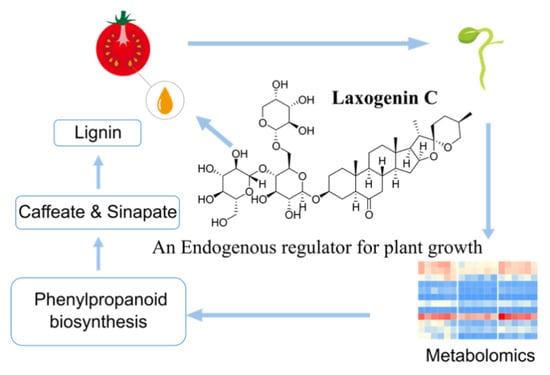
Metabolomics Mechanism and Lignin Response to Laxogenin C, a Natural Regulator of Plants Growth
Abstract
:1. Introduction
2. Results
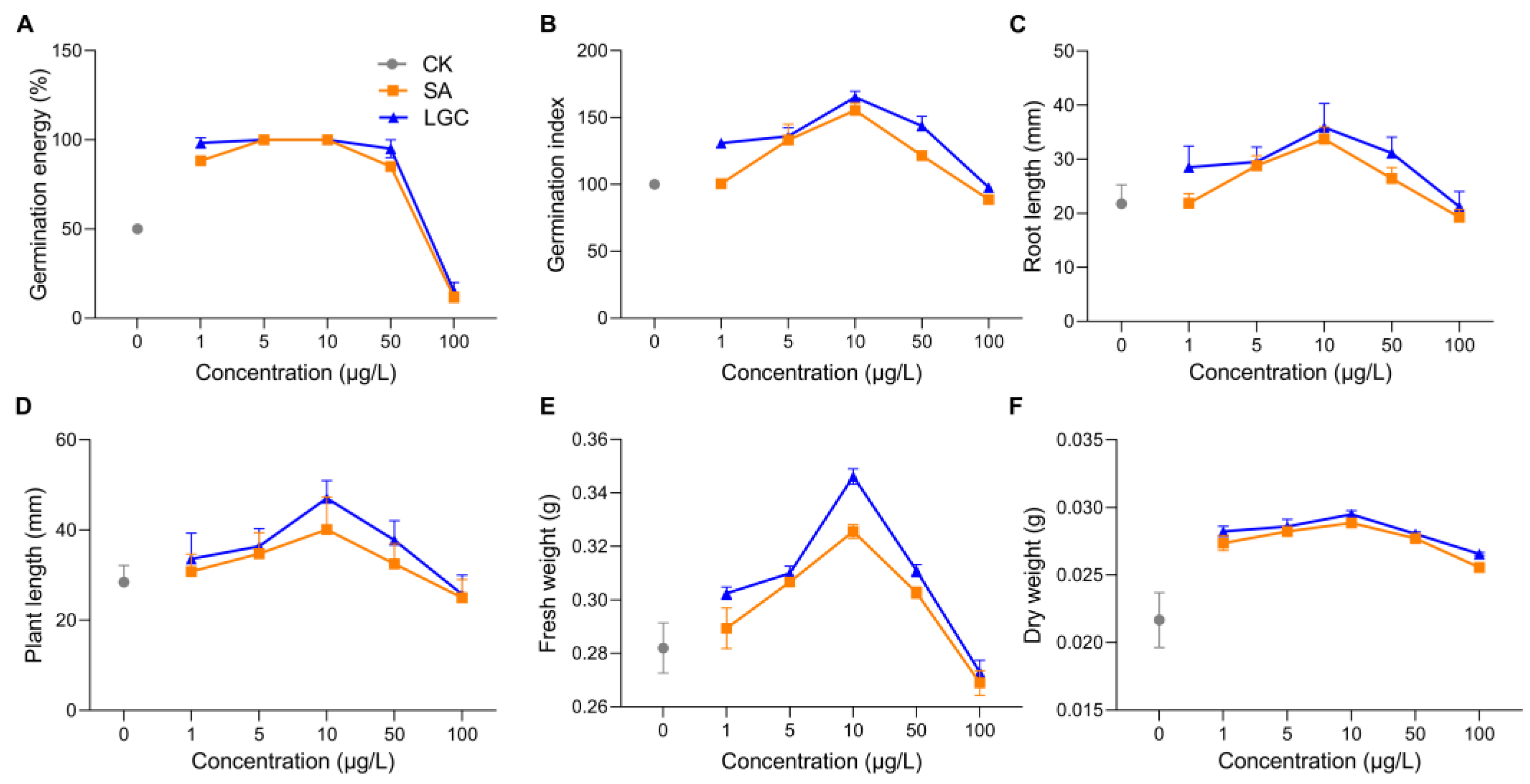
2.1. Tomato Growth Responding to LGC and SA Regulation
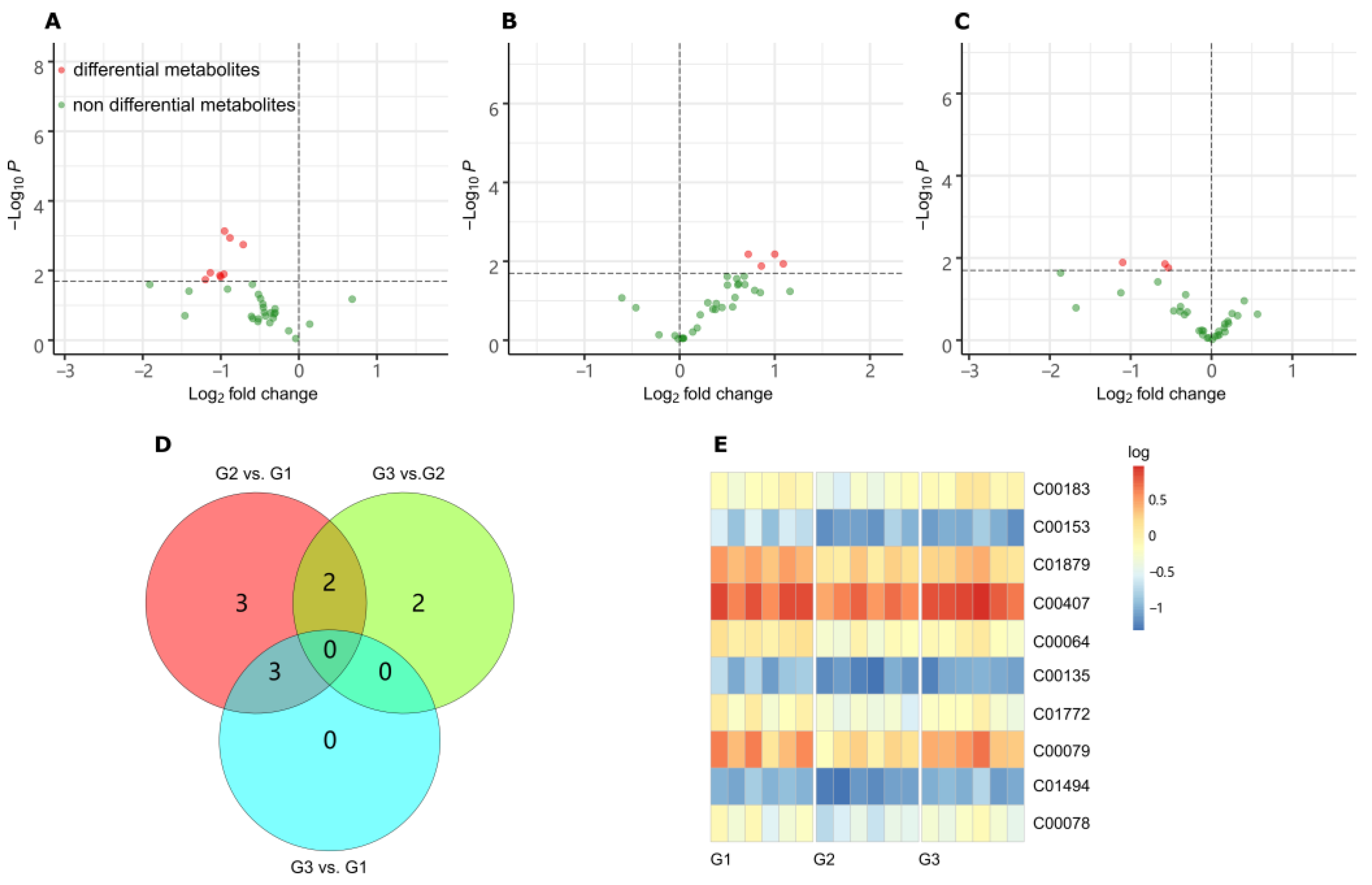
2.2. LC-MS/MS Metabolome Analysis
2.3. Pathways Responding to LGC Regulation
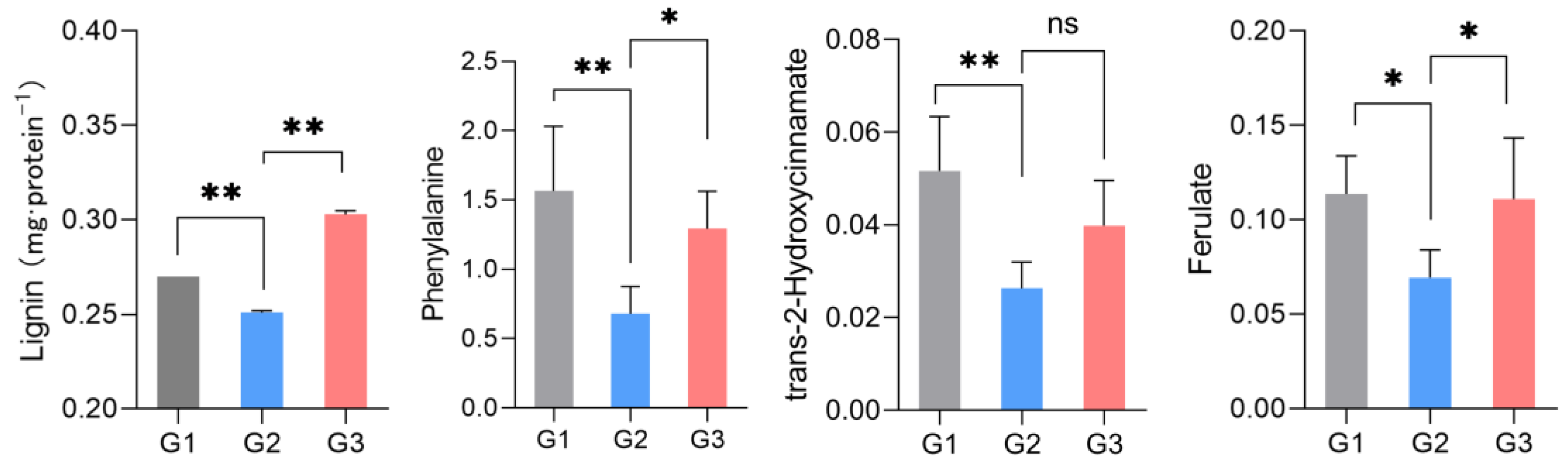
2.4. Phenotype Verification
3. Discussion
3.1. LGC Regulation on Plant Growth
3.2. Metabolome Response to LGC
3.3. Metabolic Mechanisms of LGC Regulation on Plant Growth
3.4. LGC Regulation on Lignin
4. Materials and Methods
4.1. General
4.2. Preparation and Identification of SA and LGC
4.3. Plant Culture and Growth Evaluation
4.4. LC-MS/MS Profiling of Plant Metabolome
4.5. Data Analysis and Visualization
4.6. Lignin Quantification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rademacher, W. Plant Growth Regulators: Backgrounds and Uses in Plant Production. J. Plant Growth Regul. 2015, 34, 845–872. [Google Scholar] [CrossRef]
- Steber, C.M.; McCourt, P. A Role for Brassinosteroids in Germination in Arabidopsis. Plant Physiol. 2001, 125, 763–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.; Lei, W.; He, R.; Tang, X.; Han, J.; Zou, L.; Yin, Y.; Lin, H.; Zhang, D. Brassinosteroids Regulate Root Meristem Development by Mediating BIN2-UPB1 Module in Arabidopsis. PLoS Genet. 2020, 16, e1008883. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Han, M.; Yao, W.; Wang, Y. Transcriptome Analysis Reveals the Regulation of Brassinosteroids on Petal Growth Gerbera hybrida. PeerJ 2017, 5, e3382. [Google Scholar] [CrossRef]
- Ye, H.; Liu, S.; Tang, B.; Chen, J.; Xie, Z.; Nolan, T.M.; Jiang, H.; Guo, H.; Lin, H.-Y.; Li, L.; et al. RD26 Mediates Crosstalk between Drought and Brassinosteroid Signalling Pathways. Nat. Commun. 2017, 8, 14573. [Google Scholar] [CrossRef] [Green Version]
- Hou, J.; Zhang, Q.; Zhou, Y.; Ahammed, G.J.; Zhou, Y.; Yu, J.; Fang, H.; Xia, X. Glutaredoxin GRXS16 Mediates Brassinosteroid-Induced Apoplastic H2O2 Production to Promote Pesticide Metabolism in Tomato. Environ. Pollut. 2018, 240, 227–234. [Google Scholar] [CrossRef]
- De Bock, P.; Van Bockstaele, F.; Muylle, H.; Quataert, P.; Vermeir, P.; Eeckhout, M.; Cnops, G. Yield and Nutritional Characterization of Thirteen Quinoa (Chenopodium Quinoa Willd.) Varieties Grown in North-West Europe—Part I. Plants 2021, 10, 2689. [Google Scholar] [CrossRef]
- Baik, I.-H.; Kim, K.-H.; Lee, K.-A. Antioxidant, Anti-Inflammatory and Antithrombotic Effects of Ginsenoside Compound K Enriched Extract Derived from Ginseng Sprouts. Molecules 2021, 26, 4102. [Google Scholar] [CrossRef]
- Le, N.T.M.; Cuong, D.X.; Thinh, P.V.; Minh, T.N.; Manh, T.D.; Duong, T.-H.; Minh, T.T.L.; Oanh, V.T.T. Phytochemical Screening and Evaluation of Antioxidant Properties and Antimicrobial Activity against Xanthomonas Axonopodis of Euphorbia Tirucalli Extracts in Binh Thuan Province, Vietnam. Molecules 2021, 26, 941. [Google Scholar] [CrossRef]
- Shvets, S.A.; Gutsu, O.N.; Kintia, P.K. Steroidal Glycosides from Nicotiana Tabacum L. Seeds and Their Biological Activity. In Saponins Used in Food and Agriculture; Advances in Experimental Medicine and Biology; Waller, G.R., Yamasaki, K., Eds.; Springer: Boston, MA, USA, 1996; pp. 247–257. ISBN 978-1-4613-0413-5. [Google Scholar]
- Santos Wagner, A.L.; Araniti, F.; Bruno, L.; Ishii-Iwamoto, E.L.; Abenavoli, M.R. The Steroid Saponin Protodioscin Modulates Arabidopsis Thaliana Root Morphology Altering Auxin Homeostasis, Transport and Distribution. Plants 2021, 10, 1600. [Google Scholar] [CrossRef]
- Nikaido, T.; Ohmoto, T.; Kubo, S.; Mimaki, Y.; Sashida, Y. Steroidal Saponins from the Rhizomes of Smilax sieboldii. Phytochemistry 1992, 31, 2445–2450. [Google Scholar] [CrossRef]
- Iglesias-Arteaga, M.A.; Gil, R.P.; Martínez, C.S.P.; Manchado, F.C. Spirostanic Analogues of Teasterone. Synthesis, Characterisation and Biological Activity of Laxogenin, (23S)-Hydroxylaxogenin and 23-Ketolaxogenin (23-Oxolaxogenin). J. Chem. Soc. Perkin Trans. 1 2001, 261–266. [Google Scholar] [CrossRef]
- Wang, Q.; Xu, J.; Liu, X.; Gong, W.; Zhang, C. Synthesis of Brassinosteroids Analogues from Laxogenin and Their Plant Growth Promotion. Nat. Prod. Res. 2015, 29, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.L.; Wang, D.M.; Zhu, W.; Zhang, H.L. Study on Chemical Constituents from Stem of Smilax scobinicaulis. Chin. Pharm. J. 2006, 41, 1540–1542. [Google Scholar]
- Joshi, V.; Joung, J.-G.; Fei, Z.; Jander, G. Interdependence of Threonine, Methionine and Isoleucine Metabolism in Plants: Accumulation and Transcriptional Regulation under Abiotic Stress. Amino Acids 2010, 39, 933–947. [Google Scholar] [CrossRef]
- Thangamani, S.; Eldesouky, H.E.; Mohammad, H.; Pascuzzi, P.E.; Avramova, L.; Hazbun, T.R.; Seleem, M.N. Ebselen Exerts Antifungal Activity by Regulating Glutathione (GSH) and Reactive Oxygen Species (ROS) Production in Fungal Cells. Biochimica et Biophysica Acta (BBA)—General Subjects 2017, 1861, 3002–3010. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Lv, Z.; Wei, Z.; Li, C.; Shao, Y.; Zhang, W.; Zhao, X.; Xiong, J. Microsomal Glutathione Transferase 2 Modulates LTC4 Synthesis and ROS Production in Apostichopus Japonicus. Mol Immunol 2017, 91, 114–122. [Google Scholar] [CrossRef]
- Zhang, C.; Feng, S.; Zhang, L.; Ren, Z. A New Cytotoxic Steroidal Saponin from the Rhizomes and Roots of Smilax scobinicaulis. Nat. Prod. Res. 2013, 27, 1255–1260. [Google Scholar] [CrossRef]
- Zhang, T.; Fan, S.; Xiang, Y.; Zhang, S.; Wang, J.; Sun, Q. Non-Destructive Analysis of Germination Percentage, Germination Energy and Simple Vigour Index on Wheat Seeds during Storage by Vis/NIR and SWIR Hyperspectral Imaging. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 239, 118488. [Google Scholar] [CrossRef]
- Kumar, D.S.; Kumar, P.S.; Rajendran, N.M.; Anbuganapathi, G. Compost Maturity Assessment Using Physicochemical, Solid-State Spectroscopy, and Plant Bioassay Analysis. J. Agric. Food Chem. 2013, 61, 11326–11331. [Google Scholar] [CrossRef]
- Oldiges, M.; Lütz, S.; Pflug, S.; Schroer, K.; Stein, N.; Wiendahl, C. Metabolomics: Current State and Evolving Methodologies and Tools. Appl. Microbiol. Biotechnol. 2007, 76, 495–511. [Google Scholar] [CrossRef] [PubMed]
- Meena, K.K.; Sorty, A.M.; Bitla, U.M.; Choudhary, K.; Gupta, P.; Pareek, A.; Singh, D.P.; Prabha, R.; Sahu, P.K.; Gupta, V.K.; et al. Abiotic Stress Responses and Microbe-Mediated Mitigation in Plants: The Omics Strategies. Front. Plant Sci. 2017, 8, 172. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.Q.; Xia, Y.-Q.; Bakhtiar, R. Use of a Liquid Chromatography/Ion Trap Mass Spectrometry/Triple Quadrupole Mass Spectrometry System for Metabolite Identification. Rapid Commun. Mass Spectrom. 2002, 16, 1330–1336. [Google Scholar] [CrossRef] [PubMed]
- Clarke, N.J.; Rindgen, D.; Korfmacher, W.A.; Cox, K.A. Peer Reviewed: Systematic LC/MS Metabolite Identification in Drug Discovery. Anal. Chem. 2001, 73, 430–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-Independent MS/MS Deconvolution for Comprehensive Metabolome Analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed Minimum Reporting Standards for Chemical Analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [Green Version]
- Dong, N.-Q.; Lin, H.-X. Contribution of Phenylpropanoid Metabolism to Plant Development and Plant–Environment Interactions. J. Integr. Plant Biol. 2021, 63, 180–209. [Google Scholar] [CrossRef]
- Alber, A.; Ehlting, J. Chapter 4—Cytochrome P450s in Lignin Biosynthesis. In Advances in Botanical Research; Jouanin, L., Lapierre, C., Eds.; Academic Press: Cambridge, MA, USA, 2012; Volume 61, pp. 113–143. [Google Scholar]
- Fukushima, R.S.; Hatfield, R.D. Extraction and Isolation of Lignin for Utilization as a Standard to Determine Lignin Concentration Using the Acetyl Bromide Spectrophotometric Method. J. Agric. Food Chem. 2001, 49, 3133–3139. [Google Scholar] [CrossRef]
- Boonsiriwit, A.; Lee, M.; Kim, M.; Itkor, P.; Lee, Y.S. Exogenous Melatonin Reduces Lignification and Retains Quality of Green Asparagus (Asparagus Officinalis L.). Foods 2021, 10, 2111. [Google Scholar] [CrossRef]
- Wang, W.Y.; Zhang, C.L. Effects of Laxogenin and Smilscobinoside C on Seed Germination and Seedlings Growth of Maize. Seed 2011, 30, 15–17. [Google Scholar] [CrossRef]
- Assessment of the Acute Toxicity, Uptake and Biotransformation Potential of Benzotriazoles in Zebrafish (Danio Rerio) Larvae Combining HILIC- with RPLC-HRMS for High-Throughput Identification | Environmental Science & Technology. Available online: https://pubs.acs.org/doi/10.1021/acs.est.8b01327 (accessed on 6 March 2022).
- Wang, K.; Xu, L.; Wang, X.; Chen, A.; Xu, Z. Discrimination of Beef from Different Origins Based on Lipidomics: A Comparison Study of DART-QTOF and LC-ESI-QTOF. LWT 2021, 149, 111838. [Google Scholar] [CrossRef]
- Zhang, X.-Y.; Liu, Y.-H.; Liu, D.-Z.; Xu, J.-Y.; Zhang, Q. Insulin-Mimic Components in Acer Truncatum Leaves: Bio-Guided Isolation, Annual Variance Profiling and Regulating Pathway Investigated by Omics. Pharmaceuticals 2021, 14, 662. [Google Scholar] [CrossRef] [PubMed]
- Picart-Armada, S.; Fernández-Albert, F.; Vinaixa, M.; Yanes, O.; Perera-Lluna, A. FELLA: An R Package to Enrich Metabolomics Data. BMC Bioinform. 2018, 19, 538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Picart-Armada, S.; Fernández-Albert, F.; Vinaixa, M.; Rodríguez, M.A.; Aivio, S.; Stracker, T.H.; Yanes, O.; Perera-Lluna, A. Null Diffusion-Based Enrichment for Metabolomics Data. PLoS ONE 2017, 12, e0189012. [Google Scholar] [CrossRef] [Green Version]


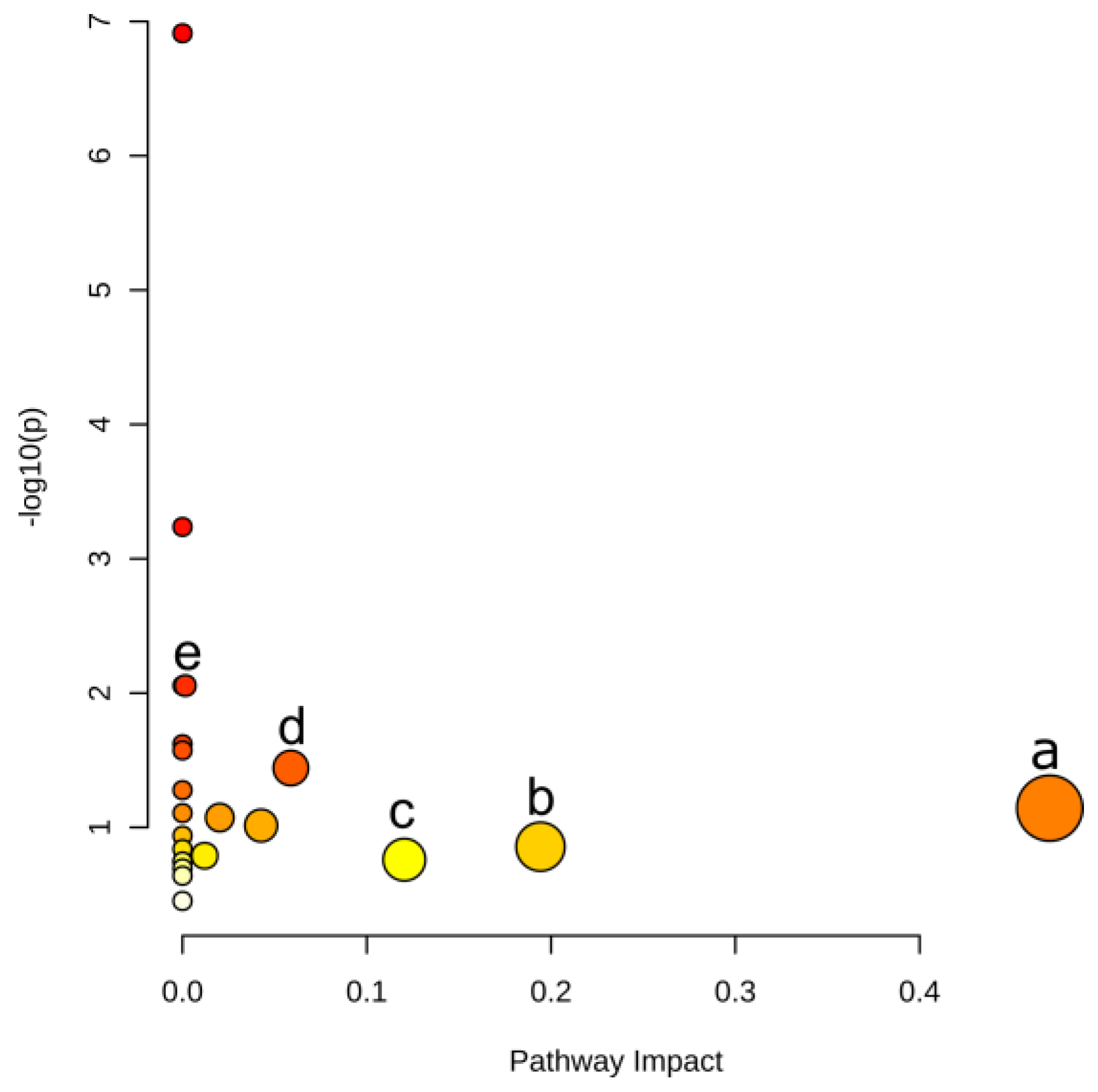
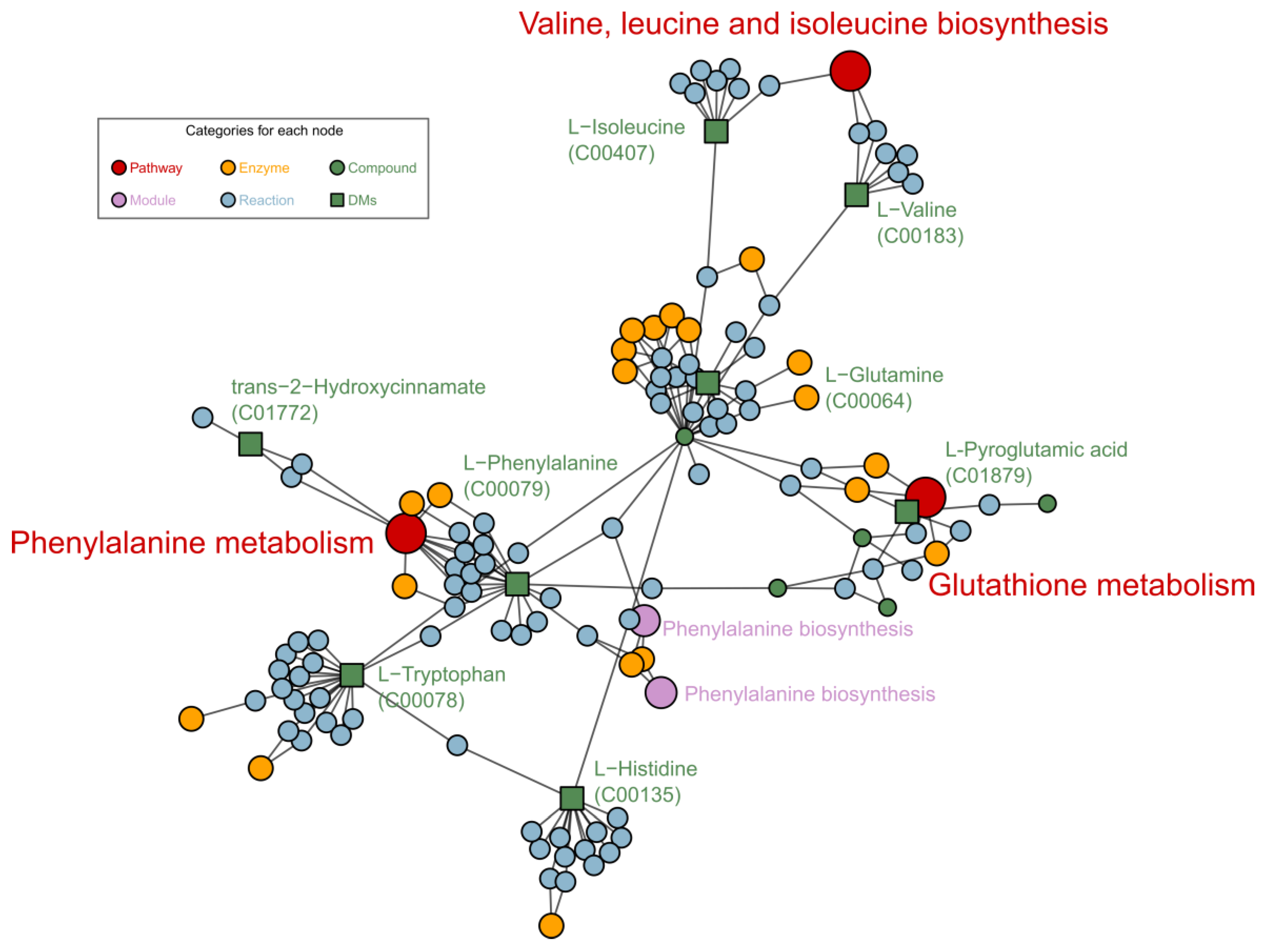
| Pathway | –logP | Impact | Matched Features |
|---|---|---|---|
| Phenylalanine metabolism | 1.14 | 0.470 | l-phenylalanine (C00079) |
| Alanine, aspartate and glutamate metabolism | 0.86 | 0.194 | l-glutamine (C00064) |
| Tryptophan metabolism | 0.76 | 0.120 | l-tryptophan (C00078) |
| Phenylpropanoid biosynthesis | 1.44 | 0.059 | l-phenylalanine (C00079); ferulate (C01494) |
| Phenylalanine, tyrosine and tryptophan biosynthesis | 2.05 | 0.002 | l-phenylalanine (C00079); l-tryptophan (C00078) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, Y.; Wang, J.; Zhang, A.; Zhu, Z.; Ren, S.; Zhang, C.; Zhang, Q. Metabolomics Mechanism and Lignin Response to Laxogenin C, a Natural Regulator of Plants Growth. Int. J. Mol. Sci. 2022, 23, 2990. https://doi.org/10.3390/ijms23062990
Deng Y, Wang J, Zhang A, Zhu Z, Ren S, Zhang C, Zhang Q. Metabolomics Mechanism and Lignin Response to Laxogenin C, a Natural Regulator of Plants Growth. International Journal of Molecular Sciences. 2022; 23(6):2990. https://doi.org/10.3390/ijms23062990
Chicago/Turabian StyleDeng, Yuchan, Jiaqi Wang, Annan Zhang, Zhaoju Zhu, Sipei Ren, Cunli Zhang, and Qiang Zhang. 2022. "Metabolomics Mechanism and Lignin Response to Laxogenin C, a Natural Regulator of Plants Growth" International Journal of Molecular Sciences 23, no. 6: 2990. https://doi.org/10.3390/ijms23062990
APA StyleDeng, Y., Wang, J., Zhang, A., Zhu, Z., Ren, S., Zhang, C., & Zhang, Q. (2022). Metabolomics Mechanism and Lignin Response to Laxogenin C, a Natural Regulator of Plants Growth. International Journal of Molecular Sciences, 23(6), 2990. https://doi.org/10.3390/ijms23062990