Melatonin Signaling Pathways Implicated in Metabolic Processes in Human Granulosa Cells (KGN)
Abstract
1. Introduction
2. Results
2.1. RNAseq Data Analysis
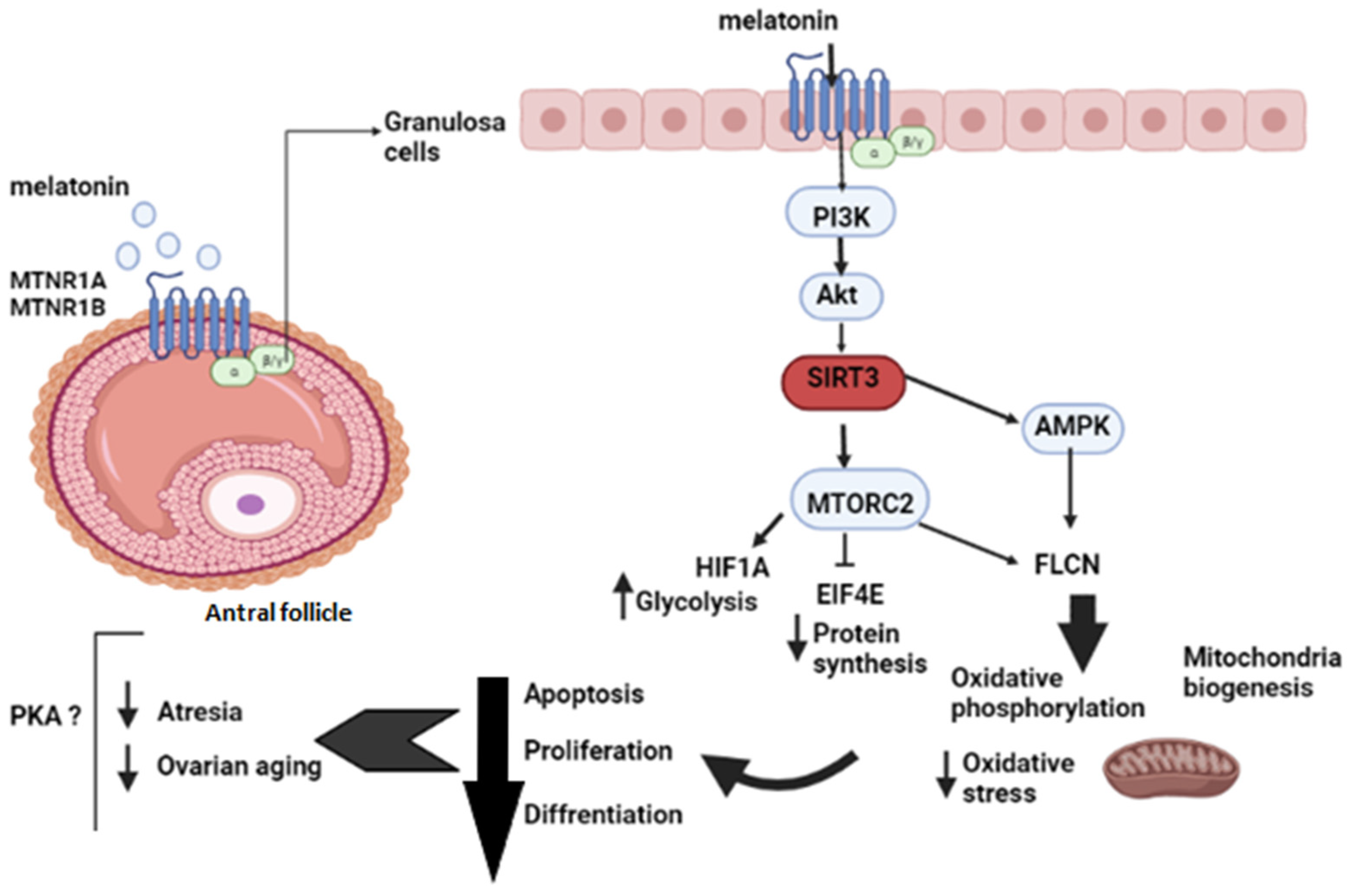
2.2. Upstream Regulators of Melatonin Effects
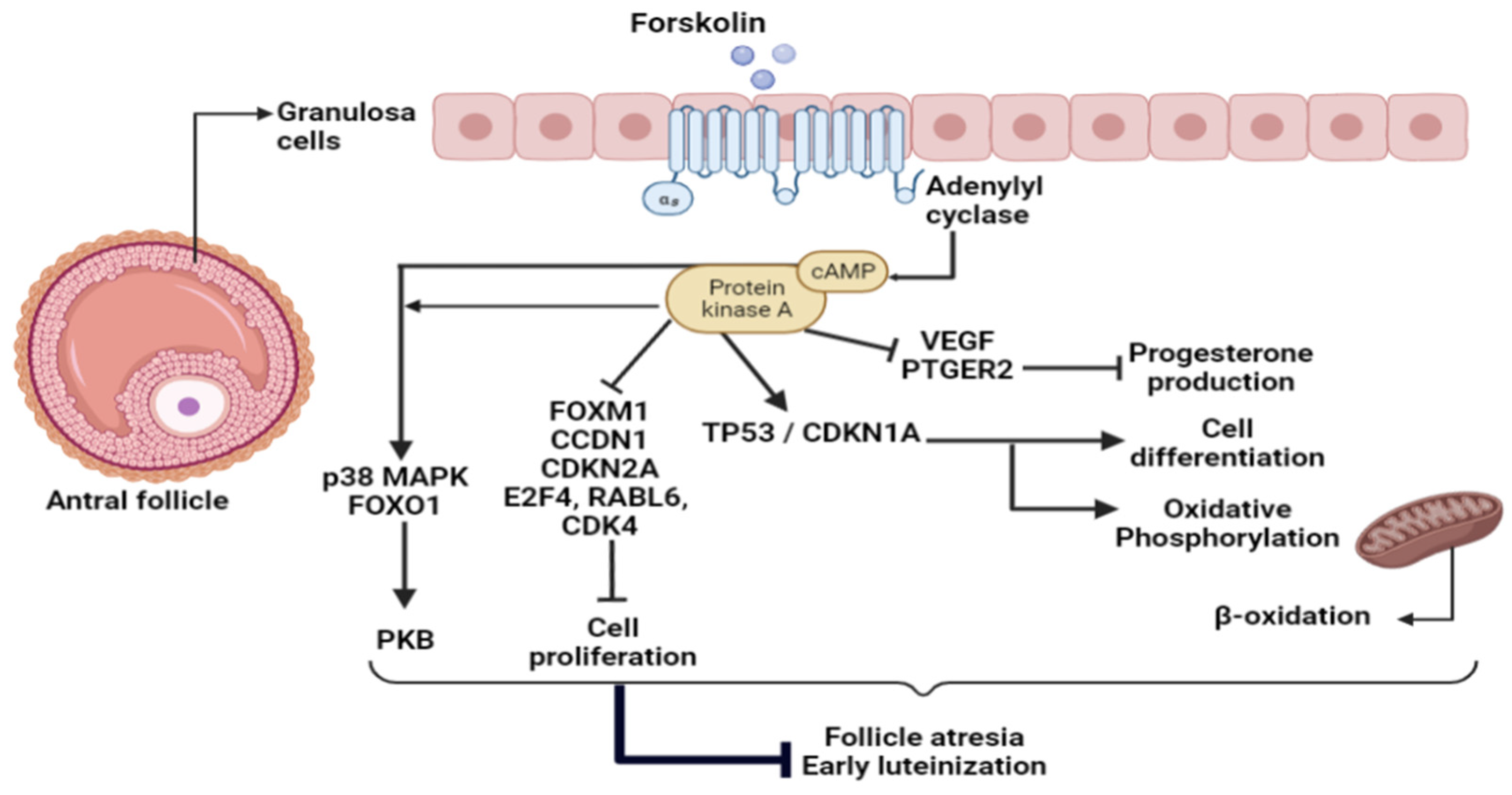
2.3. Upstream Regulators of FSK Effects
2.4. Major Canonical Pathway
3. Discussion
3.1. Melatonin Balances Oxidative Phosphorylation and Anaerobic Glycolysis Using Different Signaling Networks in Human Granulosa Cells
3.2. FSK Effects on KGN Granulosa Cells
4. Materials and Methods
4.1. Chemicals
4.2. Human Granulosa-like Tumor Cell Line (KGN)
4.3. Cell Culture
4.4. RNA Purification and Deep Sequencing
4.5. Transcriptome Assembly and Expression Level Estimate from Reading Counts
4.6. Ingenuity Pathway Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bradley, J.; Swann, K. Mitochondria and lipid metabolism in mammalian oocytes and early embryos. Int. J. Dev. Biol. 2019, 63, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.; Miyano, T. Interaction between growing oocytes and granulosa cells in vitro. Reprod. Med. Biol. 2019, 19, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Albertini, D.F. The road to maturation: Somatic cell interaction and self-organization of the mammalian oocyte. Nat. Rev. Mol. Cell Biol. 2013, 14, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Robker, R.; Hennebold, J.; Russell, D. Coordination of Ovulation and Oocyte Maturation: A Good Egg at the Right Time. Endocrinology 2018, 159, 3209–3218. [Google Scholar] [CrossRef]
- Meldrum, D.R.; Casper, R.F.; Diez-Juan, A.; Simon, C.; Domar, A.D.; Frydman, R. Aging and the environment affect gamete and embryo potential: Can we intervene? Fertil. Steril. 2016, 105, 548–559. [Google Scholar] [CrossRef]
- Niles, L.P.; Wang, J.; Shen, L.; Lobb, D.K.; Younglai, E.V. Melatonin receptor mRNA expression in human granulosa cells. Mol. Cell. Endocrinol. 1999, 156, 107–110. [Google Scholar] [CrossRef]
- Wang, S.J.; Liu, W.J.; Wu, C.J.; Ma, F.H.; Ahmad, S.; Liu, B.R.; Han, L.; Jiang, X.P.; Zhang, S.J.; Yang, L.G. Melatonin suppresses apoptosis and stimulates progesterone production by bovine granulosa cells via its receptors (MT1 and MT2). Theriogenology 2012, 78, 1517–1526. [Google Scholar] [CrossRef]
- Tamura, H.; Jozaki, M.; Tanabe, M.; Shirafuta, Y.; Mihara, Y.; Shinagawa, M.; Tamura, I.; Maekawa, R.; Sato, S.; Taketani, T.; et al. Importance of Melatonin in Assisted Reproductive Technology and Ovarian Aging. Int. J. Mol. Sci. 2020, 21, 1135. [Google Scholar] [CrossRef]
- Hong, F.; Pan, S.; Xu, P.; Xue, T.; Wang, J.; Guo, Y.; Jia, L.; Qiao, X.; Li, L.; Zhai, Y. Melatonin Orchestrates Lipid Homeostasis through the Hepatointestinal Circadian Clock and Microbiota during Constant Light Exposure. Cells 2020, 9, 489. [Google Scholar] [CrossRef]
- Cipolla-Neto, J.; Amaral, F.G.; Afeche, S.C.; Tan, D.X.; Reiter, R.J. Melatonin, energy metabolism, and obesity: A review. J. Pineal Res. 2014, 56, 371–381. [Google Scholar] [CrossRef]
- Li, Y.; Li, S.; Zhou, Y.; Meng, X.; Zhang, J.-J.; Xu, D.-P.; Li, H.-B. Melatonin for the prevention and treatment of cancer. Oncotarget 2017, 8, 39896–39921. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.-X.; Lee, S.; Taweechaipaisankul, A.; Kim, G.A.; Lee, B.C. Melatonin regulates lipid metabolism in porcine oocytes. J. Pineal Res. 2017, 62, e12388. [Google Scholar] [CrossRef]
- He, B.; Yin, C.; Gong, Y.; Liu, J.; Guo, H.; Zhao, R. Melatonin-induced increase of lipid droplets accumulation and in vitro maturation in porcine oocytes is mediated by mitochondrial quiescence. J. Cell. Physiol. 2018, 233, 302–312. [Google Scholar] [CrossRef] [PubMed]
- Nishi, Y.; Yanase, T.; Mu, Y.-M.; Oba, K.; Ichino, I.; Saito, M.; Nomura, M.; Mukasa, C.; Okabe, T.; Goto, K.; et al. Establishment and Characterization of a Steroidogenic Human Granulosa-Like Tumor Cell Line, KGN, That Expresses Functional Follicle-Stimulating Hormone Receptor. Endocrinology 2001, 142, 437–445. [Google Scholar] [CrossRef]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2013, 30, 523–530. [Google Scholar] [CrossRef]
- Doridot, L.; Châtre, L.; Ducat, A.; Vilotte, J.-L.; Lombès, A.; Méhats, C.; Barbaux, S.; Calicchio, R.; Ricchetti, M.; Vaiman, D. Nitroso-Redox Balance and Mitochondrial Homeostasis Are Regulated by STOX1, a Pre-Eclampsia-Associated Gene. Antioxid. Redox Signal. 2014, 21, 819–834. [Google Scholar] [CrossRef]
- Thulluru, H.; Park, C.; Dufort, D.; Kleiverda, G.; Oudejans, C.; Van Dijk, M. Maternal Nodal inversely affects NODAL and STOX1 expression in the fetal placenta. Front. Genet. 2013, 4, 170. [Google Scholar] [CrossRef][Green Version]
- Sun, N.; Yun, J.; Liu, J.; Malide, D.; Liu, C.; Rovira, I.I.; Holmström, K.M.; Fergusson, M.M.; Yoo, Y.H.; Combs, C.A.; et al. Measuring In Vivo Mitophagy. Mol. Cell 2015, 60, 685–696. [Google Scholar] [CrossRef]
- Fung, S.; Nishimura, T.; Sasarman, F.; Shoubridge, E.A. The conserved interaction of C7orf30 with MRPL14 promotes biogenesis of the mitochondrial large ribosomal subunit and mitochondrial translation. Mol. Biol. Cell 2012, 24, 184–193. [Google Scholar] [CrossRef]
- Scott, C.C.; Vossio, S.; Rougemont, J.; Gruenberg, J. TFAP2 transcription factors are regulators of lipid droplet biogenesis. eLife 2018, 7, e36330. [Google Scholar] [CrossRef]
- Osborne, B.; Bentley, N.L.; Montgomery, M.K.; Turner, N. The role of mitochondrial sirtuins in health and disease. Free Radic. Biol. Med. 2016, 100, 164–174. [Google Scholar] [CrossRef] [PubMed]
- De Bock, K.; Georgiadou, M.; Carmeliet, P. Role of Endothelial Cell Metabolism in Vessel Sprouting. Cell Metab. 2013, 18, 634–647. [Google Scholar] [CrossRef] [PubMed]
- Sharif, T.; Martell, E.; Dai, C.; Singh, S.K.; Gujar, S. Regulation of the proline regulatory axis and autophagy modulates stemness in TP73/p73 deficient cancer stem-like cells. Autophagy 2019, 15, 934–936. [Google Scholar] [CrossRef] [PubMed]
- Nam, S.O.; Yotsumoto, F.; Miyata, K.; Fukagawa, S.; Yamada, H.; Kuroki, M.; Miyamoto, S. Warburg effect regulated by amphiregulin in the development of colorectal cancer. Cancer Med. 2015, 4, 575–587. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.H.; Zhou, M.; Liu, H.; Ding, Y.; Khong, H.T.; Yu, D.; Fodstad, O.; Tan, M. Upregulation of lactate dehydrogenase A by ErbB2 through heat shock factor 1 promotes breast cancer cell glycolysis and growth. Oncogene 2009, 28, 3689–3701. [Google Scholar] [CrossRef]
- Krisher, R.; Prather, R. A Role for the Warburg Effect in Preimplantation Embryo Development: Metabolic Modification to Support Rapid Cell Proliferation. Mol. Reprod. Dev. 2012, 79, 311–320. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef]
- Fontana, R.; Torre, S. The Deep Correlation between Energy Metabolism and Reproduction: A View on the Effects of Nutrition for Women Fertility. Nutrients 2016, 8, 87. [Google Scholar] [CrossRef]
- Torre, S.D.; Benedusi, V.; Fontana, R.; Maggi, A. Energy metabolism and fertility—A balance preserved for female health. Nat. Rev. Endocrinol. 2014, 10, 13–23. [Google Scholar] [CrossRef]
- Woo, M.M.M.; Tai, C.-J.; Kang, S.K.; Nathwani, P.S.; Pang, S.F.; Leung, P.C.K. Direct Action of Melatonin in Human Granulosa-Luteal Cells. J. Clin. Endocrinol. Metab. 2001, 86, 4789–4797. [Google Scholar] [CrossRef]
- Acuña-Castroviejo, D.; Escames, G.; Venegas, C.; Díaz-Casado, M.E.; Lima-Cabello, E.; López, L.C.; Rosales-Corral, S.; Tan, D.-X.; Reiter, R.J. Extrapineal melatonin: Sources, regulation, and potential functions. Cell. Mol. Life Sci. 2014, 71, 2997–3025. [Google Scholar] [CrossRef] [PubMed]
- Dragojevic Dikic, S.; Jovanovic, A.M.; Dikic, S.; Jovanovic, T.; Jurisic, A.; Dobrosavljevic, A. Melatonin: A “Higgs boson” in human reproduction. Gynecol. Endocrinol. 2015, 31, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Gbahou, F.; Cecon, E.; Viault, G.; Gerbier, R.; Jean-Alphonse, F.; Karamitri, A.; Guillaumet, G.; Delagrange, P.; Friedlander, R.M.; Vilardaga, J.-P.; et al. Design and validation of the first cell-impermeant melatonin receptor agonist. Br. J. Pharmacol. 2017, 174, 2409–2421. [Google Scholar] [CrossRef] [PubMed]
- Oishi, A.; Karamitri, A.; Gerbier, R.; Lahuna, O.; Ahmad, R.; Jockers, R. Orphan GPR61, GPR62 and GPR135 receptors and the melatonin MT2 receptor reciprocally modulate their signaling functions. Sci. Rep. 2017, 7, 8990. [Google Scholar] [CrossRef]
- Cecon, E.; Oishi, A.; Jockers, R. Melatonin receptors: Molecular pharmacology and signalling in the context of system bias. Br. J. Pharmacol. 2018, 175, 3263–3280. [Google Scholar] [CrossRef]
- Guo, Q.; Wang, Z.; Dong, Y.; Cao, J.; Chen, Y. Physiological crosstalk between the AC/PKA and PLC/PKC pathways modulates melatonin-mediated, monochromatic-light-induced proliferation of T-lymphocytes in chickens. Cell Tissue Res. 2017, 369, 555–565. [Google Scholar] [CrossRef]
- Kim, H.-R.; Chae, H.-J.; Thomas, M.; Miyazaki, T.; Monosov, A.; Monosov, E.; Krajewska, M.; Krajewski, S.; Reed, J.C. Mammalian dap3 is an essential gene required for mitochondrial homeostasis in vivo and contributing to the extrinsic pathway for apoptosis. FASEB J. 2007, 21, 188–196. [Google Scholar] [CrossRef]
- Miyazaki, T.; Shen, M.; Fujikura, D.; Tosa, N.; Kim, H.-R.; Kon, S.; Uede, T.; Reed, J.C. Functional Role of Death-associated Protein 3 (DAP3) in Anoikis. J. Biol. Chem. 2004, 279, 44667–44672. [Google Scholar] [CrossRef]
- Santos, A.N.; Langner, J.; Herrmann, M.; Riemann, D. Aminopeptidase N/CD13 Is Directly Linked to Signal Transduction Pathways in Monocytes. Cell. Immunol. 2000, 201, 22–32. [Google Scholar] [CrossRef]
- Lendeckel, U.A.M.; Fran, K.; Spiess, A.; Reinhold, D. Ansorge, S. Modulation of WNT-5At Eexpression by Actinonin: Linkage of APN to the WNT-Pathway? Cell. Pept. Immune Funct. Dis. 2. Adv. Exp. Med. Biol. 2002, 477, 35–41. [Google Scholar]
- Richter, U.; Lahtinen, T.; Marttinen, P.; Suomi, F.; Battersby, B.J. Quality control of mitochondrial protein synthesis is required for membrane integrity and cell fitness. J. Cell Biol. 2015, 211, 373–389. [Google Scholar] [CrossRef] [PubMed]
- Sina, A.; Lord-Dufour, S.; Annabi, B. Cell-based evidence for aminopeptidase N/CD13 inhibitor actinonin targeting of MT1-MMP-mediated proMMP-2 activation. Cancer Lett. 2009, 279, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, P.G.; Sirard, M.-A. Gene analysis of major signaling pathways regulated by gonadotropins in human ovarian granulosa tumor cells (KGN). Biol. Reprod. 2020, 103, 583–598. [Google Scholar] [CrossRef] [PubMed]
- Berisha, B.; Schams, D.; Rodler, D.; Sinowatz, F. Expression pattern of HIF1alpha and vasohibins during follicle maturation and corpus luteum function in the bovine ovary. Reprod. Domest. Anim. 2016, 52, 130–139. [Google Scholar] [CrossRef]
- Henríquez, S.; Kohen, P.; Muñoz, A.; Godoy, A.; Orge, F.; Strauss, J.F., III; Devoto, L. In-vitro study of gonadotrophin signaling pathways in human granulosa cells in relation to progesterone receptor expression. Reprod. BioMed. Online 2017, 35, 363–371. [Google Scholar] [CrossRef]
- Fischer, B.; Kleinstein, W.K.J.; Gips, H. Oxygen tension in follicular fluid falls with follicle maturation Eur. J. Obstet. Gynecol. Reprod. Biol. 1992, 43, 39–43. [Google Scholar] [CrossRef]
- Zhang, Z.H.; Chen, L.Y.; Wang, F.; Wu, Y.Q.; Su, J.Q.; Huang, X.H.; Wang, Z.C.; Cheng, Y. Expression of hypoxia-inducible factor-1α during ovarian follicular growth and development in Sprague-Dawley rats Genet. Mol. Res. 2015, 14, 5896–5909. [Google Scholar] [CrossRef]
- Tam, K.K.Y.; Russell, D.L.; Peet, D.J.; Bracken, C.P.; Rodgers, R.J.; Thompson, J.G.; Kind, K.L. Hormonally regulated follicle differentiation and luteinization in the mouse is associated with hypoxia inducible factor activity. Mol. Cell. Endocrinol. 2010, 327, 47–55. [Google Scholar] [CrossRef]
- Movafagh, S.; Crook, S.; Vo, K. Regulation of hypoxia-inducible factor-1a by reactive oxygen species: New developments in an old debate. J. Cell. Biochem. 2015, 116, 696–703. [Google Scholar] [CrossRef]
- Semenza, G.L. HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J. Clin. Investig. 2013, 123, 3664–3671. [Google Scholar] [CrossRef]
- Fadhillah; Yoshioka, S.; Nishimura, R.; Yamamoto, Y.; Kimura, K.; Okuda, K. Hypoxia-inducible factor 1 mediates hypoxia-enhanced synthesis of progesterone during luteinization of granulosa cells. J. Reprod. Dev. 2017, 63, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Baddela, V.S.; Sharma, A.; Michaelis, M.; Vanselow, J. HIF1 driven transcriptional activity regulates steroidogenesis and proliferation of bovine granulosa cells. Sci. Rep. 2020, 10, 3906. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Bagchi, I.C.; Bagchi, M.K. Signaling by hypoxia-inducible factors is critical for ovulation in mice. Endocrinology 2009, 150, 3392–3400. [Google Scholar] [CrossRef] [PubMed]
- Meléndez-Rodríguez, F.; Urrutia, A.A.; Lorendeau, D.; Rinaldi, G.; Roche, O.; Böğürcü-Seidel, N.; Ortega Muelas, M.; Mesa-Ciller, C.; Turiel, G.; Bouthelier, A.; et al. HIF1α Suppresses Tumor Cell Proliferation through Inhibition of Aspartate Biosynthesis. Cell Rep. 2019, 26, 2257–2265. [Google Scholar] [CrossRef] [PubMed]
- Vaupel, P.; Schmidberger, H.; Mayer, A. The Warburg effect: An essential part of metabolic reprogramming and central contributor to cancer progression. Int. J. Radiat. Biol. 2019, 95, 912–919. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.-C.; Quintin, J.; Cramer, R.A.; Shepardson, K.M.; Saeed, S.; Kumar, V.; Giamarellos-Bourboulis, E.J.; Martens, J.H.A.; Rao, N.A.; Aghajanirefah, A.; et al. mTOR- and HIF-1α–mediated aerobic glycolysis as metabolic basis for trained immunity. Science 2014, 345, 1250684. [Google Scholar] [CrossRef] [PubMed]
- Pacella-Ince, L.; Zander-Fox, D.L.; Lane, M. Mitochondrial SIRT3 and its target glutamate dehydrogenase are altered in follicular cells of women with reduced ovarian reserve or advanced maternal age. Hum. Reprod. 2014, 29, 1490–1499. [Google Scholar] [CrossRef]
- Akemi Nishigaki, T.K.; Kida, N.; Kakita-Kobayashi, M.; Tsubokura, H.; Hisamatsu, Y.; Okada, H. Resveratrol protects mitochondrial quantity by activating SIRT1/PGC-1α expression during ovarian hypoxia. Reprod. Med. Biol. 2020, 19, 189–197. [Google Scholar] [CrossRef]
- Lombard, D.; Zwaans, B. SIRT3: As simple as it seems? Gerontology 2013, 60, 56–64. [Google Scholar] [CrossRef]
- Fu, H.; Wada-Hiraike, O.; Hirano, M.; Kawamura, Y.; Sakurabashi, A.; Shirane, A.; Morita, Y.; Isono, W.; Oishi, H.; Koga, K.; et al. SIRT3 Positively Regulates the Expression of Folliculogenesis- and Luteinization-Related Genes and Progesterone Secretion by Manipulating Oxidative Stress in Human Luteinized Granulosa Cells. Endocrinology 2014, 155, 3079–3087. [Google Scholar] [CrossRef]
- Chen, L.; Vasoya, R.P.; Toke, N.H.; Parthasarathy, A.; Luo, S.; Chiles, E.; Flores, J.; Gao, N.; Bonder, E.M.; Su, X.; et al. HNF4 Regulates Fatty Acid Oxidation and Is Required for Renewal of Intestinal Stem Cells in Mice. Gastroenterology 2020, 158, 985–999. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Peng, W.; Yin, S.; Zhao, J.; Fu, B.; Zhang, J.; Mao, T.; Wu, H.; Zhang, Y. Melatonin improves age-induced fertility decline and attenuates ovarian mitochondrial oxidative stress in mice. Sci. Rep. 2016, 6, 35165. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Han, L.; Yu, J.; Li, H.; Li, Q. miR-224 aggravates cancer-associated fibroblast-induced progression of non-small cell lung cancer by modulating a positive loop of the SIRT3/AMPK/mTOR/HIF-1α axis. Aging 2021, 13, 10431–10449. [Google Scholar] [CrossRef] [PubMed]
- Yaba, A.; Demir, N. The mechanism of mTOR (mammalian target of rapamycin) in a mouse model of polycystic ovary syndrome (PCOS). J. Ovarian Res. 2012, 5, 38. [Google Scholar] [CrossRef]
- Chen, Z.; Kang, X.; Wang, L.; Dong, H.; Wang, C.; Xiong, Z.; Zhao, W.; Jia, C.; Lin, J.; Zhang, W.; et al. Rictor/mTORC2 Pathway in Oocytes Regulates Folliculogenesis, and Its Inactivation Causes Premature Ovarian Failure. J. Biol. Chem. 2015, 290, 6387–6396. [Google Scholar] [CrossRef]
- Yaba, A.; Bianchi, V.; Borini, A.; Johnson, J. A Putative Mitotic Checkpoint Dependent on mTOR Function Controls Cell Proliferation and Survival in Ovarian Granulosa Cells. Reprod. Sci. 2008, 15, 128–138. [Google Scholar] [CrossRef]
- Hagiwara, A.; Cornu, M.; Cybulski, N.; Polak, P.; Betz, C.; Trapani, F.; Terracciano, L.; Heim, M.H.; Rüegg, M.A.; Hall, M.N. Hepatic mTORC2 Activates Glycolysis and Lipogenesis through Akt, Glucokinase, and SREBP1c. Cell Metab. 2012, 15, 725–738. [Google Scholar] [CrossRef]
- Kazyken, D.; Magnuson, B.; Bodur, C.; Acosta-Jaquez, H.A.; Zhang, D.; Tong, X.; Barnes, T.M.; Steinl, G.K.; Patterson, N.E.; Altheim, C.H.; et al. AMPK directly activates mTORC2 to promote cell survival during acute energetic stress. Sci. Signal. 2019, 12, eaav3249. [Google Scholar] [CrossRef]
- Mao, Z.; Zhang, W. Role of mTOR in Glucose and Lipid Metabolism. Int. J. Mol. Sci. 2018, 19, 2043. [Google Scholar] [CrossRef]
- Ruggero, D. The role of Myc-induced protein synthesis in cancer. Cancer Res. 2009, 69, 8839–8843. [Google Scholar] [CrossRef]
- Isono, Y.; Furuya, M.; Kuwahara, T.; Sano, D.; Suzuki, K.; Jikuya, R.; Mitome, T.; Otake, S.; Kawahara, T.; Ito, Y.; et al. FLCN alteration drives metabolic reprogramming towards nucleotide synthesis and cyst formation in salivary gland. Biochem. Biophys. Res. Commun. 2020, 522, 931–938. [Google Scholar] [CrossRef] [PubMed]
- Wada, S.; Neinast, M.; Jang, C.; Ibrahim, Y.H.; Lee, G.; Babu, A.; Li, J.; Hoshino, A.; Rowe, G.C.; Rhee, J.; et al. The tumor suppressor FLCN mediates an alternate mTOR pathway to regulate browning of adipose tissue. Genes Dev. 2016, 30, 2551–2564. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Gingras, M.-C.; Dunlop, E.A.; Nouët, Y.; Dupuy, F.; Jalali, Z.; Possik, E.; Coull, B.J.; Kharitidi, D.; Dydensborg, A.B.; et al. The tumor suppressor folliculin regulates AMPK-dependent metabolic transformation. J. Clin. Investig. 2014, 124, 2640–2650. [Google Scholar] [CrossRef] [PubMed]
- François, C.M.; Wargnier, R.; Petit, F.; Goulvent, T.; Rimokh, R.; Treilleux, I.; Ray-Coquard, I.; Zazzu, V.; Cohen-Tannoudji, J.; Guigon, C.J. 17β-estradiol inhibits spreading of metastatic cells from granulosa cell tumors through a non-genomic mechanism involving GPER1. Carcinogenesis 2015, 36, 564–573. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.; Thomas, P. Role of G protein-coupled estrogen receptor 1, GPER, in inhibition of oocyte maturation by endogenous estrogens in zebrafish. Dev. Biol. 2010, 342, 194–206. [Google Scholar] [CrossRef]
- Chan, Q.K.Y.; Lam, H.M.; Ng, C.F.; Lee, A.Y.Y.; Chan, E.S.Y.; Ng, H.K.; Ho, S.M.; Lau, K.M. Activation of GPR30 inhibits the growth of prostate cancer cells through sustained activation of Erk1/2, c-jun/c-fos-dependent upregulation of p21, and induction of G2 cell-cycle arrest. Cell Death Differ. 2010, 17, 1511–1523. [Google Scholar] [CrossRef]
- Liang, S.; Chen, Z.; Jiang, G.; Zhou, Y.; Liu, Q.; Su, Q.; Wei, W.; Du, J.; Wang, H. Activation of GPER suppresses migration and angiogenesis of triple negative breast cancer via inhibition of NF-κB/IL-6 signals. Cancer Lett. 2017, 386, 12–23. [Google Scholar] [CrossRef]
- Chowdhury, I.; Thompson, W.E.; Thomas, K. Prohibitins Role in Cellular Survival Through Ras-Raf-MEK-ERK Pathway. J. Cell. Physiol. 2014, 229, 998–1004. [Google Scholar] [CrossRef]
- Tao, L.; Zhu, Y. Melatonin regulates CRE-dependent gene transcription underlying osteoblast proliferation by activating Src and PKA in parallel. Am. J. Transl. Res. 2018, 10, 86–100. [Google Scholar]
- Gazarini, M.L.; Beraldo, F.H.; Almeida, F.M.; Bootman, M.; Da Silva, A.M.; Garcia, C.R.S. Melatonin triggers PKA activation in the rodent malaria parasite Plasmodium chabaudi. J. Pineal Res. 2011, 50, 64–70. [Google Scholar] [CrossRef]
- Shinagawa, M.; Tamura, I.; Maekawa, R.; Sato, S.; Shirafuta, Y.; Mihara, Y.; Okada-Matsumoto, M.; Taketani, T.; Asada, H.; Tamura, H.; et al. C/EBPβ regulates Vegf gene expression in granulosa cells undergoing luteinization during ovulation in female rats. Sci. Rep. 2019, 9, 714. [Google Scholar] [CrossRef] [PubMed]
- Fraser, H.M.; Wulff, C. Angiogenesis in the primate ovary. Reprod. Fertil. Dev. 2002, 13, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Lei, L.; Liu, D.; Jovin, I.; Russell, R.; Johnson, R.S.; Di Lorenzo, A.; Giordano, F.J. Normal glucose uptake in the brain and heart requires an endothelial cell-specific HIF-1α-dependent function. Proc. Natl. Acad. Sci. USA 2012, 109, 17478–17483. [Google Scholar] [CrossRef]
- Doyle, L.K.; Walker, C.A.; Donadeu, F.X. VEGF modulates the effects of gonadotropins in granulosa cells. Domest. Anim. Endocrinol. 2010, 38, 127–137. [Google Scholar] [CrossRef]
- Grasselli, F.; Basini, G.; Bussolati, S.; Tamanini, C. Effects of VEGF and bFGF on Proliferation and Production of Steroids and Nitric Oxide in Porcine Granulosa Cells. Reprod. Domest. Anim. 2002, 37, 362–368. [Google Scholar] [CrossRef]
- Hernández-Coronado, C.G.; Guzmán, A.; Rodríguez, A.; Mondragón, J.A.; Romano, M.C.; Gutiérrez, C.G.; Rosales-Torres, A.M. Sphingosine-1-phosphate, regulated by FSH and VEGF, stimulates granulosa cell proliferation. Gen. Comp. Endocrinol. 2016, 236, 1–8. [Google Scholar] [CrossRef]
- Bogan, R.L.; Murphy, M.J.; Stouffer, R.L.; Hennebold, J.D. Prostaglandin Synthesis, Metabolism, and Signaling Potential in the Rhesus Macaque Corpus Luteum throughout the Luteal Phase of the Menstrual Cycle. Endocrinology 2008, 149, 5861–5871. [Google Scholar] [CrossRef][Green Version]
- Chandras, C.; Harris, T.E.; Bernal, A.L.P.; Abayasekara, D.R.E.; Michael, A.E. PTGER1 and PTGER2 receptors mediate regulation of progesterone synthesis and type 1 11β-hydroxysteroid dehydrogenase activity by prostaglandin E2 in human granulosa–lutein cells. J. Endocrinol. 2007, 194, 595. [Google Scholar] [CrossRef]
- Shrestha, K.; Meidan, R. The cAMP-EPAC Pathway Mediates PGE2-Induced FGF2 in Bovine Granulosa Cells. Endocrinology 2018, 159, 3482–3491. [Google Scholar] [CrossRef]
- Kruiswijk, F.; Labuschagne, C.F.; Vousden, K.H. p53 in survival, death and metabolic health: A lifeguard with a licence to kill. Nat. Rev. Mol. Cell Biol. 2015, 16, 393–405. [Google Scholar] [CrossRef]
- Zhang, X.-d.; Qin, Z.-h.; Wang, J. The role of p53 in cell metabolism. Acta Pharm. Sin. 2010, 31, 1208–1212. [Google Scholar] [CrossRef] [PubMed]
- Sirotkin, A.V.; Benčo, A.; Tandlmajerova, A.; Vašíček, D.; Kotwica, J.; Darlak, K.; Valenzuela, F. Transcription factor p53 can regulate proliferation, apoptosis and secretory activity of luteinizing porcine ovarian granulosa cell cultured with and without ghrelin and FSH. Reproduction 2008, 136, 611–618. [Google Scholar] [CrossRef] [PubMed]
- Douville, G.; Sirard, M.-A. Changes in granulosa cells gene expression associated with growth, plateau and atretic phases in medium bovine follicles. J. Ovarian Res. 2014, 7, 50. [Google Scholar] [CrossRef] [PubMed]
- Hatzirodos, N.; Hummitzsch, K.; Irving-Rodgers, H.F.; Harland, M.L.; Morris, S.E.; Rodgers, R.J. Transcriptome profiling of granulosa cells from bovine ovarian follicles during atresia. BMC Genom. 2014, 15, 40. [Google Scholar] [CrossRef]
- Tremblay, P.G.; Sirard, M.-A. Transcriptomic analysis of gene cascades involved in protein kinase A and C signaling in the KGN line of human ovarian granulosa tumor cells. Biol. Reprod. 2017, 96, 855–865. [Google Scholar] [CrossRef]
- Parrott, J.A.; Skinner, M.K. Developmental and Hormonal Regulation of Hepatocyte Growth Factor Expression and Action in the Bovine Ovarian Follicle1. Biol. Reprod. 1998, 59, 553–560. [Google Scholar] [CrossRef]
- Taniguchi, F.; Harada, T.; Deura, I.; Iwabe, T.; Tsukihara, S.; Terakawa, N. Hepatocyte growth factor promotes cell proliferation and inhibits progesterone secretion via PKA and MAPK pathways in a human granulosa cell line. Mol. Reprod. Dev. 2004, 68, 335–344. [Google Scholar] [CrossRef]
- Hunzicker-Dunn, M.E.; Lopez-Biladeau, B.; Law, N.C.; Fiedler, S.E.; Carr, D.W.; Maizels, E.T. PKA and GAB2 play central roles in the FSH signaling pathway to PI3K and AKT in ovarian granulosa cells. Proc. Natl. Acad. Sci. USA 2012, 109, E2979–E2988. [Google Scholar] [CrossRef]
- Herndon, M.K.; Law, N.C.; Donaubauer, E.M.; Kyriss, B.; Hunzicker-Dunn, M. Forkhead box O member FOXO1 regulates the majority of follicle-stimulating hormone responsive genes in ovarian granulosa cells. Mol. Cell. Endocrinol. 2016, 434, 116–126. [Google Scholar] [CrossRef]
- Ding, Y.; Liu, Z.; Desai, S.; Zhao, Y.; Liu, H.; Pannell, L.K.; Yi, H.; Wright, E.R.; Owen, L.B.; Dean-Colomb, W.; et al. Receptor tyrosine kinase ErbB2 translocates into mitochondria and regulates cellular metabolism. Nat. Commun. 2012, 3, 1271. [Google Scholar] [CrossRef]
- Lv, X.; He, C.; Huang, C.; Wang, H.; Hua, G.; Wang, Z.; Zhou, J.; Chen, X.; Ma, B.; Timm, B.K.; et al. Timely expression and activation of YAP1 in granulosa cells is essential for ovarian follicle development. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2019, 33, 10049–10064. [Google Scholar] [CrossRef] [PubMed]
- Girard, A.; Dufort, I.; Douville, G.; Sirard, M.-A. Global gene expression in granulosa cells of growing, plateau and atretic dominant follicles in cattle. Reprod. Biol. Endocrinol. 2015, 13, 17. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
| Upstream Regulator | Predicted Activation State | Activation z-Score | p-Value of Overlap |
|---|---|---|---|
| DAP3 | Inhibited | −2.449 | 2.64 × 10−6 |
| MT-TE | 4.96 × 10−5 | ||
| LONP1 | −0.522 | 7.91 × 10−5 | |
| STOX1 | 8.31 × 10−5 | ||
| Actinonin | Activated | 2.236 | 2.35 × 10−4 |
| HIF1A | 0.520 | 6.20 × 10−4 | |
| MALSU1 | 8.09 × 10−4 | ||
| AP2α | 8.09 × 10−4 | ||
| MRPL14 | 1.38 × 10−3 | ||
| SIRT3 | Activated | 2.020 | 1.99 × 10−3 |
| Upstream Regulator | Predicted Activation State | Activation z-Score | p-Value of Overlap |
|---|---|---|---|
| ZBTB17 | 1.55 × 10−26 | ||
| Vegf | Inhibited | −2.090 | 5.96 × 10−18 |
| PTGER2 | Inhibited | −3.394 | 1.09 × 10−15 |
| TP53 | 1.729 | 5.11 × 10−15 | |
| HGF | Inhibited | −2.072 | 7.72 × 10−15 |
| CDKN1A | 1.907 | 8.50 × 10−15 | |
| FOXM1 | Inhibited | −3.168 | 8.99 × 10−14 |
| NR1H3 | 1.10 × 10−13 | ||
| AREG | Inhibited | −3.357 | 3.02 × 10−13 |
| Canonical Pathways | Upstream Regulators | z-Score |
|---|---|---|
| Melatonin | ||
| Oxidative Phosphorylation | ATP5PF, COX11, COX4I1, MT-ATP6, MT-CO3, MT-CYB, MT-ND1, MT-ND4, NDUFA2, NDUFA6, NDUFB8, NDUFV3, UQCRFS1 | −1.941 |
| Mitochondrial Dysfunction | ATP5PF, COX11, COX4I1, FIS1, HSD17B10, MT-ATP6, MT-CO3, MT-CYB, MT-ND1, MT-ND4, NDUFA2, NDUFA6, NDUFB8, NDUFV3, PDHA1, UQCRFS1 | |
| Sirtuin Signaling Pathway | ATG14, ATG16L1, ATP5PF, CPS1, H3F3A/H3F3B, LDHD, MT-ATP6, MT-CYB, MT-ND1, MT-ND4, NDUFA2, NDUFA6, NDUFB8, NDUFV3, PDHA1, POLR1E, PPARA, TIMM13, TOMM40L, UQCRFS1 | 1.941 |
| TGF-β Signaling | ACVR1C, MAP3K7, MAPK11, PIAS4, RALB, RUNX2, TGFB2, TGIF1 | 1.134 |
| Thiamin Salvage III | TPK1 | |
| FSK | ||
| tRNA Splicing | PDE1A, PDE3A, PDE4B, PDE4D, PDE7B | 2.236 |
| Protein Kinase A Signaling | ADCY1, CDKN3, DUSP5, GNB3, LEF1, MYLK2, PDE1A, PDE3A, PDE4B, PDE4D, PDE7B, PLCL1, PTPRN, PTPRR | 1.732 |
| Glutamate Receptor Signaling | GNB3, GRIA4, SLC1A3, SLC1A7 | |
| TGF-β Signaling | BCL2, INHA, PITX2, RASD1, VDR | |
| Methylglyoxal Degradation VI | LDHD |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asma, A.; Marc-André, S. Melatonin Signaling Pathways Implicated in Metabolic Processes in Human Granulosa Cells (KGN). Int. J. Mol. Sci. 2022, 23, 2988. https://doi.org/10.3390/ijms23062988
Asma A, Marc-André S. Melatonin Signaling Pathways Implicated in Metabolic Processes in Human Granulosa Cells (KGN). International Journal of Molecular Sciences. 2022; 23(6):2988. https://doi.org/10.3390/ijms23062988
Chicago/Turabian StyleAsma, Arjoune, and Sirard Marc-André. 2022. "Melatonin Signaling Pathways Implicated in Metabolic Processes in Human Granulosa Cells (KGN)" International Journal of Molecular Sciences 23, no. 6: 2988. https://doi.org/10.3390/ijms23062988
APA StyleAsma, A., & Marc-André, S. (2022). Melatonin Signaling Pathways Implicated in Metabolic Processes in Human Granulosa Cells (KGN). International Journal of Molecular Sciences, 23(6), 2988. https://doi.org/10.3390/ijms23062988






