Sunflower WRINKLED1 Plays a Key Role in Transcriptional Regulation of Oil Biosynthesis
Abstract
:1. Introduction
2. Results
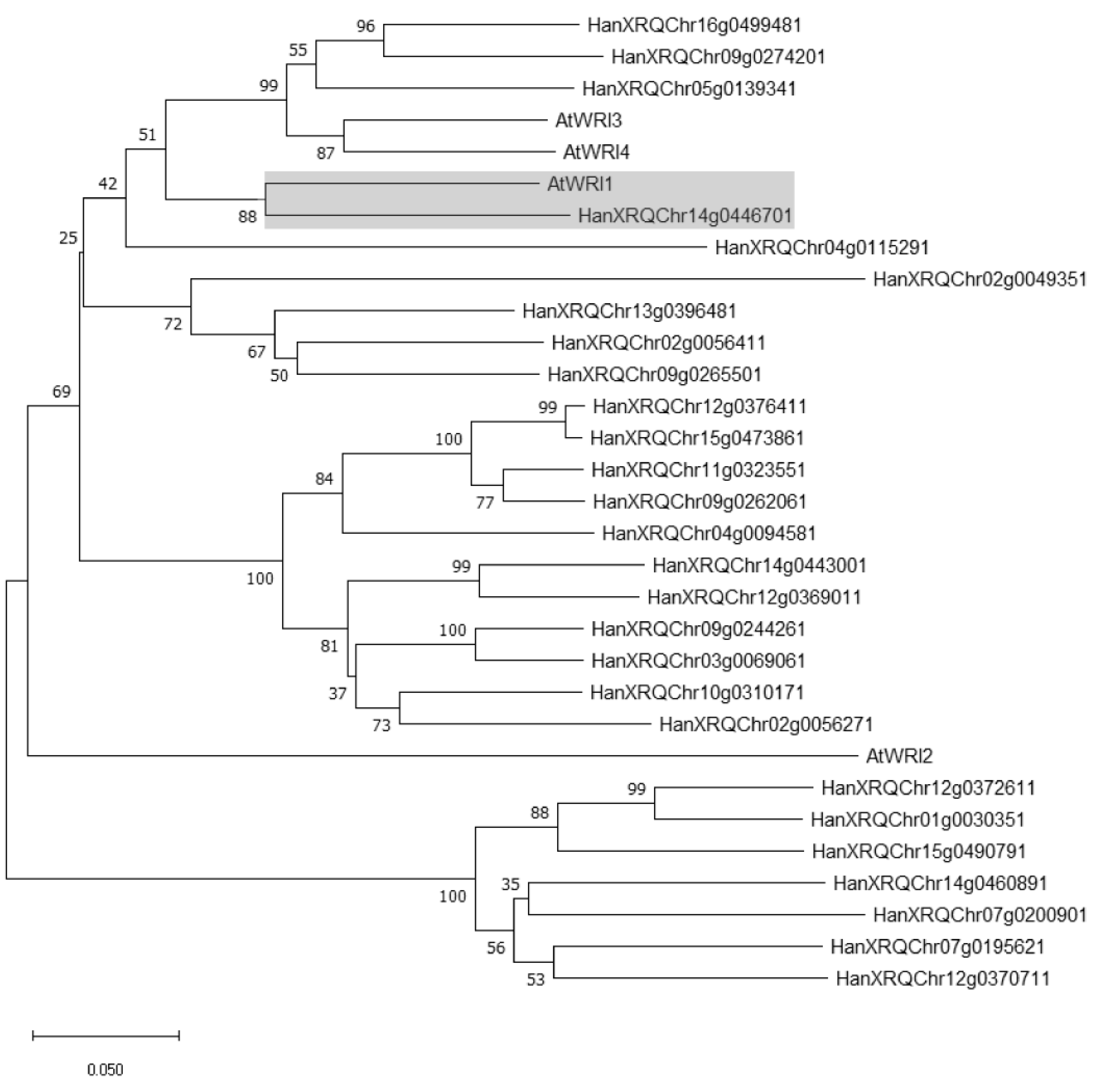
2.1. Identification of WRI1 Ortholog in H. annuus
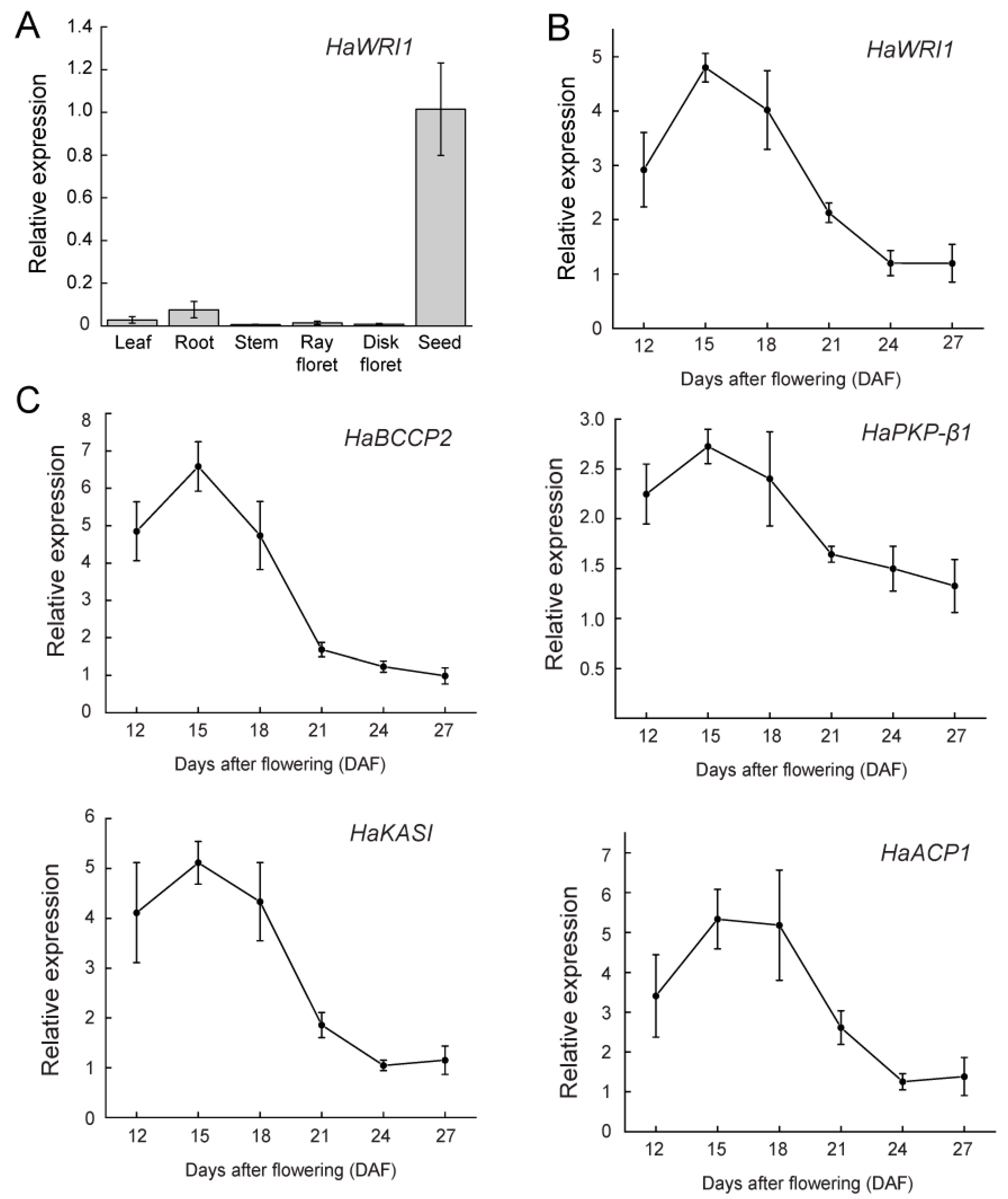
2.2. HaWRI1 Is Highly Expressed in Developing Seeds
2.3. Subcellular Localization of HaWRI1
2.4. Transient Production of HaWRI1 Stimulates TAG Biosynthesis in N. benthamiana Leaves
2.5. HaWRI1 Binds to and Activates proBCCP2 and proPKP-β1
2.6. TAD Is Located at the C-terminus of HaWRI1
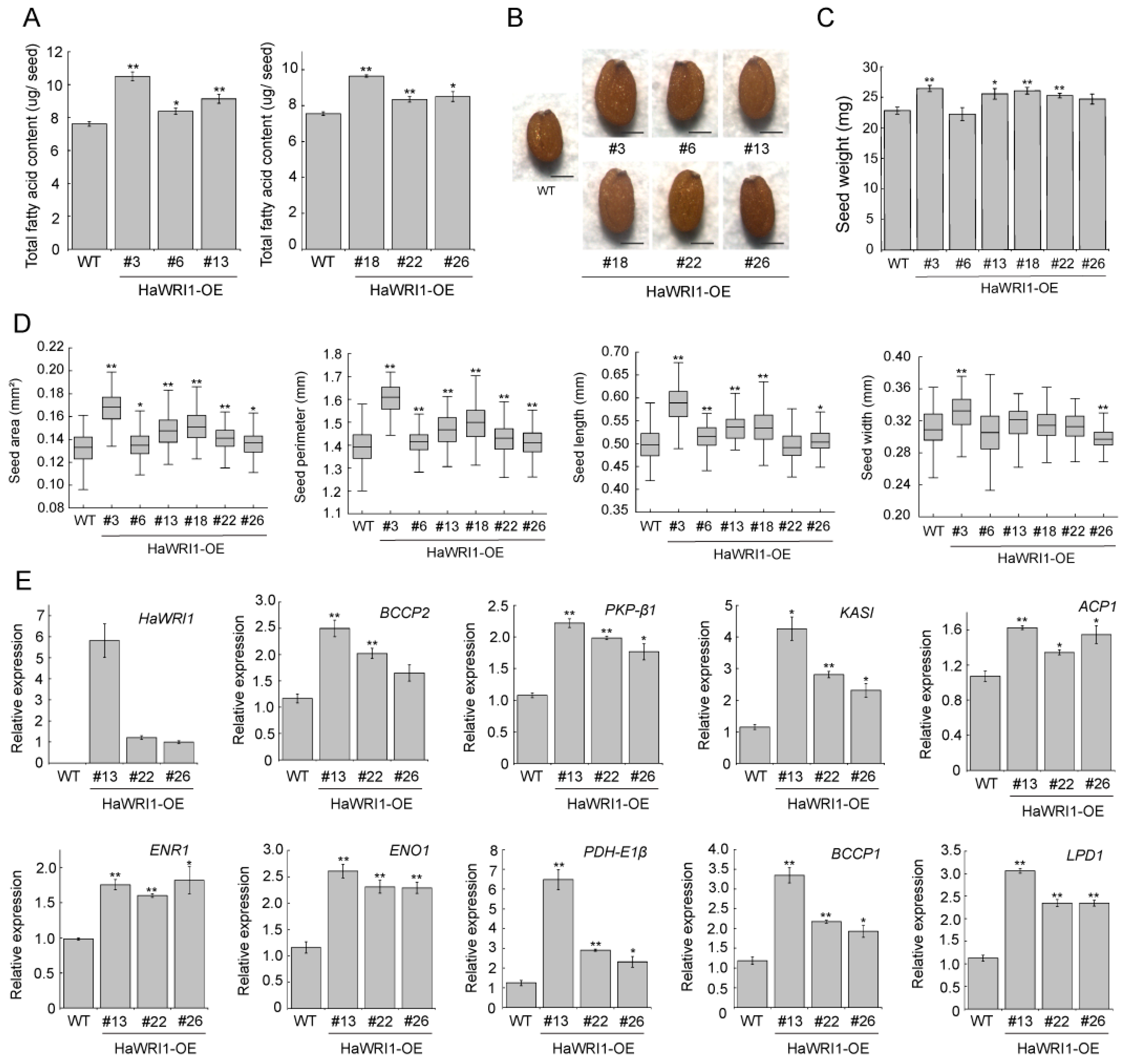
2.7. Overexpression of HaWRI1 in Arabidopsis Leads to Enhanced Seed Oil Accumulation
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Bioinformatic Analysis
4.3. Plasmid Construction
4.4. Transient Expression in N. benthamiana and Confocal Microscopy
4.5. BODIPY Staining
4.6. Yeast Transactivation Assay
4.7. Recombinant Protein Production and EMSA
4.8. Measurement of Seed Weight and Size
4.9. Gene Expression Analysis
4.10. Dual-LUC Assay
4.11. Fatty Acid Analysis
4.12. Accession Numbers
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Durrett, T.P.; Benning, C.; Ohlrogge, J. Plant triacylglycerols as feedstocks for the production of biofuels. Plant J. 2008, 54, 593–607. [Google Scholar] [CrossRef] [PubMed]
- Chapman, K.D.; Ohlrogge, J.B. Compartmentation of triacylglycerol accumulation in plants. J. Biol. Chem. 2012, 287, 2288–2294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Covas, M.I.; de la Torre, R.; Fito, M. Virgin olive oil: A key food for cardiovascular risk protection. Br. J. Nutr. 2015, 113 (Suppl. 2), S19–S28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, J.M.; Zhang, Y.; Zhou, Z.W.; Lu, S.; Ma, W.; Lu, C.; Chen, L.L.; Guo, L. Oil plant genomes: Current state of the science. J. Exp. Bot. 2021, in press. [Google Scholar] [CrossRef] [PubMed]
- Kong, Q.; Yuan, L.; Ma, W. WRINKLED1, a “Master Regulator” in Transcriptional Control of Plant Oil Biosynthesis. Plants 2019, 8, 238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, S.; Ge, Y.; Na Jom, K. A review of phytochemistry, metabolite changes, and medicinal uses of the common sunflower seed and sprouts (Helianthus annuus L.). Chem. Cent. J. 2017, 11, 95. [Google Scholar] [CrossRef] [PubMed]
- Aznar-Moreno, J.A.; Sanchez, R.; Gidda, S.K.; Martinez-Force, E.; Moreno-Perez, A.J.; Caleron, M.V.; Garces, R.; Mullen, R.T.; Salas, J.J. New Insights into Sunflower (Helianthus annuus L.) FatA and FatB Thioesterases, Their Regulation, Structure and Distribution. Front. Plant Sci. 2018, 9, 1496. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Force, E.; Cantisan, S.; Serrano-Vega, M.J.; Garces, R. Acyl-acyl carrier protein thioesterase activity from sunflower (Helianthus annuus L.) seeds. Planta 2000, 211, 673–678. [Google Scholar] [CrossRef]
- Bates, P.D.; Stymne, S.; Ohlrogge, J. Biochemical pathways in seed oil synthesis. Curr. Opin. Plant Biol. 2013, 16, 358–364. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.; Shanklin, J. Triacylglycerol Metabolism, Function, and Accumulation in Plant Vegetative Tissues. Annu. Rev. Plant Biol. 2016, 67, 179–206. [Google Scholar] [CrossRef]
- Li-Beisson, Y.; Shorrosh, B.; Beisson, F.; Andersson, M.X.; Arondel, V.; Bates, P.D.; Baud, S.; Bird, D.; Debono, A.; Durrett, T.P.; et al. Acyl-lipid metabolism. Arabidopsis Book 2013, 11, e0161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miray, R.; Kazaz, S.; To, A.; Baud, S. Molecular Control of Oil Metabolism in the Endosperm of Seeds. Int. J. Mol. Sci. 2021, 22, 1621. [Google Scholar] [CrossRef] [PubMed]
- Kong, Q.; Yang, Y.; Guo, L.; Yuan, L.; Ma, W. Molecular Basis of Plant Oil Biosynthesis: Insights Gained from Studying the WRINKLED1 Transcription Factor. Front. Plant Sci. 2020, 11, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cernac, A.; Benning, C. WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J. 2004, 40, 575–585. [Google Scholar] [CrossRef] [PubMed]
- Masaki, T.; Mitsui, N.; Tsukagoshi, H.; Nishii, T.; Morikami, A.; Nakamura, K. ACTIVATOR of Spomin::LUC1/WRINKLED1 of Arabidopsis thaliana transactivates sugar-inducible promoters. Plant Cell Physiol. 2005, 46, 547–556. [Google Scholar] [CrossRef]
- Baud, S.; Mendoza, M.S.; To, A.; Harscoet, E.; Lepiniec, L.; Dubreucq, B. WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J. 2007, 50, 825–838. [Google Scholar] [CrossRef]
- Maeo, K.; Tokuda, T.; Ayame, A.; Mitsui, N.; Kawai, T.; Tsukagoshi, H.; Ishiguro, S.; Nakamura, K. An AP2-type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW-box sequence conserved among proximal upstream regions of genes involved in fatty acid synthesis. Plant J. 2009, 60, 476–487. [Google Scholar] [CrossRef]
- Focks, N.; Benning, C. wrinkled1: A novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol. 1998, 118, 91–101. [Google Scholar] [CrossRef] [Green Version]
- Ruuska, S.A.; Girke, T.; Benning, C.; Ohlrogge, J.B. Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 2002, 14, 1191–1206. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Liu, X.; Li, N.; Du, C.; Wang, K.; Zhao, C.; Wang, Z.; Hu, Y.; Zhang, M. WRINKLED1 homologs highly and functionally express in oil-rich endosperms of oat and castor. Plant Sci. 2019, 287, 110193. [Google Scholar] [CrossRef]
- Grimberg, A.; Carlsson, A.S.; Marttila, S.; Bhalerao, R.; Hofvander, P. Transcriptional transitions in Nicotiana benthamiana leaves upon induction of oil synthesis by WRINKLED1 homologs from diverse species and tissues. BMC Plant Biol. 2015, 15, 192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Munz, J.; Cass, C.; Zienkiewicz, A.; Kong, Q.; Ma, W.; Sedbrook, J.; Benning, C. Ectopic Expression of WRINKLED1 Affects Fatty Acid Homeostasis in Brachypodium distachyon Vegetative Tissues. Plant Physiol. 2015, 169, 1836–1847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Hua, W.; Zhan, G.; Wei, F.; Wang, X.; Liu, G.; Wang, H. Increasing seed mass and oil content in transgenic Arabidopsis by the overexpression of wri1-like gene from Brassica napus. Plant Physiol. Biochem. 2010, 48, 9–15. [Google Scholar] [CrossRef] [PubMed]
- An, D.; Kim, H.; Ju, S.; Go, Y.S.; Kim, H.U.; Suh, M.C. Expression of Camelina WRINKLED1 Isoforms Rescue the Seed Phenotype of the Arabidopsis wri1 Mutant and Increase the Triacylglycerol Content in Tobacco Leaves. Front. Plant Sci. 2017, 8, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourgis, F.; Kilaru, A.; Cao, X.; Ngando-Ebongue, G.F.; Drira, N.; Ohlrogge, J.B.; Arondel, V. Comparative transcriptome and metabolite analysis of oil palm and date palm mesocarp that differ dramatically in carbon partitioning. Proc. Natl. Acad. Sci. USA 2011, 108, 12527–12532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, W.; Kong, Q.; Arondel, V.; Kilaru, A.; Bates, P.D.; Thrower, N.A.; Benning, C.; Ohlrogge, J.B. Wrinkled1, a ubiquitous regulator in oil accumulating tissues from Arabidopsis embryos to oil palm mesocarp. PLoS ONE 2013, 8, e68887. [Google Scholar] [CrossRef]
- Chen, B.; Zhang, G.; Li, P.; Yang, J.; Guo, L.; Benning, C.; Wang, X.; Zhao, J. Multiple GmWRI1s are redundantly involved in seed filling and nodulation by regulating plastidic glycolysis, lipid biosynthesis and hormone signalling in soybean (Glycine max). Plant Biotechnol. J. 2020, 18, 155–171. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Wang, C.; Sun, Y.; Qu, J.; Mao, H.; Chua, N.H. Overexpression of a Transcription Factor Increases Lipid Content in a Woody Perennial Jatropha curcas. Front. Plant Sci. 2018, 9, 1479. [Google Scholar] [CrossRef] [Green Version]
- Mano, F.; Aoyanagi, T.; Kozaki, A. Atypical Splicing Accompanied by Skipping Conserved Micro-exons Produces Unique WRINKLED1, An AP2 Domain Transcription Factor in Rice Plants. Plants 2019, 8, 207. [Google Scholar] [CrossRef] [Green Version]
- Pouvreau, B.; Baud, S.; Vernoud, V.; Morin, V.; Py, C.; Gendrot, G.; Pichon, J.P.; Rouster, J.; Paul, W.; Rogowsky, P.M. Duplicate maize Wrinkled1 transcription factors activate target genes involved in seed oil biosynthesis. Plant Physiol. 2011, 156, 674–686. [Google Scholar] [CrossRef] [Green Version]
- Shen, B.; Allen, W.B.; Zheng, P.; Li, C.; Glassman, K.; Ranch, J.; Nubel, D.; Tarczynski, M.C. Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol. 2010, 153, 980–987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanjaya; Durrett, T.P.; Weise, S.E.; Benning, C. Increasing the energy density of vegetative tissues by diverting carbon from starch to oil biosynthesis in transgenic Arabidopsis. Plant Biotechnol. J. 2011, 9, 874–883. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Lee, J.H.; Weber, H.; Tohge, T.; Witt, S.; Roje, S.; Fernie, A.R.; Hellmann, H. Arabidopsis BPM Proteins Function as Substrate Adaptors to a CULLIN3-Based E3 Ligase to Affect Fatty Acid Metabolism in Plants. Plant Cell 2013, 25, 2253–2264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, W.; Kong, Q.; Grix, M.; Mantyla, J.J.; Yang, Y.; Benning, C.; Ohlrogge, J.B. Deletion of a C-terminal intrinsically disordered region of WRINKLED1 affects its stability and enhances oil accumulation in Arabidopsis. Plant J. 2015, 83, 864–874. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Kong, Q.; Mantyla, J.J.; Yang, Y.; Ohlrogge, J.B.; Benning, C. 14-3-3 protein mediates plant seed oil biosynthesis through interaction with AtWRI1. Plant J. 2016, 88, 228–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhai, Z.; Liu, H.; Shanklin, J. Phosphorylation of WRINKLED1 by KIN10 Results in Its Proteasomal Degradation, Providing a Link between Energy Homeostasis and Lipid Biosynthesis. Plant Cell 2017, 29, 871–889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhai, Z.; Keereetaweep, J.; Liu, H.; Feil, R.; Lunn, J.E.; Shanklin, J. Trehalose 6-Phosphate Positively Regulates Fatty Acid Synthesis by Stabilizing WRINKLED1. Plant Cell 2018, 30, 2616–2627. [Google Scholar] [CrossRef] [Green Version]
- Kong, Q.; Singh, S.K.; Mantyla, J.J.; Pattanaik, S.; Guo, L.; Yuan, L.; Benning, C.; Ma, W. TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR4 Interacts with WRINKLED1 to Mediate Seed Oil Biosynthesis. Plant Physiol. 2020, 184, 658–665. [Google Scholar] [CrossRef]
- Zhang, M.; Cao, X.; Jia, Q.; Ohlrogge, J. FUSCA3 activates triacylglycerol accumulation in Arabidopsis seedlings and tobacco BY2 cells. Plant J. 2016, 88, 95–107. [Google Scholar] [CrossRef]
- Vanhercke, T.; El Tahchy, A.; Shrestha, P.; Zhou, X.R.; Singh, S.P.; Petrie, J.R. Synergistic effect of WRI1 and DGAT1 coexpression on triacylglycerol biosynthesis in plants. FEBS Lett. 2013, 587, 364–369. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, P.J.; Tjian, R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science 1989, 245, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.; Silva, J.E.; Podicheti, R.; Macrander, J.; Yang, W.; Nazarenus, T.J.; Nam, J.W.; Jaworski, J.G.; Lu, C.; Scheffler, B.E.; et al. Camelina seed transcriptome: A tool for meal and oil improvement and translational research. Plant Biotechnol. J. 2013, 11, 759–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno-Perez, A.J.; Santos-Pereira, J.M.; Martins-Noguerol, R.; DeAndres-Gil, C.; Troncoso-Ponce, M.A.; Venegas-Caleron, M.; Sanchez, R.; Garces, R.; Salas, J.J.; Tena, J.J.; et al. Genome-Wide Mapping of Histone H3 Lysine 4 Trimethylation (H3K4me3) and Its Involvement in Fatty Acid Biosynthesis in Sunflower Developing Seeds. Plants 2021, 10, 706. [Google Scholar] [CrossRef] [PubMed]
- Salas, J.J.; Martinez-Force, E.; Harwood, J.L.; Venegas-Caleron, M.; Aznar-Moreno, J.A.; Moreno-Perez, A.J.; Ruiz-Lopez, N.; Serrano-Vega, M.J.; Graham, I.A.; Mullen, R.T.; et al. Biochemistry of high stearic sunflower, a new source of saturated fats. Prog. Lipid Res. 2014, 55, 30–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchive, C.; Nikovics, K.; To, A.; Lepiniec, L.; Baud, S. Transcriptional regulation of fatty acid production in higher plants: Molecular bases and biotechnological outcomes. Eur. J. Lipid Sci. Technol. 2014, 116, 1332–1343. [Google Scholar] [CrossRef]
- Wang, F.; Perry, S.E. Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiol. 2013, 161, 1251–1264. [Google Scholar] [CrossRef] [Green Version]
- Pelletier, J.M.; Kwong, R.W.; Park, S.; Le, B.H.; Baden, R.; Cagliari, A.; Hashimoto, M.; Munoz, M.D.; Fischer, R.L.; Goldberg, R.B.; et al. LEC1 sequentially regulates the transcription of genes involved in diverse developmental processes during seed development. Proc. Natl. Acad. Sci. USA 2017, 114, E6710–E6719. [Google Scholar] [CrossRef] [Green Version]
- Kong, Q.; Ma, W. WRINKLED1 transcription factor: How much do we know about its regulatory mechanism? Plant Sci. 2018, 272, 153–156. [Google Scholar] [CrossRef]
- Mu, J.; Tan, H.; Zheng, Q.; Fu, F.; Liang, Y.; Zhang, J.; Yang, X.; Wang, T.; Chong, K.; Wang, X.J.; et al. LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol. 2008, 148, 1042–1054. [Google Scholar] [CrossRef] [Green Version]
- An, D.; Suh, M.C. Overexpression of Arabidopsis WRI1 enhanced seed mass and storage oil content in Camelina sativa. Plant Biotechnol. Rep. 2015, 9, 137–148. [Google Scholar] [CrossRef]
- Sun, J.; Chen, T.; Liu, M.; Zhao, D.; Tao, J. Analysis and Functional Verification of PoWRI1 Gene Associated with Oil Accumulation Process in Paeonia ostii. Int. J. Mol. Sci. 2021, 22, 6996. [Google Scholar] [CrossRef] [PubMed]
- Broun, P. Transcription factors as tools for metabolic engineering in plants. Curr. Opin. Plant Biol. 2004, 7, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Grotewold, E. Transcription factors for predictive plant metabolic engineering: Are we there yet? Curr. Opin. Biotechnol. 2008, 19, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.; Vucetic, S.; Radivojac, P.; Brown, C.J.; Dunker, A.K.; Obradovic, Z. Optimizing long intrinsic disorder predictors with protein evolutionary information. J. Bioinform. Comput. Biol. 2005, 3, 35–60. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Kolde, R.; Kolde, M. Package ‘Pheatmap’. 2018. Available online: https://cran.r-project.org/web/packages/pheatmap/index.html (accessed on 15 January 2022).
- Earley, K.W.; Haag, J.R.; Pontes, O.; Opper, K.; Juehne, T.; Song, K.; Pikaard, C.S. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006, 45, 616–629. [Google Scholar] [CrossRef] [PubMed]
- Kong, Q.; Pattanaik, S.; Feller, A.; Werkman, J.R.; Chai, C.; Wang, Y.; Grotewold, E.; Yuan, L. Regulatory switch enforced by basic helix-loop-helix and ACT-domain mediated dimerizations of the maize transcription factor R. Proc. Natl. Acad. Sci. USA 2012, 109, E2091–E2097. [Google Scholar] [CrossRef] [Green Version]
- Hellens, R.P.; Allan, A.C.; Friel, E.N.; Bolitho, K.; Grafton, K.; Templeton, M.D.; Karunairetnam, S.; Gleave, A.P.; Laing, W.A. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 2005, 1, 13. [Google Scholar] [CrossRef] [Green Version]
- Fatihi, A.; Zbierzak, A.M.; Dormann, P. Alterations in seed development gene expression affect size and oil content of Arabidopsis seeds. Plant Physiol. 2013, 163, 973–985. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Yu, X.; Li, K.; Klejnot, J.; Yang, H.; Lisiero, D.; Lin, C. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 2008, 322, 1535–1539. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Li, G.; Gao, S.; Martinez, C.; He, G.; Zhou, Z.; Huang, X.; Lee, J.H.; Zhang, H.; Shen, Y.; et al. Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome A signaling. Plant Cell 2010, 22, 3634–3649. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lim, A.R.Q.; Kong, Q.; Singh, S.K.; Guo, L.; Yuan, L.; Ma, W. Sunflower WRINKLED1 Plays a Key Role in Transcriptional Regulation of Oil Biosynthesis. Int. J. Mol. Sci. 2022, 23, 3054. https://doi.org/10.3390/ijms23063054
Lim ARQ, Kong Q, Singh SK, Guo L, Yuan L, Ma W. Sunflower WRINKLED1 Plays a Key Role in Transcriptional Regulation of Oil Biosynthesis. International Journal of Molecular Sciences. 2022; 23(6):3054. https://doi.org/10.3390/ijms23063054
Chicago/Turabian StyleLim, Audrey R. Q., Que Kong, Sanjay K. Singh, Liang Guo, Ling Yuan, and Wei Ma. 2022. "Sunflower WRINKLED1 Plays a Key Role in Transcriptional Regulation of Oil Biosynthesis" International Journal of Molecular Sciences 23, no. 6: 3054. https://doi.org/10.3390/ijms23063054
APA StyleLim, A. R. Q., Kong, Q., Singh, S. K., Guo, L., Yuan, L., & Ma, W. (2022). Sunflower WRINKLED1 Plays a Key Role in Transcriptional Regulation of Oil Biosynthesis. International Journal of Molecular Sciences, 23(6), 3054. https://doi.org/10.3390/ijms23063054






