GhENODL6 Isoforms from the Phytocyanin Gene Family Regulated Verticillium Wilt Resistance in Cotton
Abstract
:1. Introduction
2. Results
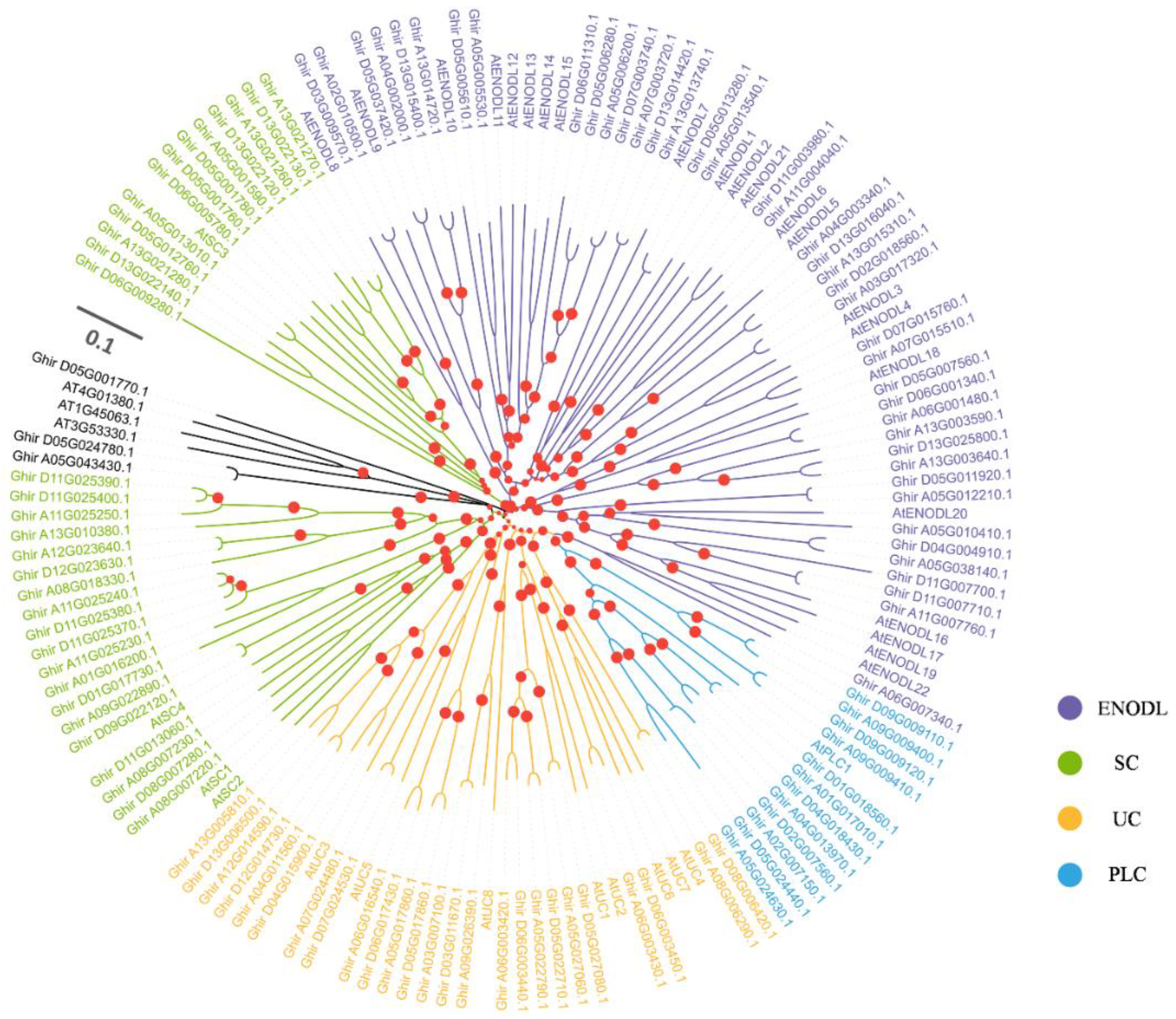
2.1. Identification, Sequence Features, and Phylogeny of the Cotton GhPCs Gene Family
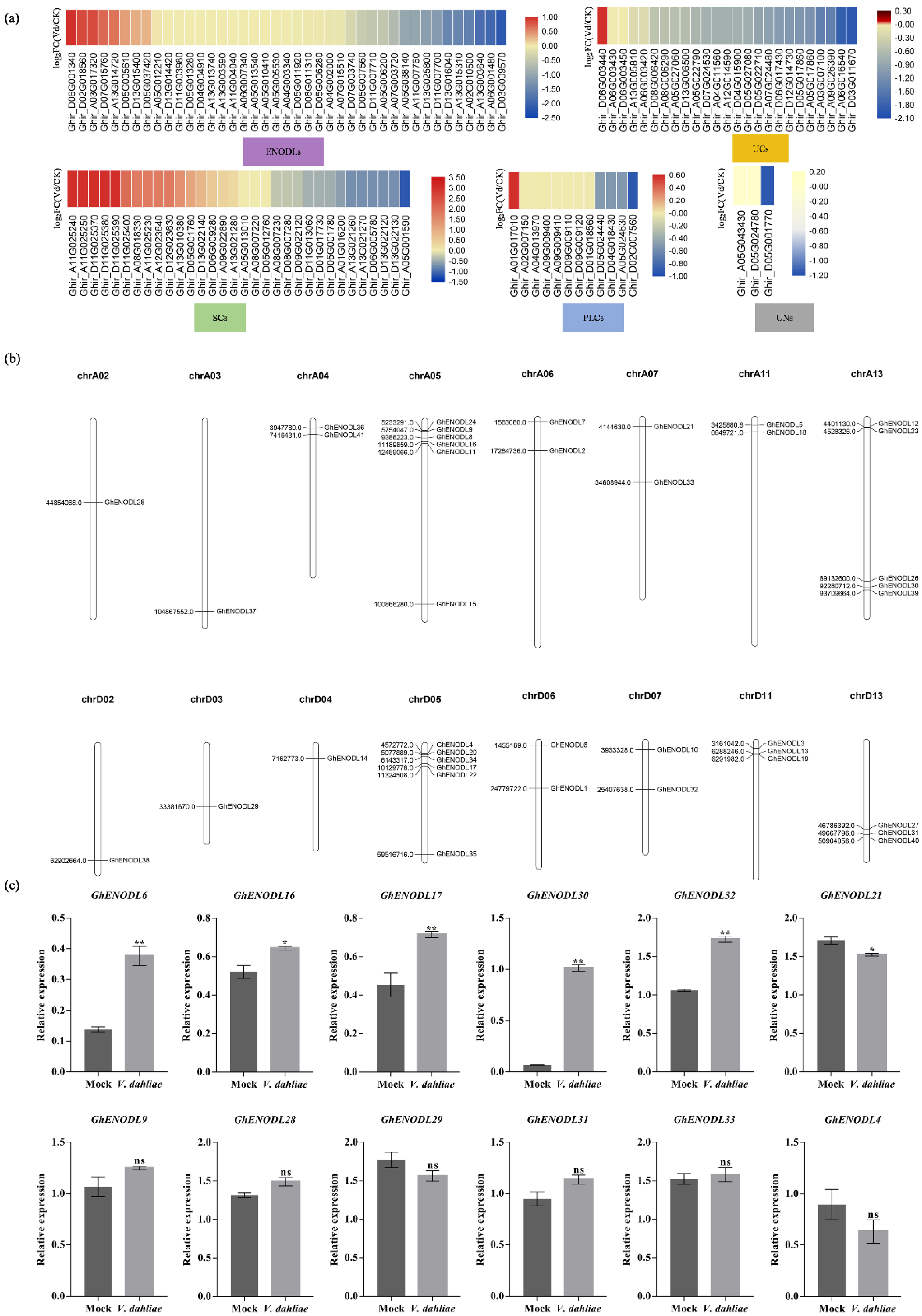
2.2. GhPC Genes Have Distinct Expression Patterns upon V. dahliae Stress
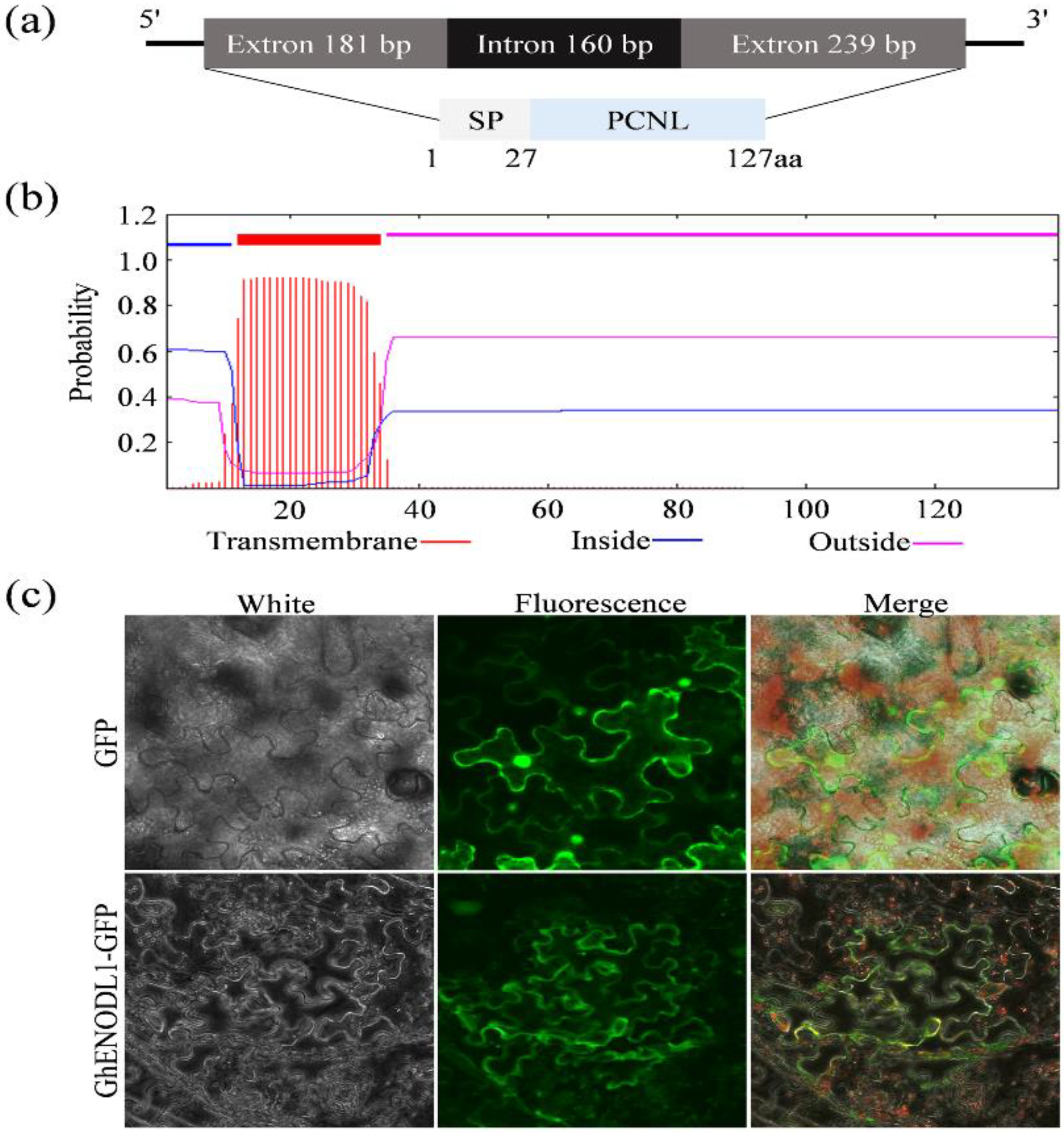
2.3. GhENODL6 Is a Plasma Membrane Protein
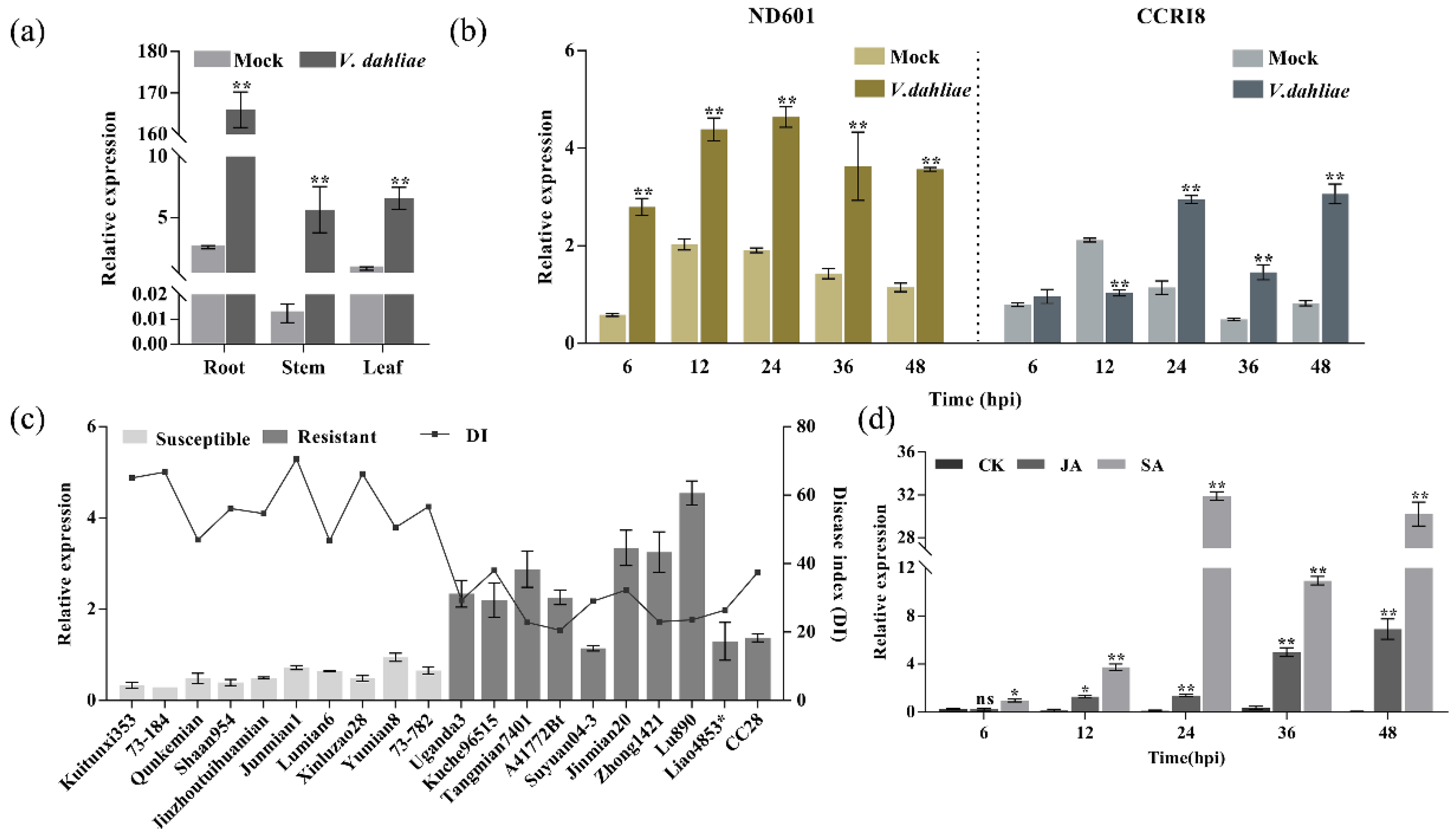
2.4. GhENODL6 Expression Is Upregulated in Response to V. dahliae and Hormonal Signal in Cotton
2.5. GhENODL6 Positively Regulated Verticillium Wilt Resistance in Plants
2.6. GhENODL6 Regulates Defense Genes Involving in SA Signaling
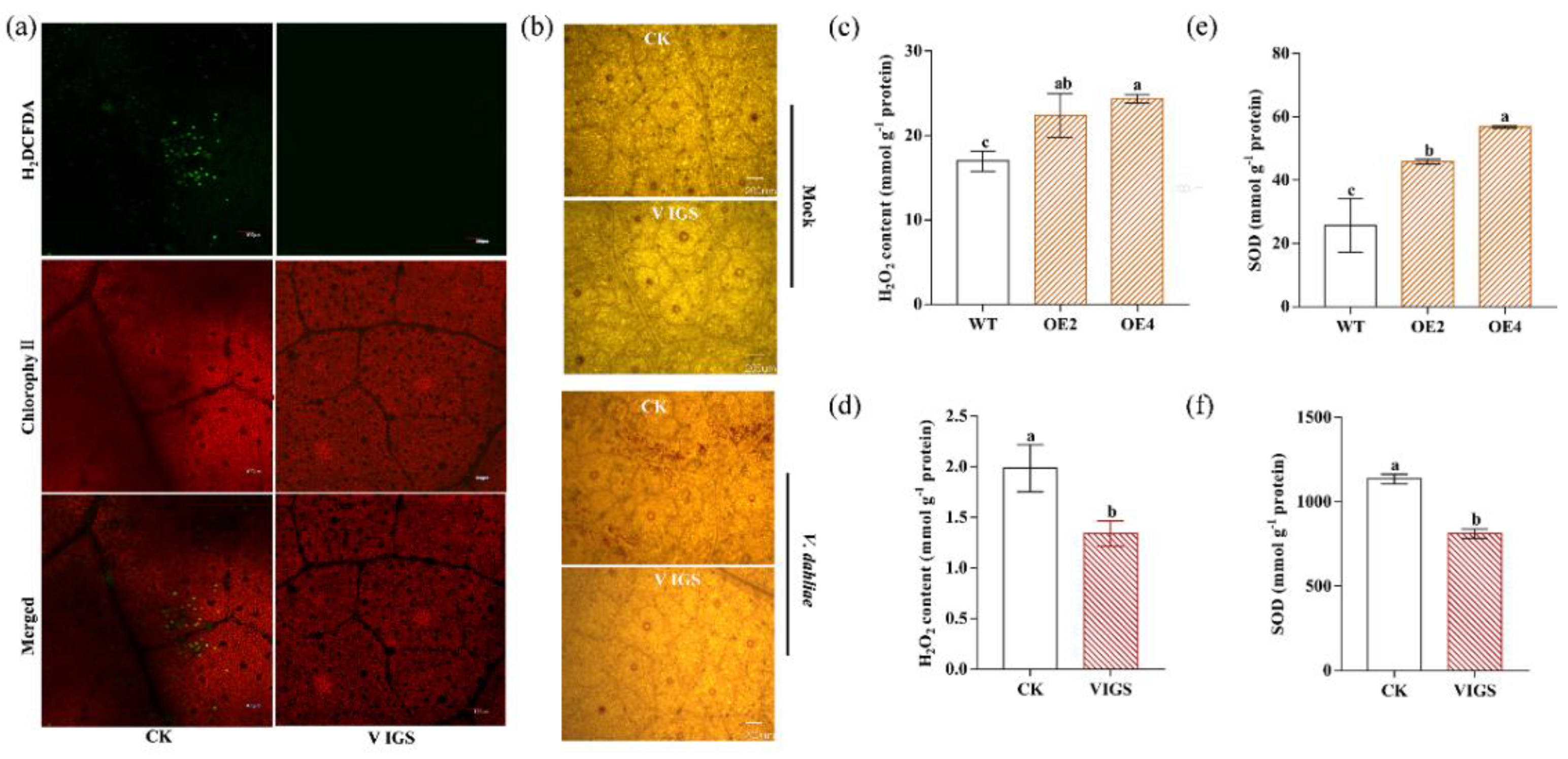
2.7. GhENODL6 Contributed to the Production of ROS
3. Discussion
4. Methods
4.1. Plant Growth and V. dahliae Culture
4.2. Identification of GhPCs and Bioinformatics Analysis
4.3. Multiple Sequence Alignment of PCNLs and Phylogenetic Analysis
4.4. Expression Analysis of GhPC Genes
4.5. Gene Cloning and Bioinformatics Analysis
4.6. Real-Time PCR and Expression Analysis
4.7. Subcellular Location
4.8. Generation and Analysis of Transgenic Arabidopsis
4.9. VIGS Experiment in Cotton
4.10. ROS Dyeing
4.11. Determination of H2O2 and SOD Content
4.12. Determination of SA Content
4.13. Transcriptome Sequencing
4.14. Primers for Gene Cloning, Vector Construction and Expression Analysis
4.15. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mo, H.; Wang, X.; Zhang, Y.; Zhang, G.; Zhang, J.; Ma, Z. Cotton polyamine oxidase is required for spermine and camalexin signalling in the defence response to Verticillium dahliae. Plant J. 2015, 83, 962–975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaban, M.; Miao, Y.; Ullah, A.; Khan, A.Q.; Menghwar, H.; Khan, A.H.; Ahmed, M.M.; Tabassum, M.A.; Zhu, L. Physiological and molecular mechanism of defense in cotton against Verticillium dahliae. Plant Physiol. Biochem. 2018, 125, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Sanogo, S.; Flynn, R.; Baral, J.B.; Bajaj, S.; Hughs, S.E.; Percy, R.G. Germplasm evaluation and transfer of Verticillium wilt resistance from Pima (Gossypium barbadense) to Upland cotton (G. hirsutum). Euphytica 2011, 187, 147–160. [Google Scholar] [CrossRef]
- Tong, S.; Yuan, M.; Liu, Y.; Li, X.; Jin, D.; Cheng, X.; Lin, D.; Ling, H.; Yang, D.; Wang, Y. Ergosterol-targeting fusion antifungal peptide significantly increases the Verticillium wilt resistance of cotton. Plant Biotechnol. J. 2020, 19, 926–936. [Google Scholar] [CrossRef]
- Ma, Z.; Zhang, Y.; Wu, L.; Zhang, G.; Sun, Z.; Li, Z.; Jiang, Y.; Ke, H.; Chen, B.; Liu, Z. High-quality genome assembly and resequencing of modern cotton cultivars provide resources for crop improvement. Nat. Genet. 2021, 53, 1385–1391. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, G.; Wu, L.; Wang, X.; Zhang, Y.; Wu, J.; Ma, Z. New cotton variety Nong da 601 with disease resistance: Insect resisitance and high yield. Seed 2015, 34, 3. [Google Scholar]
- Andersen, E.J.; Ali, S.; Byamukama, E.; Yen, Y.; Nepal, M.P. Disease resistance mechanisms in plants. Genes 2018, 9, 339. [Google Scholar] [CrossRef] [Green Version]
- Bari, R.; Jones, J.D. Role of plant hormones in plant defence responses. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef]
- Vlot, A.C.; Klessig, D.F.; Park, S.W. Systemic acquired resistance: The elusive signal(s). Curr. Opin. Plant Biol. 2008, 11, 436–442. [Google Scholar] [CrossRef] [Green Version]
- Cheng, H.Q.; Han, L.B.; Yang, C.L.; Wu, X.M.; Zhong, N.Q.; Wu, J.H.; Wang, F.X.; Wang, H.Y.; Xia, G.X. The cotton MYB108 forms a positive feedback regulation loop with CML11 and participates in the defense response against Verticillium dahliae infection. J. Exp. Bot. 2016, 67, 1935–1950. [Google Scholar] [CrossRef] [Green Version]
- Kouzai, Y.; Kimura, M.; Watanabe, M.; Kusunoki, K.; Osaka, D.; Suzuki, T.; Matsui, H.; Yamamoto, M.; Ichinose, Y.; Toyoda, K. Salicylic acid-dependent immunity contributes to resistance against Rhizoctonia solani, a necrotrophic fungal agent of sheath blight, in rice and Brachypodium distachyon. New Phytol. 2018, 217, 771–783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinay, K.; Tushar, K.; Mansi, S.; Shabir, H.W. ROS-induced signaling and gene expression in crops under salinity stress. In Reactive Oxygen Species and Antioxidant Systems in Plants. Role and Regulation under Abiotic Stress; Springer: Singapore, 2017; pp. 159–184. [Google Scholar]
- Camejo, D.; Guzman-Cedeno, A.; Moreno, A. Reactive oxygen species, essential molecules, during plant-pathogen interactions. Plant Physiol. Biochem. 2016, 103, 10–23. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, H. Programmed cell death of tracheary elements as a paradigm in plants. Plant Mol. Biol. Rep. 2000, 44, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Han, L.B.; Li, Y.B.; Wang, F.X.; Wang, W.Y.; Liu, J.; Wu, J.H.; Zhong, N.Q.; Wu, S.J.; Jiao, G.L.; Wang, H.Y. The cotton apoplastic protein CRR1 stabilizes chitinase 28 to facilitate defense against the fungal pathogen Verticillium dahliae. Plant Cell 2019, 31, 520–536. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, L.; Wang, X.; Chen, B.; Zhao, J.; Cui, J.; Li, Z.; Yang, J.; Wu, L.; Wu, J. The cotton laccase gene GhLAC15 enhances Verticillium wilt resistance via an increase in defence-induced lignification and lignin components in the cell walls of plants. Mol. Plant Pathol. 2019, 20, 309–322. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Xu, J.; Li, L.; Guo, Z.; Si, Q.; Zhu, G.; Wang, X.; Guo, W. GbCYP86A1-1 from Gossypium barbadense positively regulates defence against Verticillium dahliae by cell wall modification and activation of immune pathways. Plant Biotechnol. J. 2020, 18, 222–238. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Yu, X.; Zhang, C.; Zhang, Q.; Sun, Y.; Zhu, H.; Tang, C. Pectin lyase enhances cotton resistance to Verticillium wilt by inducing cell apoptosis of Verticillium dahliae. J. Hazard. Mater. 2021, 404, 124029. [Google Scholar] [CrossRef]
- Jun, Z.; Zhang, Z.; Gao, Y.; Zhou, L.; Fang, L.; Chen, X.; Ning, Z.; Chen, T.; Guo, W.; Zhang, T. Overexpression of GbRLK, a putative receptor-like kinase gene, improved cotton tolerance to Verticillium wilt. Sci. Rep. 2015, 5, 15048. [Google Scholar] [CrossRef] [Green Version]
- Gilroy, S.; Suzuki, N.; Miller, G. A tidal wave of signals: Calcium and ROS at the forefront of rapid systemic signaling. Trends Plant Sci. 2014, 19, 623–630. [Google Scholar] [CrossRef]
- Levine, A.; Tenhaken, R.; Dixon, R.; Lamb, C. H2O2 from the oxidative burst orchestrates the plant hypersensitive diseases resistance response. Cell 1994, 79, 583. [Google Scholar] [CrossRef]
- Yalpani, N.; Silverman, P.; Wilson, T. Salicylic acid is a systemic signal and an inducer of pathogenesis-related proteins in virus-infected tobacco. Plant Cell 1991, 3, 809–818. [Google Scholar] [PubMed] [Green Version]
- Leon, J. Hydrogen peroxide stimulates salicylic acid biosynthesis in tobacco. Plant Physiol. 1995, 108, 1673–1678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Wang, X.F.; Ding, Z.G.; Ma, Q.; Zhang, G.R.; Zhang, S.L.; Li, Z.K.; Wu, L.Q.; Zhang, G.Y.; Ma, Z.Y. Transcriptome profiling of Gossypium barbadense inoculated with Verticillium dahliae provides a resource for cotton improvement. BMC Genom. 2013, 14, 637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Gao, G.; Zhang, T.; Wu, X. The putative phytocyanin genes in Chinese cabbage (Brassica rapa L.) genome-wide identification, classification and expression analysis. Mol. Genet. Genom. 2013, 288, 1–20. [Google Scholar] [CrossRef]
- Yoshizaki, M.; Furumoto, T.; Hata, S.; Shinozaki, M.; Izui, K. Characterization of a novel gene encoding a phytocyanin-related protein in morning glory (Pharbitis nil). Biochem. Biophys. Res. Commun. 2000, 268, 466–470. [Google Scholar] [CrossRef]
- Cao, J.; Li, X.; Lv, Y.; Ding, L. Comparative analysis of the phytocyanin gene family in 10 plant species. a focus on Zea mays. Front. Plant Sci. 2015, 6, 515. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Wang, X.J.; Wang, T.; Li, L.B. Genome-wide identification.; classification.; and expression analysis of the phytocyanin gene family in Phalaenopsis equestris. Biol. Plant. 2017, 61, 445–452. [Google Scholar] [CrossRef]
- Ma, H.; Zhao, H.; Liu, Z.; Zhao, J. The phytocyanin gene family in rice (Oryza sativa L.): Genome-wide identification.; classification and transcriptional analysis. PLoS ONE 2011, 6, e25184. [Google Scholar] [CrossRef]
- Xu, Q.; Dunbrack, R.L., Jr. Assignment of protein sequences to existing domain and family classification systems: Pfam and the PDB. Bioinformatics 2012, 28, 2763–2772. [Google Scholar] [CrossRef]
- Mashiguchi, K.; Asami, T.; Suzuki, Y. Genome-wide identification.; structure and expression studies, and mutant collection of 22 early nodulin-like protein genes in Arabidopsis. Biosci. Biotech. Biochem. 2009, 3, 2452–2459. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Wang, X.; Zhang, Y.; Yang, J.; Li, Z.; Wu, L.; Wu, J.; Wu, N.; Liu, L.; Liu, Z.; et al. Dynamic characteristics and functional analysis provide new insights into long non-coding RNA responsive to Verticillium dahliae infection in Gossypium hirsutum. BMC Plant Biol. 2021, 21, 68. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; He, S.; Wang, X.; Sun, J.; Zhang, Y.; Zhang, G.; Wu, L.; Li, Z.; Liu, Z.; Sun, G. Resequencing a core collection of upland cotton identifies genomic variation and loci influencing fiber quality and yield. Nat. Genet. 2018, 50, 803–813. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Zhang, Y.; Yang, J.; Zhang, M.; Ma, Z. The G-protein α subunit GhGPA positively regulates Gossypium hirsutum resistance to Verticillium dahliae via induction of SA and JA signaling pathways and ROS accumulation. Crop J. 2020, 9, 11. [Google Scholar] [CrossRef]
- Mo, S.; Zhang, Y.; Wang, X.; Yang, J.; Sun, Z.; Zhang, D.; Chen, B.; Wang, G.; Ke, H.; Liu, Z. Cotton GhSSI2 isoforms from the stearoyl acyl carrier protein fatty acid desaturase family regulate Verticillium wilt resistance. Mol. Plant Pathol. 2021, 22, 1041–1056. [Google Scholar] [CrossRef]
- Khan, J.A.; Wang, Q.; Sjolund, R.D.; Schulz, A.; Thompson, G.A. An early nodulin-like protein accumulates in the sieve element plasma membrane of Arabidopsis. Plant Physiol. 2007, 143, 1576–1589. [Google Scholar] [CrossRef] [Green Version]
- Serova, T.A.; Tsyganova, A.V.; Tsyganov, V.E. Early nodule senescence is activated in symbiotic mutants of pea (Pisum sativum L.) forming ineffective nodules blocked at different nodule developmental stages. Protoplasma 2018, 255, 1443–1459. [Google Scholar] [CrossRef]
- Hou, Y.; Guo, X.; Cyprys, P.; Zhang, Y.; Bleckmann, A.; Cai, L.; Huang, Q.; Luo, Y.; Gu, H.; Dresselhaus, T. Maternal ENODLs are required for pollen tube reception in Arabidopsis. Curr. Biol. 2016, 26, 2343–2350. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Shen, Y.; Hu, Y.; Tan, S.; Lin, Z. A phytocyanin-related early nodulin-like gene, BcBCP1, cloned from Boea crassifolia enhances osmotic tolerance in transgenic tobacco. Plant Physiol. 2011, 168, 935–943. [Google Scholar] [CrossRef]
- Fu, Z.Q.; Yan, S.; Saleh, A.; Wang, W.; Ruble, J.; Oka, N.; Mohan, R.; Spoel, S.H.; Tada, Y.; Zheng, N. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 2012, 486, 228–232. [Google Scholar] [CrossRef] [Green Version]
- Cui, H.; Gobbato, E.; Kracher, B.; Qiu, J.; Bautor, J.; Parker, J.E. A core function of EDS1 with PAD4 is to protect the salicylic acid defense sector in Arabidopsis immunity. New Phytol. 2016, 213, 1802–1817. [Google Scholar] [CrossRef] [Green Version]
- Weigel, R.R.; Pfitzner, U.M.; Gatz, C. Interaction of NIMIN1 with NPR1 modulates PR gene expression in Arabidopsis. Plant Cell 2005, 17, 1279–1291. [Google Scholar] [CrossRef] [Green Version]
- Rietz, S.; Stamm, A.; Malonek, S. Different roles of enhanced disease susceptibility1 (EDS1) bound to and dissociated from phytoalexin deficient4 (PAD4) in Arabidopsis immunity. New Phytol. 2011, 191, 107–119. [Google Scholar] [CrossRef]
- Ding, Y.; Sun, T.; Ao, K.; Peng, Y.; Zhang, Y.; Li, X.; Zhang, Y. Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell 2018, 173, 1454–1467. [Google Scholar] [CrossRef] [PubMed]
- Sachdev, S.; Ansari, S.A.; Ansari, M.I.; Fujita, M.; Hasanuzzaman, M. Abiotic stress and reactive oxygen species. generation, signaling, and defense mechanisms. Antioxidants 2021, 10, 277. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Cheng, Z.; Ahmad, H.; Hayat, S. Reactive oxygen species (ROS) as defenses against a broad range of plant fungal infections and case study on ROS employed by crops against Verticillium dahliae wilts. J. Plant Interact. 2018, 13, 353–363. [Google Scholar] [CrossRef] [Green Version]
- Wirthmueller, L.; Zhang, Y.; Jones, J.D.; Parker, J.E. Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr. Biol. 2007, 17, 2023–2029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Li, Y.; Hu, Y.; Yang, Y.; Zhang, W.; He, M.; Li, X.; Zhang, C.; Kong, F.; Liu, X. EDS1-interacting J protein 1 is an essential negative regulator of plant innate immunity in Arabidopsis. Plant Cell 2021, 33, 153–171. [Google Scholar] [CrossRef]
- Yang, J.; Ke, H.F.; Zhang, Y.; Ji, L.L.; Huang, L.Z.; Zhang, C.Y.; Wang, X.F.; Ma, Z.Y. Genome-wide identification of cyclophilin genes in Gossypium hirsutum and functional characterization of a CYP with antifungal activity against Verticillium dahliae. BMC Plant Biol. 2019, 19, 272. [Google Scholar] [CrossRef]
- Yang, J.; Ji, L.; Wang, X.; Zhang, Y.; Wu, L.; Yang, Y.; Ma, Z. Overexpression of 3-deoxy-7-phosphoheptulonate synthase gene from Gossypium hirsutum enhances Arabidopsis resistance to Verticillium wilt. Plant Cell Rep. 2015, 34, 1429–1441. [Google Scholar] [CrossRef]
- Bendtsen, J.D.; Nielsen, H.; Hei, G.; Brunak, S.S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Cell Biol. 2004, 340, 783–795. [Google Scholar]
- Eisenhaber, B.; Wildpaner, M.; Schultz, C.J.; Borner, G.; Eisenhaber, D.F. Glycosylphosphatidylinositol lipid anchoring of plant proteins: Sensitive prediction from sequence-and genome-wide studies for Arabidopsis and rice. Plant Physiol. 2003, 133, 1691–1701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, R.; Brunak, S. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac. Symp. Biocomput. 2002, 7, 310–322. [Google Scholar]
- Schultz, C.J.; Rumsewicz, M.P.; Johnson, K.L.; Jones, B.J.; Gaspar, Y.M.; Bacic, A. Using genomic resources to guide research directions: The arabinogalactan protein gene family as a test case. Plant Physiol. 2002, 129, 1448–1463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larkin, M. Clustal W and Clustal X v. 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Wang, X.; Sun, Z.; Zhang, Y.; Meng, C.; Chen, B.; Wang, G.; Ke, H.; Wu, J.; Yan, Y. Evolution, expression and functional analysis of cultivated allotetraploid cotton DIR genes. BMC Plant Biol. 2021, 21, 89. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Ma, Q.; Zhang, Y.; Wang, X.; Zhang, G.; Ma, Z. Molecular cloning and functional analysis of GbRvd, a gene in Gossypium barbadense that plays an important role in conferring resistance to Verticillium wilt. Gene 2016, 575, 687–694. [Google Scholar] [CrossRef]
- Yang, Y.; Ling, X.; Chen, T.; Cai, L.; Liu, T.; Wang, J.; Fan, X.; Ren, Y.; Yuan, H.; Zhu, W. A cotton Gbvdr5 gene encoding a leucine-rich-repeat receptor-like protein confers resistance to Verticillium dahliae in transgenic Arabidopsis and upland cotton. Plant Mol. Biol. Rep. 2014, 33, 987–1001. [Google Scholar] [CrossRef]
- Smedley, M.A.; Harwood, W.A. Gateway(R)-compatible plant transformation vectors. Methods Mol. Biol. 2015, 1, 3–16. [Google Scholar]
- Sree, V.S.; Manoharan, M.; Ranjit, K.T.; Savtthri, H.S.; Lakshmi, S.G. Agrobacterium-mediated transformation of tomato (Lycopersicon esculentum var. Pusa Ruby) with coat-protein gene of Physalis Mottle Tymovirus. J. Plant Physiol. 2000, 156, 106–110. [Google Scholar] [CrossRef]
- Sparkes, I.A.; Runions, J.; Kearns, A.; Hawes, C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006, 1, 2019–2025. [Google Scholar] [CrossRef]
- Bent, S.J.; Ca, A.F. Floral dip a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar]
- Zhang, X.; Henriques, R.; Lin, S.S.; Niu, Q.W.; Chua, N.H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; He, X.; Luo, X.; Xu, L.; Liu, L.; Min, L.; Jin, L.; Zhu, L.; Zhang, X. Cotton WRKY1 mediates the plant defense-to-development transition during infection of cotton by Verticillium dahliae by activating JASMONATE ZIM-DOMAIN1 expression. Plant Physiol. 2014, 166, 2179–2194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.; Yang, Y.; Chen, T.; Yu, W.; Liu, T.; Li, H.; Fan, X.; Ren, Y.; Shen, D.; Liu, L. Island cotton Gbve1 gene encoding a receptor-like protein confers resistance to both defoliating and non-defoliating isolates of Verticillium dahliae. PLoS ONE 2012, 7, e51091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghorbel, M.; Zribi, I.; Missaoui, K.; Drira-Fakhfekh, M.; Azzouzi, B.; Brini, F. Differential regulation of the durum wheat Pathogenesis-related protein (PR1) by Calmodulin TdCaM1.3 protein. Mol. Biol. Rep. 2021, 48, 347–362. [Google Scholar] [CrossRef]
- Reyes, M.I.; Flores-Vergara, M.A.; Guerra-Peraza, O.; Rajabu, C.; Desai, J.; Hiromoto-Ruiz, Y.H.; Ndunguru, J.; Hanley-Bowdoin, L.; Kjemtrup, S.; Ascencio-Ibanez, J.T. A VIGS screen identifies immunity in the Arabidopsis Pla-1 accession to viruses in two different genera of the Geminiviridae. Plant J. 2017, 92, 796–807. [Google Scholar] [CrossRef] [Green Version]
- Ren, F.; Zhang, Z.; Fan, X. Construction of a plant expression vector of the Grapevine virus B coat protein gene and transformation into Nicotiana tabacum. Plant Protect. 2018, 44, 11–16. [Google Scholar]
- Gao, X.; Wheeler, T.; Li, Z.; Kenerley, C.M.; He, P.; Shan, L. Silencing GhNDR1 and GhMKK2 compromises cotton resistance to Verticillium wilt. Plant J. 2011, 66, 293–305. [Google Scholar] [CrossRef] [Green Version]
- Athira, S.S.; Biby, E.T.; Mohanan, P.V. Dextran stabilized fullerene soot induced toxicity on alveolar epithelial cells (A549 cells). Environ. Res. 2020, 188, 109716. [Google Scholar] [CrossRef]
- Chai, Q.; Shang, X.; Wu, S.; Zhu, G.; Cheng, C.; Cai, C.; Wang, X.; Guo, W. 5-Aminolevulinic acid dehydratase gene dosage affects programmed cell death and immunity. Plant Physiol. 2017, 175, 511–528. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Liu, N.; Sun, Y.; Wang, P.; Hou, Y. The cotton GhWIN2 gene activates the cuticle biosynthesis pathway and influences the salicylic and jasmonic acid biosynthesis pathways. BMC Plant Biol. 2019, 19, 379. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Zhou, X.; Ling, Y.; Zhang, Z.; Su, Z. agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010, 38, W64–W70. [Google Scholar] [CrossRef] [PubMed] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, M.; Wang, X.; Yang, J.; Wang, Z.; Chen, B.; Zhang, X.; Zhang, D.; Sun, Z.; Wu, J.; Ke, H.; et al. GhENODL6 Isoforms from the Phytocyanin Gene Family Regulated Verticillium Wilt Resistance in Cotton. Int. J. Mol. Sci. 2022, 23, 2913. https://doi.org/10.3390/ijms23062913
Zhang M, Wang X, Yang J, Wang Z, Chen B, Zhang X, Zhang D, Sun Z, Wu J, Ke H, et al. GhENODL6 Isoforms from the Phytocyanin Gene Family Regulated Verticillium Wilt Resistance in Cotton. International Journal of Molecular Sciences. 2022; 23(6):2913. https://doi.org/10.3390/ijms23062913
Chicago/Turabian StyleZhang, Man, Xingfen Wang, Jun Yang, Zhicheng Wang, Bin Chen, Xinyu Zhang, Dongmei Zhang, Zhengwen Sun, Jinhua Wu, Huifeng Ke, and et al. 2022. "GhENODL6 Isoforms from the Phytocyanin Gene Family Regulated Verticillium Wilt Resistance in Cotton" International Journal of Molecular Sciences 23, no. 6: 2913. https://doi.org/10.3390/ijms23062913
APA StyleZhang, M., Wang, X., Yang, J., Wang, Z., Chen, B., Zhang, X., Zhang, D., Sun, Z., Wu, J., Ke, H., Wu, L., Zhang, G., Zhang, Y., & Ma, Z. (2022). GhENODL6 Isoforms from the Phytocyanin Gene Family Regulated Verticillium Wilt Resistance in Cotton. International Journal of Molecular Sciences, 23(6), 2913. https://doi.org/10.3390/ijms23062913





