] [CrossRef] [PubMed]
- Villa, C.; Lavitrano, M.; Combi, R. Long non-coding RNAs and related molecular pathways in the pathogenesis of epilepsy. Int. J. Mol. Sci. 2019, 20, 4898. [Google Scholar] [CrossRef] [Green Version]
- Beermann, J.; Piccoli, M.T.; Viereck, J.; Thum, T. Non-coding RNAs in development and disease: Background, mechanisms, and therapeutic approaches. Physiol Rev. 2016, 96, 1297–1325. [Google Scholar] [CrossRef] [Green Version]
- Rajman, M.; Schratt, G. MicroRNAs in neural development: From master regulators to fine-tuners. Development 2017, 144, 2310–2322. [Google Scholar] [CrossRef] [Green Version]
- Abdolmaleki, A.; Ferdowsi, S.; Asadi, A.; Panahi, Y. Long Non-coding RNAs Associated with Brain Disorders: A Literature Review. Gene Cell Tissue 2021, 8, e111802. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Rong, D.; Sun, H.; Li, Z.; Liu, S.; Dong, C.; Fu, K.; Tang, W.; Cao, H. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget 2017, 8, 73271–73281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venø, M.T.; Reschke, C.R.; Morris, G.; Connolly, N.M.C.; Su, J.; Yan, Y.; Engel, T.; Jimenez-Mateos, E.M.; Harder, L.M.; Pultz, D.; et al. A systems approach delivers a functional microRNA catalog and expanded targets for seizure suppression in temporal lobe epilepsy. Proc. Natl. Acad. Sci. USA 2020, 117, 15977–15988. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Wu, Z.; Guo, Z.; Chen, L.; Ma, Y.; Wang, Z.; Xiao, W.; Wang, Y. Systems-level analysis identifies key regulators driving epileptogenesis in temporal lobe epilepsy. Genomics 2020, 112, 1768–1780. [Google Scholar] [CrossRef] [PubMed]
- Gong, G.H.; An, F.M.; Wang, Y.; Bian, M.; Wang, D.; Wei, C.X. Comprehensive circular RNA profiling reveals the regulatory role of the CircRNA-0067835/miR-155 pathway in temporal lobe epilepsy. Cell. Physiol. Biochem. 2018, 51, 1399–1409. [Google Scholar] [CrossRef]
- Gray, L.G.; Mills, J.D.; Curry-Hyde, A.; Devore, S.; Friedman, D.; Thom, M.; Scott, C.; Thijs, R.D.; Aronica, E.; Devinsky, O.; et al. Identification of specific circular RNA expression patterns and MicroRNA interaction networks in mesial temporal lobe epilepsy. Front. Genet. 2020, 11, 564301. [Google Scholar] [CrossRef] [PubMed]
- Catalanotto, C.; Cogoni, C.; Zardo, G. MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions. Int. J. Mol. Sci. 2016, 17, 1712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.M.; Li, X.M.; Zheng, X.P.; Yu, J.T.; Tan, L. MicroRNAs dysregulation in epilepsy. Brain Res. 2014, 1584, 94–104. [Google Scholar] [CrossRef]
- Korotkov, A.; Mills, J.D.; Gorter, J.A.; van Vliet, E.A.; Aronica, E. Systematic review and meta-analysis of differentially expressed miRNAs in experimental and human temporal lobe epilepsy. Sci Rep. 2017, 7, 11592. [Google Scholar] [CrossRef]
- Nudelman, A.S.; Di Rocco, D.P.; Lambert, T.J.; Garelick, M.G.; Le, J.; Nathanson, N.M.; Storm, D.R. Neuronal activity rapidly induces transcription of the CREB-regulated microRNA-132, in vivo. Hippocampus 2010, 20, 492–498. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.J.; Tian, X.B.; Zhang, S.; Zhang, Y.X.; Li, X.; Li, D.; Cheng, Y.; Zhang, J.N.; Kang, C.S.; Zhao, W. Temporal lobe epilepsy induces differential expression of hippocampal miRNAs including let-7e and miR-23a/b. Brain Res. 2011, 1387, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Bencurova, P.; Baloun, J.; Musilova, K.; Radova, L.; Tichy, B.; Pail, M.; Zeman, M.; Brichtova, E.; Hermanova, M.; Pospisilova, S.; et al. MicroRNA and mesial temporal lobe epilepsy with hippocampal sclerosis: Whole miRNome profiling of human hippocampus. Epilepsia 2017, 58, 1782–1793. [Google Scholar] [CrossRef] [Green Version]
- Vezzani, A.; French, J.; Bartfai, T.; Baram, T.Z. The role of inflammation in epilepsy. Nat. Rev. Neurol. 2011, 7, 31–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Zhao, J. MicroRNA Dysregulation in Epilepsy: From Pathogenetic Involvement to Diagnostic Biomarker and Therapeutic Agent Development. Front. Mol. Neurosci. 2021, 14, 650372. [Google Scholar] [CrossRef]
- Aronica, E.; Fluiter, K.; Iyer, A.; Zurolo, E.; Vreijling, J.; van Vliet, E.A.; Baayen, J.C.; Gorter, J.A. Expression pattern of miR-146a, an inflammation-associated microRNA, in experimental and human temporal lobe epilepsy. Eur. J. Neurosci. 2010, 31, 1100–1107. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Liu, B.; Meng, Q.; Sun, Y.; Wang, W.; Wang, C. Modulation of miR-146a/complement factor H-mediated inflammatory responses in a rat model of temporal lobe epilepsy. Biosci. Rep. 2016, 36, e00433. [Google Scholar] [CrossRef] [PubMed]
- Ashhab, M.U.; Omran, A.; Kong, H.; Gan, N.; He, F.; Peng, J.; Yin, F. Expressions of tumor necrosis factor alpha and microRNA-155 in immature rat model of status epilepticus and children with mesial temporal lobe epilepsy. J. Mol. Neurosci. 2013, 51, 950–958. [Google Scholar] [CrossRef] [PubMed]
- Tivnan, A.; Tracey, L.; Buckley, P.G.; Alcock, L.C.; Davidoff, A.M.; Stallings, R.L. MicroRNA-34a is a potent tumor suppressor molecule in vivo in neuroblastoma. BMC Cancer 2011, 25, 11–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, K.; Xie, Y.Y.; Zhang, C.; Ouyang, D.S.; Long, H.Y.; Sun, D.N.; Long, L.L.; Feng, L.; Li, Y.; Xiao, B. MicroRNA expression profile of the hippocampus in a rat model of temporal lobe epilepsy and miR-34a-targeted neuroprotection against hippocampal neurone cell apoptosis post-status epilepticus. BMC Neurosci. 2012, 22, 13–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, J.; Omran, A.; Ashhab, M.U.; Kong, H.; Gan, N.; He, F.; Yin, F. Expression patterns of miR-124, miR-134, miR-132, and miR-21 in an immature rat model and children with mesial temporal lobe epilepsy. J. Mol. Neurosci. 2013, 50, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Zhou, N.; Yang, P.; Deng, L.; Liu, G. MicroRNA-27a-3p Downregulation Inhibits Inflammatory Response and Hippocampal Neuronal Cell Apoptosis by Upregulating Mitogen-Activated Protein Kinase 4 (MAP2K4) Expression in Epilepsy: In Vivo and In Vitro Studies. Med. Sci. Monit. 2019, 25, 8499–8508. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Wang, W.; Li, W.; Li, X. miR-15a inhibits cell apoptosis and inflammation in a temporal lobe epilepsy model by downregulating GFAP. Mol. Med. Rep. 2020, 22, 3504–3512. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Hu, J.; Cao, S. The Clinical Significance of miR-135b-5p and Its Role in the Proliferation and Apoptosis of Hippocampus Neurons in Children with Temporal Lobe Epilepsy. Dev. Neurosci. 2020, 42, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Bot, A.M.; Dębski, K.J.; Lukasiuk, K. Alterations in miRNA Levels in the Dentate Gyrus in Epileptic Rats. PLoS ONE 2013, 8, e76051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dudek, F.E.; Sutula, T.P. Epileptogenesis in the dentate gyrus: A critical perspective. Prog. Brain Res. 2007, 163, 755–773. [Google Scholar]
- Cramer, J.A.; Baker, G.A.; Jacoby, A. Development of a new seizure severity questionnaire: Initial reliability and validity testing. Epilepsy Res. 2002, 48, 187–189. [Google Scholar] [CrossRef]
- Schratt, G.M.; Tuebing, F.; Nigh, E.A.; Kane, C.G.; Sabatini, M.E.; Kiebler, M.; Greenberg, M.E. A brain-specific microRNA regulates dendritic spine development. Nature 2006, 439, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Kaalund, S.S.; Venø, M.T.; Bak, M.; Møller, R.S.; Laursen, H.; Madsen, F.; Broholm, H.; Quistorff, B.; Uldall, P.; Tommerup, N.; et al. Aberrant expression of miR-218 and miR-204 in human mesial temporal lobe epilepsy and hippocampal sclerosis-convergence on axonal guidance. Epilepsia 2014, 55, 2017–2027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, J.; Huang, H.; Zhou, X.; Liu, X.; Ou, S.; Xu, T.; Li, R.; Ma, L.; Chen, Y. MicroRNA-132 Interact with p250GAP/Cdc42 Pathway in the Hippocampal Neuronal Culture Model of Acquired Epilepsy and Associated with Epileptogenesis Process. Neural Plast. 2016, 2016, 5108489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sohel, M.H. Extracellular/Circulating MicroRNAs: Release Mechanisms, Functions and Challenges. Achiev. Life Sci. 2016, 10, 175–186. [Google Scholar] [CrossRef] [Green Version]
- Manna, I.; Quattrone, A.; De Benedittis, S.; Vescio, B.; Iaccino, E.; Quattrone, A. Exosomal miRNA as peripheral biomarkers in Parkinson’s disease and progressive supranuclear palsy: A pilot study. Parkinsonism Relat Disord. 2021, 93, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Manna, I.; De Benedittis, S.; Quattrone, A.; Maisano, D.; Iaccino, E.; Quattrone, A. Exosomal miRNAs as Potential Diagnostic Biomarkers in Alzheimer’s Disease. Pharmaceuticals 2020, 13, 243. [Google Scholar] [CrossRef]
- Cava, C.; Manna, I.; Gambardella, A.; Bertoli, G.; Castiglioni, I. Potential Role of miRNAs as Theranostic Biomarkers of Epilepsy. Mol. Ther. Nucleic Acids 2018, 13, 275–290. [Google Scholar] [CrossRef] [Green Version]
- Avansini, S.H.; de Sousa Lima, B.P.; Secolin, R.; Santos, M.L.; Coan, A.C.; Vieira, A.S.; Torres, F.R.; Carvalho, B.S.; Alvim, M.K.; Morita, M.E.; et al. MicroRNA hsa-miR-134 is a circulating biomarker for mesial temporal lobe epilepsy. PLoS ONE 2017, 12, e0173060. [Google Scholar] [CrossRef] [PubMed]
- Raoof, R.; Bauer, S.; El Naggar, H.; Connolly, N.M.C.; Brennan, G.P.; Brindley, E.; Hill, T.; McArdle, H.; Spain, E.; Forster, R.J.; et al. Dual-center, dual-platform microRNA profiling identifies potential plasma biomarkers of adult temporal lobe epilepsy. EBioMedicine 2018, 38, 127–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antônio, L.G.L.; Freitas-Lima, P.; Pereira-da-Silva, G.; Assirati, J.A.; Matias, C.M.; Cirino, M.L.A.; Tirapelli, L.F.; Velasco, T.R.; Sakamoto, A.C.; Carlotti, C.G.; et al. Expression of MicroRNAs miR-145, miR-181c, miR-199a and miR-1183 in the Blood and Hippocampus of Patients with Mesial Temporal Lobe Epilepsy. J. Mol. Neurosci. 2019, 69, 580–587. [Google Scholar] [CrossRef] [PubMed]
- Ioriatti, E.S.; Cirino, M.L.A.; Lizarte Neto, F.S.; Velasco, T.R.; Sakamoto, A.C.; Freitas-Lima, P.; Tirapelli, D.P.C.; Carlotti, C.G. Expression of circulating microRNAs as predictors of diagnosis and surgical outcome in patients with mesial temporal lobe epilepsy with hippocampal sclerosis. Epilepsy Res. 2020, 166, 106373. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tan, L.; Tan, L.; Tian, Y.; Ma, J.; Tan, C.C.; Wang, H.F.; Liu, Y.; Tan, M.S.; Jiang, T.; et al. Circulating microRNAs are promising novel biomarkers for drug-resistant epilepsy. Sci. Rep. 2015, 5, 10201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leontariti, M.; Avgeris, M.; Katsarou, M.S.; Drakoulis, N.; Siatouni, A.; Verentzioti, A.; Alexoudi, A.; Fytraki, A.; Patrikelis, P.; Vassilacopoulou, D.; et al. Circulating miR-146a and miR-134 in predicting drug-resistant epilepsy in patients with focal impaired awareness seizures. Epilepsia 2020, 61, 959–970. [Google Scholar] [CrossRef] [PubMed]
- Brennan, G.P.; Bauer, S.; Engel, T.; Jimenez-Mateos, E.M.; Del Gallo, F.; Hill, T.D.M.; Connolly, N.M.C.; Costard, L.S.; Neubert, V.; Salvetti, B.; et al. Genome-wide microRNA profiling of plasma from three different animal models identifies biomarkers of temporal lobe epilepsy. Neurobiol. Dis. 2020, 144, 105048. [Google Scholar] [CrossRef] [PubMed]
- De Benedittis, S.; Fortunato, F.; Cava, C.; Gallivanone, F.; Iaccino, E.; Caligiuri, M.E.; Castiglioni, I.; Bertoli, G.; Manna, I.; Labate, A.; et al. Circulating microRNA: The Potential Novel Diagnostic Biomarkers to Predict Drug Resistance in Temporal Lobe Epilepsy, a Pilot Study. Int. J. Mol. Sci. 2021, 22, 702. [Google Scholar] [CrossRef] [PubMed]
- Bohosova, J.; Vajcner, J.; Jabandziev, P.; Oslejskova, H.; Slaby, O.; Aulicka, S. MicroRNAs in the development of resistance to antiseizure drugs and their potential as biomarkers in pharmacoresistant epilepsy. Epilepsia 2021, 62, 2573–2588. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.; O’Brien, D.; Henshall, D.C. Opportunities and challenges for microRNA-targeting therapeutics for epilepsy. Trends Pharmacol. Sci. 2021, 42, 605–616. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Mateos, E.M.; Engel, T.; Merino-Serrais, P.; McKiernan, R.C.; Tanaka, K.; Mouri, G.; Sano, T.; O’Tuathaigh, C.; Waddington, J.L.; Prenter, S.; et al. Silencing microRNA-134 produces neuroprotective and prolonged seizure- suppressive effects. Nat. Med. 2012, 18, 1087–1094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jimenez-Mateos, E.M.; Engel, T.; Merino-Serrais, P.; Fernaud-Espinosa, I.; Rodriguez-Alvarez, N.; Reynolds, J.; Reschke, C.R.; Conroy, R.M.; McKiernan, R.C.; de Felipe, J.; et al. Antagomirs targeting microRNA-134 increase hippocampal pyramidal neuron spine volume in vivo and protect against pilocarpine-induced status epilepticus. Brain Struct. Funct. 2015, 220, 2387–2399. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Mateos, E.M.; Bray, I.; Sanz-Rodriguez, A.; Engel, T.; McKiernan, R.C.; Mouri, G.; Tanaka, K.; Sano, T.; Saugstad, J.A.; Simon, R.P.; et al. miRNA expression profile after status epilepticus and hippocampal neuroprotection by targeting miR-132. Am. J. Pathol. 2011, 179, 2519–2532. [Google Scholar] [CrossRef] [PubMed]
- Tao, H.; Zhao, J.; Liu, T.; Cai, Y.; Zhou, X.; Xing, H.; Wang, Y.; Yin, M.; Zhong, W.; Liu, Z.; et al. Intranasal Delivery of miR-146a Mimics Delayed Seizure Onset in the Lithium-Pilocarpine Mouse Model. Mediat. Inflamm. 2017, 2017, 6512620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Guo, Y.; He, L.; Chen, C.; Luo, J.; Ma, Y.; Li, J.; Yang, Y.; Yang, Q.; Du, C.; et al. Overexpression of miRNA-137 in the brain suppresses seizure activity and neuronal excitability: A new potential therapeutic strategy for epilepsy. Neuropharmacology 2018, 138, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.Y.; Moon, J.; Lee, S.T.; Jung, K.H.; Park, D.K.; Yoo, J.S.; Sunwoo, J.S.; Byun, J.I.; Lim, J.A.; Kim, T.J.; et al. Dysregulation of long non-coding RNAs in mouse models of localization-related epilepsy. Biochem. Biophys. Res. Commun. 2015, 462, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.; Moon, J.; Lee, S.T.; Jun, J.S.; Kim, T.J.; Lim, J.A.; Park, B.S.; Yu, J.S.; Park, D.K.; Yang, A.R.; et al. Dysregulated long non-coding RNAs in the temporal lobe epilepsy mouse model. Seizure 2018, 58, 110–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, Z.; Zhang, X.; Song, H.; Yang, F.; Feng, S.; Feng, L.; Ling, Z.; Pan, L.; Liang, S.; Mao, Z.; et al. Differential long non-coding RNA (lncRNA) profiles associated with hippocampal sclerosis in human mesial temporal lobe epilepsy. Int. J. Clin. Exp. Pathol. 2019, 12, 259–266. [Google Scholar] [PubMed]
- Vezzani, A.; Ravizza, T.; Balosso, S.; Aronica, E. Glia as a source of cytokines: Implications for neuronal excitability and survival. Epilepsia 2008, 49, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, S.; Fitzgerald, K.A. Cytokines and Long Noncoding RNAs. Cold Spring Harb. Perspect. Biol. 2018, 10, a028589. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Yang, Z.Q. LncRNA NEAT1 affects inflammatory response by targeting miR-129–5p and regulating Notch signaling pathway in epilepsy. Cell Cycle 2020, 19, 419–431. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Tao, J.; Zhang, S.; Lv, X. LncRNA MEG3 Reduces Hippocampal Neuron Apoptosis via the PI3K/AKT/mTOR Pathway in a Rat Model of Temporal Lobe Epilepsy. Neuropsychiatr. Dis. Treat. 2020, 16, 2519–2528. [Google Scholar] [CrossRef] [PubMed]
- Dutra, M.R.H.; Feliciano, R.D.S.; Jacinto, K.R.; Gouveia, T.L.F.; Brigidio, E.; Morris, M.; Naah-Mazzacoratti, M.D.G.; Silva, J.A. Protective role of UCP2 in oxidative stress and apoptosis during the silent phase of an experimental model of epilepsy induced by pilocarpine. Oxid. Med. Cell. Longev. 2018, 2018, 6736721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, N.; Gao, Y.; Yan, N.; Liu, C.; Zhang, J.G.; Xing, W.M.; Kong, D.M.; Meng, F.G. High-frequency stimulation of the hippocampus protects against seizure activity and hippocampal neuronal apoptosis induced by kainic acid administration in macaques. Neuroscience 2014, 256, 370–378. [Google Scholar] [CrossRef] [PubMed]
- Han, C.L.; Liu, Y.P.; Zhao, X.M.; Wang, K.L.; Chen, N.; Hu, W.; Zhang, J.G.; Ge, M.; Meng, F.G. Whole-transcriptome screening reveals the regulatory targets and functions of long non-coding RNA H19 in epileptic rats. Biophys. Res. Commun. 2017, 489, 262–269. [Google Scholar] [CrossRef] [PubMed]
- Han, C.L.; Ge, M.; Liu, Y.P.; Zhao, X.M.; Wang, K.L.; Chen, N.; Hu, W.; Zhang, J.G.; Li, L.; Meng, F.G. Long non-coding RNA H19 contributes to apoptosis of hippocampal neurons by inhibiting let-7b in a rat model of temporal lobe epilepsy. Cell Death Dis. 2018, 9, 617. [Google Scholar] [CrossRef] [PubMed]
- Han, C.L.; Ge, M.; Liu, Y.P.; Zhao, X.M.; Wang, K.L.; Chen, N.; Meng, W.J.; Hu, W.; Zhang, J.G.; Li, L.; et al. LncRNA H19 contributes to hippocampal glial cell activation via JAK/STAT signaling in a rat model of temporal lobe epilepsy. J. Neuroinflammation 2018, 15, 103. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.K.; Yan, H.; Wang, K.; Wang, J. Dynamic regulation effect of long non-coding RNA-UCA1 on NF-kB in hippocampus of epilepsy rats. Eur. Rev. Med. Pharm. Sci. 2017, 21, 3113–3119. [Google Scholar]
- Wang, H.; Yao, G.; Li, L.; Ma, Z.; Chen, J.; Chen, W. LncRNA-UCA1 inhibits the astrocyte activation in the temporal lobe epilepsy via regulating the JAK/STAT signaling pathway. J. Cell. Biochem. 2020, 121, 4261–4270. [Google Scholar] [CrossRef] [PubMed]
- Maag, J.L.; Panja, D.; Sporild, I.; Patil, S.; Kaczorowski, D.C.; Bramham, C.R.; Dinger, M.E.; Wibrand, K. Dynamic expression of long noncoding RNAs and repeat elements in synaptic plasticity. Front. Neurosci. 2015, 9, 351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lipovich, L.; Dachet, F.; Cai, J.; Bagla, S.; Balan, K.; Jia, H.; Loeb, J.A. Activity-dependent human brain coding/noncoding gene regulatory networks. Genetics 2012, 192, 1133–1148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Hamblin, M.H.; Yin, K.J. The long noncoding RNA Malat1: Its physiological and pathophysiological functions. RNA Biol. 2017, 14, 1705–1714. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Yi, X. Down-regulation of Long Noncoding RNA MALAT1 Protects Hippocampal Neurons Against Excessive Autophagy and Apoptosis via the PI3K/Akt Signaling Pathway in Rats with Epilepsy. J. Mol. Neurosci. 2018, 65, 234–245. [Google Scholar] [CrossRef] [PubMed]
- Akers, J.C.; Gonda, D.; Kim, R.; Carter, B.S.; Chen, C.C. Biogenesis of extracellular vesicles (EV): Exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J. Neurooncol. 2013, 113, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashemian, F.; Ghafouri-Fard, S.; Arsang-Jang, S.; Mirzajani, S.; Fallah, H.; Mehvari Habibabadi, J.; Sayad, A.; Taheri, M. Epilepsy is associated with dysregulation of long non-coding RNAs in the peripheral blood. Front. Mol. Biosci. 2019, 6, 113. [Google Scholar] [CrossRef]
- Cai, X.; Long, L.; Zeng, C.; Ni, G.; Meng, Y.; Guo, Q.; Chen, Z.; Li, Z. LncRNA ILF3-AS1 mediated the occurrence of epilepsy through suppressing hippocampal miR-212 expression. Aging 2020, 12, 8413–8422. [Google Scholar] [CrossRef]
- Mirzajani, S.; Ghafouri-Fard, S.; Habibabadi, J.M.; Arsang-Jang, S.; Omrani, M.D.; Fesharaki, S.S.H.; Sayad, A.; Taheri, M. Expression Analysis of lncRNAs in Refractory and Non-Refractory Epileptic Patients. J. Mol. Neurosci. 2020, 70, 689–698. [Google Scholar] [CrossRef]
- Kobow, K.; Reid, C.A.; van Vliet, E.A.; Becker, A.J.; Carvill, G.L.; Goldman, A.M.; Hirose, S.; Lopes-Cendes, I.; Khiari, H.M.; Poduri, A.; et al. Epigenetics explained: A topic “primer” for the epilepsy community by the ILAE Genetics/Epigenetics Task Force. Epileptic Disord. 2020, 22, 127–141. [Google Scholar] [CrossRef] [PubMed]
- Modarresi, F.; Faghihi, M.A.; Lopez-Toledano, M.A.; Fatemi, R.P.; Magistri, M.; Brothers, S.P.; van der Brug, M.P.; Wahlestedt, C. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotechnol. 2012, 30, 453–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsiao, J.; Yuan, T.Y.; Tsai, M.S.; Lu, C.Y.; Lin, Y.C.; Lee, M.L.; Lin, S.W.; Chang, F.C.; Liu, P.H.; Olive, C.; et al. Upregulation of haploinsufficient gene expression in the brain by targeting a long non-coding RNA improves seizure phenotype in a model of Dravet syndrome. EBioMedicine 2016, 9, 257–277. [Google Scholar] [CrossRef] [Green Version]
- Thomas, L.F.; Sætrom, P. Circular RNAs are depleted of polymorphisms at microRNA binding sites. Bioinformatics 2014, 30, 2243–2246. [Google Scholar] [CrossRef] [PubMed]
- You, X.; Vlatkovic, I.; Babic, A.; Will, T.; Epstein, I.; Tushev, G.; Akbalik, G.; Wang, M.; Glock, C.; Quedenau, C.; et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci 2015, 18, 603–610. [Google Scholar] [CrossRef] [Green Version]
- Lee, W.J.; Moon, J.; Jeon, D.; Kim, T.J.; Yoo, J.S.; Park, D.K.; Lee, S.T.; Jung, K.H.; Park, K.I.; Jung, K.Y.; et al. Possible epigenetic regulatory effect of dysregulated circular RNAs in epilepsy. PLoS ONE 2018, 13, e0209829. [Google Scholar] [CrossRef] [Green Version]
- Gomes-Duarte, A.; Bauer, S.; Venø, M.T.; Norwood, B.A.; Henshall, D.C.; Kjems, J.; Rosenow, F.; Vangoor, V.R.; Pasterkamp, R.J. Enrichment of Circular RNA Expression Deregulation at the Transition to Recurrent Spontaneous Seizures in Experimental Temporal Lobe Epilepsy. Front. Genet. 2021, 12, 627907. [Google Scholar] [CrossRef]
- Gong, X.W.; Li, J.B.; Lu, Q.C.; Liang, P.J.; Zhang, P.M. Effective connectivity of hippocampal neural network and its alteration in Mg2+-free epilepsy model. PLoS ONE 2014, 9, e92961. [Google Scholar] [CrossRef]
- Chen, F.; Zheng, H.; Zhang, W.; Kang, J.; Liu, Q.; Pu, J.; Yang, L. circ_0003170 aggravates human hippocampal neuron injuries by regulating the miR-421/CCL2 axis in cells models of epilepsy. Gen. Physiol. Biophys. 2021, 40, 115–126. [Google Scholar] [CrossRef]
- Zhu, Z.; Wang, S.; Cao, Q.; Li, G. CircUBQLN1 Promotes Proliferation but Inhibits Apoptosis and Oxidative Stress of Hippocampal Neurons in Epilepsy via the miR-155-Mediated SOX7 Upregulation. J. Mol. Neurosci. 2021, 71, 1933–1943. [Google Scholar] [CrossRef] [PubMed]
- Hanan, M.; Soreq, H.M.; Kadener, S. CircRNAs in the brain. RNA Biol. 2017, 14, 1028–1034. [Google Scholar] [CrossRef] [Green Version]
- Cavazos, J.E.; Cross, D.J. The role of synaptic reorganization in mesial temporal lobe epilepsy. Epilepsy Behav. 2006, 8, 483–493. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Lin, H.; Sun, Z.; Kong, G.; Yan, X.; Wang, Y.; Wang, X.; Wen, Y.; Liu, X.; Zheng, H.; et al. High-Throughput Data of Circular RNA Profiles in Human Temporal Cortex Tissue Reveals Novel Insights into Temporal Lobe Epilepsy. Cell. Physiol. Biochem. 2018, 45, 677–691. [Google Scholar] [CrossRef]
- Zheng, D.; Li, M.; Li, G.; Hu, J.; Jiang, X.; Wang, Y.; Sun, Y. Circular RNA circ_DROSHA alleviates the neural damage in a cell model of temporal lobe epilepsy through regulating miR-106b-5p/MEF2C axis. Cell. Signal. 2021, 80, 109901. [Google Scholar] [CrossRef]
- Lemcke, H.; Steinhoff, G.; David, R. Gap junctional shuttling of miRNA--A novel pathway of intercellular gene regulation and its prospects in clinical application. Cell. Signal. 2015, 27, 2506–2514. [Google Scholar] [CrossRef]
- Enright, N.; Simonato, M.; Henshall, D.C. Discovery and validation of blood microRNAs as molecular biomarkers of epilepsy: Ways to close current knowledge gaps. Open Epilepsia 2018, 3, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Volnova, A.; Tsytsarev, V.; Ganina, O.; Vélez-Crespo, G.E.; Alves, J.M.; Ignashchenkova, A.; Inyushin, M. The Anti-Epileptic Effects of Carbenoxolone In Vitro and In Vivo. Int. J. Mol. Sci. 2022, 8, 663. [Google Scholar] [CrossRef] [PubMed]
- Van Vliet, E.A.; Puhakka, N.; Mills, J.D.; Srivastava, P.K.; Johnson, M.R.; Roncon, P.; Das Gupta, S.; Karttunen, J.; Simonato, M.; Lukasiuk, K.; et al. Standardization procedure for plasma biomarker analysis in rat models of epileptogenesis: Focus on circulating microRNAs. Epilepsia 2017, 58, 2013–2024. [Google Scholar] [CrossRef] [PubMed]
- Corey, D.R. Nusinersen, an antisense oligonucleotide drug for spinal muscular atrophy. Nat. Neurosci. 2017, 20, 497–499. [Google Scholar] [CrossRef] [PubMed]
| Significantly Dysregulated miRNAs | Source | Cohort Composition | Methods | Regulation | p-Value, AUC | Reference |
|---|---|---|---|---|---|---|
| miR-134 | plasma | 14 TLE 16 HC 65 TLE * 83 HC * | qRT-PCR | ↓ ↓ | 0.018, 75% 0.0003, 67% | [49] |
| miR-27a-3p miR-328-3p miR-654-3p miR-654-3p miR-27a-3p miR-328-3p | plasma | 64 refractory focal epilepsy 32 HC 102 TLE * vs. 110 HC * | OpenArray qRT-PCR | ↑ ↑ ↑ ↑ ↑ ↑ | <0.05, 63% >0.05 0.05, 63% 0.05, 63% | [50] |
| miR-145 miR-181c miR-199a miR-1183 | blood | 10 mTLE-HS 20 HC | qRT-PCR | ↑ ↑ ↑ ↑ | 0.005 0.03 0.01 0.001 | [51] |
| miR-328-3p | serum | 28 TLE 11 HC | qRT-PCR | ↑ | <0.001, 93% | [52] |
| 3 miRNAs 12 miRNAs miR-301a-3p miR-194-5p miR-30b-5p miR-342-5p miR-4446-3p | serum serum | 30 DRE vs 30 DSE 77 DRE vs. 45 HC 77 DRE vs. 81 DSE | NGS qRT-PCR | ↑ ↓ ↓ ↓ ↓ ↓ ↓ ↓ | <0.05 <0.0001, 89% <0.0001, 74% <0.0001, 68% <0.033, 72% <0.004, 70% <0.0001 | [53] |
| miR-146a-5p miR-134 | serum | 86 DRE vs. 76 DSE | qRT-PCR | ↑ ↑ | 0.002, 64% 0.01, 61% | [54] |
| miR-93-5p miR-199a-3p miR-574-3p | plasma | 20 intractable TLE 16 HC | qRT-PCR | ↑ ↑ ↑ | <0.05, 77% <0.05, 80% <0.05, 79% | [55] |
| miR-142-5p miR-146a-5p miR-223-3p miR-142-5p miR-223-3p | serum | 27 TLE vs. 20 HC 10 DRE vs. 17 DSE | qRT-PCR qRT-PCR | ↑ ↑ ↑ ↑ ↑ | 0.001 0.02 < 0.001 <0.001, 80% <0.001, 75% | [56] |
| Pathaways | miRNAs | lncRNAs | circRNAs |
|---|---|---|---|
| Neuroinflammation | miR-146a [30,31] | lncNEAT1 [69] | |
| miR-155 [32] | lncMEG3 [70] | ||
| lncH19 [75] | |||
| lncUCA1 [76,77] | |||
| Apoptosis/Neuronal loss | miR-34a [33,34] | lncH19 [74] | circ_0003170 [94] |
| miR-21 [35] | lncMALAT1 [81] | circ_UBQLN1 [95] | |
| miR-27a-3p [36] | circ_DROSHA [99] | ||
| miR-15a [37] | |||
| miR-135b-5p [38] | |||
| Synaptic plasticity | miR-134 [41,42] | lncBDNF-AS [79] | |
| miR-218 [43] | lncMALAT1 [80] | ||
| miR-132 [44] |
| Significantly Dysregulated lncRNAs | Source | Cohort Composition | Methods | Regulation | p-Value | Referenece |
|---|---|---|---|---|---|---|
| HOXA-AS2 SPRY4-IT1 | blood | 40 TLE vs. 40 controls | qRT-PCR | ↑ ↑ | 0.001 0.02 | [83] |
| ILF3-AS1 | blood temporal cortex (lesionectomy) control tissues (intracranial hematoma) | 23 TLE vs. 18 controls | qRT-PCR | ↑ ↑ | 0.001 0.001 | [84] |
| UCA1 NKILA ANRIL THRIL | blood | 40 DRE vs. 40 HC 40 DSE vs. 40 HC | qRT-PCR | ↑ ↑ ↑ ↓ | 0.003 <0.0001 0.018 0.006 <0.0001 0.019 0.04 <0.0001 | [85] |
| Significantly dysregulated circRNA | ||||||
| circ-EFCAB2 circ-DROSHA | temporal cortices | 17 TLE 17 HC | qRT-PCR | ↑ ↓ | <0.05 <0.05 | [98] |
| circRNA-0067835 | temporal cortex (lobectomy) control tissues (temporal neocortical) plasma | 5 TLE 5 HC 22 TLE 22 HC | Microarray qRT-PCR | ↓ ↓ | <0.01 <0.01 | [20] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manna, I.; Fortunato, F.; De Benedittis, S.; Sammarra, I.; Bertoli, G.; Labate, A.; Gambardella, A. Non-Coding RNAs: New Biomarkers and Therapeutic Targets for Temporal Lobe Epilepsy. Int. J. Mol. Sci. 2022, 23, 3063. https://doi.org/10.3390/ijms23063063
Manna I, Fortunato F, De Benedittis S, Sammarra I, Bertoli G, Labate A, Gambardella A. Non-Coding RNAs: New Biomarkers and Therapeutic Targets for Temporal Lobe Epilepsy. International Journal of Molecular Sciences. 2022; 23(6):3063. https://doi.org/10.3390/ijms23063063
Chicago/Turabian StyleManna, Ida, Francesco Fortunato, Selene De Benedittis, Ilaria Sammarra, Gloria Bertoli, Angelo Labate, and Antonio Gambardella. 2022. "Non-Coding RNAs: New Biomarkers and Therapeutic Targets for Temporal Lobe Epilepsy" International Journal of Molecular Sciences 23, no. 6: 3063. https://doi.org/10.3390/ijms23063063
APA StyleManna, I., Fortunato, F., De Benedittis, S., Sammarra, I., Bertoli, G., Labate, A., & Gambardella, A. (2022). Non-Coding RNAs: New Biomarkers and Therapeutic Targets for Temporal Lobe Epilepsy. International Journal of Molecular Sciences, 23(6), 3063. https://doi.org/10.3390/ijms23063063






