Post-Transcriptional Regulation of Molecular Determinants during Cardiogenesis
Abstract
1. Growth Factor Signalling in Cardiac Morphogenesis
2. Transcriptional Control of Cardiac Morphogenesis
3. The Emergence of a Novel Layer of Gene Regulation: Post-Transcriptional Regulation by Non-Coding RNAs
4. Biogenesis and Function of microRNAs and lncRNAs
5. Post-Transcriptional Control of Precardiac Mesoderm Formation by ncRNAs
6. Post-Transcriptional Control of Heart Fields Deployment by ncRNAs
7. Post-Transcriptional Control of Sidedness and Cardiac Looping by ncRNAs
8. Post-Transcriptional Control of Proepicardium/Epicardium Formation by ncRNAs
9. Post-Transcriptional Control of Conduction System Development by ncRNAs
10. Post-Transcriptional Control of Chamber Morphogenesis and Valve Development by ncRNAs
11. Post-Transcriptional Control of Outflow Tract and Atrioventricular Septation by ncRNAs
12. Post-Transcriptional Control of Aortic Arch Development by ncRNAs
13. Conclusions and Future Perspectives
Funding
Conflicts of Interest
References
- Van Wijk, B.; Moorman, A.F.M.; van den Hoff, M.J.B. Role of bone morphogenetic proteins in cardiac differentiation. Cardiovasc. Res. 2007, 74, 244–255. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.N.; Green, J.; Wang, Z.; Deng, Y.; Qiao, M.; Peabody, M.; Zhang, Q.; Ye, J.; Yan, Z.; Denduluri, S.; et al. Bone Morphogenetic Protein (BMP) signaling in development and human diseases. Genes Dis. 2014, 1, 87–105. [Google Scholar] [CrossRef] [PubMed]
- Khosravi, F.; Ahmadvand, N.; Bellusci, S.; Sauer, H. The Multifunctional Contribution of FGF Signaling to Cardiac Development, Homeostasis, Disease and Repair. Front. Cell Dev. Biol. 2021, 9, 672935. [Google Scholar] [CrossRef] [PubMed]
- Kruithof, B.P.; van Wijk, B.; Somi, S.; Julio, M.K.-D.; Pomares, J.M.P.; Weesie, F.; Wessels, A.; Moorman, A.F.; Hoff, M.J.V.D. BMP and FGF regulate the differentiation of multipotential pericardial mesoderm into the myocardial or epicardial lineage. Dev. Biol. 2006, 295, 507–522. [Google Scholar] [CrossRef] [PubMed]
- Van Wijk, B.; van den Berg, G.; Abu-Issa, R.; Barnett, P.; van der Velden, S.; Schmidt, M.; Ruijter, J.M.; Kirby, M.L.; Moorman, A.F.; Hoff, M.J.V.D. Epicardium and Myocardium Separate From a Common Precursor Pool by Crosstalk Between Bone Morphogenetic Protein– and Fibroblast Growth Factor–Signaling Pathways. Circ. Res. 2009, 105, 431–441. [Google Scholar] [CrossRef]
- Torlopp, A.; Schlueter, J.; Brand, T. Role of fibroblast growth factor signaling during proepicardium formation in the chick embryo. Dev. Dyn. 2010, 239, 2393–2403. [Google Scholar] [CrossRef]
- Ishii, Y.; Garriock, R.J.; Navetta, A.M.; Coughlin, L.E.; Mikawa, T. BMP Signals Promote Proepicardial Protrusion Necessary for Recruitment of Coronary Vessel and Epicardial Progenitors to the Heart. Dev. Cell 2010, 19, 307–316. [Google Scholar] [CrossRef]
- Andrés-Delgado, L.; Ernst, A.; Galardi-Castilla, M.; Bazaga, D.; Peralta, M.; Münch, J.; González-Rosa, J.M.; Marques, I.; Tessadori, F.; de la Pompa, J.L.; et al. Actin dynamics and the Bmp pathway drive apical extrusion of proepicardial cells. Development 2019, 146, dev174961. [Google Scholar] [CrossRef]
- Andrés-Delgado, L.; Galardi-Castilla, M.; Münch, J.; Peralta, M.; Ernst, A.; González-Rosa, J.M.; Tessadori, F.; Santamaría, L.; Bakkers, J.; Vermot, J.; et al. Notch and Bmp signaling pathways act coordinately during the formation of the proepicardium. Dev. Dyn. 2020, 249, 1455–1469. [Google Scholar] [CrossRef]
- Yamagishi, T.; Ando, K.; Nakamura, H. Roles of TGFβ and BMP during valvulo–septal endocardial cushion formation. Anat. Sci. Int. 2009, 84, 77–87. [Google Scholar] [CrossRef]
- Kruithof, B.P.; Duim, S.N.; Moerkamp, A.T.; Goumans, M.-J. TGFβ and BMP signaling in cardiac cushion formation: Lessons from mice and chicken. Differentiation 2012, 84, 89–102. [Google Scholar] [CrossRef]
- Saxon, J.G.; Baer, D.R.; Barton, J.A.; Hawkins, T.; Wu, B.; Trusk, T.C.; Harris, S.E.; Zhou, B.; Mishina, Y.; Sugi, Y. BMP2 expression in the endocardial lineage is required for AV endocardial cushion maturation and remodeling. Dev. Biol. 2017, 430, 113–128. [Google Scholar] [CrossRef]
- Inai, K.; Burnside, J.L.; Hoffman, S.; Toole, B.P.; Sugi, Y. BMP-2 Induces Versican and Hyaluronan That Contribute to Post-EMT AV Cushion Cell Migration. PLoS ONE 2013, 8, e77593. [Google Scholar] [CrossRef]
- Zhang, J.; Chang, J.Y.-F.; Huang, Y.; Lin, X.; Luo, Y.; Schwartz, R.J.; Martin, J.F.; Wang, F. The FGF-BMP Signaling Axis Regulates Outflow Tract Valve Primordium Formation by Promoting Cushion Neural Crest Cell Differentiation. Circ. Res. 2010, 107, 1209–1219. [Google Scholar] [CrossRef]
- Sugi, Y.; Ito, N.; Szebenyi, G.; Myers, K.; Fallon, J.F.; Mikawa, T.; Markwald, R.R. Fibroblast growth factor (FGF)-4 can induce proliferation of cardiac cushion mesenchymal cells during early valve leaflet formation. Dev. Biol. 2003, 258, 252–263. [Google Scholar] [CrossRef]
- Nakajima, Y.; Yamagishi, T.; Hokari, S.; Nakamura, H. Mechanisms involved in valvuloseptal endocardial cushion formation in early cardiogenesis: Roles of transforming growth factor (TGF)-beta and bone morphogenetic protein (BMP). Anat. Rec. 2000, 258, 119–127. [Google Scholar] [CrossRef]
- Todorovic, V.; Frendewey, D.; Gutstein, D.E.; Chen, Y.; Freyer, L.; Finnegan, E.; Liu, F.; Murphy, A.; Valenzuela, D.; Yancopoulos, G.; et al. Long form of latent TGF-β binding protein 1 (Ltbp1L) is essential for cardiac outflow tract septation and remodeling. Development 2007, 134, 3723–3732. [Google Scholar] [CrossRef]
- Zhang, R.; Cao, P.; Yang, Z.; Wang, Z.; Wu, J.-L.; Chen, Y.; Pan, Y. Heparan Sulfate Biosynthesis Enzyme, Ext1, Contributes to Outflow Tract Development of Mouse Heart via Modulation of FGF Signaling. PLoS ONE 2015, 10, e0136518. [Google Scholar] [CrossRef]
- Beppu, H.; Malhotra, R.; Beppu, Y.; Lepore, J.J.; Parmacek, M.S.; Bloch, K.D. BMP type II receptor regulates positioning of outflow tract and remodeling of atrioventricular cushion during cardiogenesis. Dev. Biol. 2009, 331, 167–175. [Google Scholar] [CrossRef]
- Wang, J.; Liu, S.; Heallen, T.; Martin, J.F. The Hippo pathway in the heart: Pivotal roles in development, disease, and regeneration. Nat. Rev. Cardiol. 2018, 15, 672–684. [Google Scholar] [CrossRef]
- Heallen, T.; Zhang, M.; Wang, J.; Bonilla-Claudio, M.; Klysik, E.; Johnson, R.L.; Martin, J.F. Hippo Pathway Inhibits Wnt Signaling to Restrain Cardiomyocyte Proliferation and Heart Size. Science 2011, 332, 458–461. [Google Scholar] [CrossRef]
- Zhou, Q.; Li, L.; Zhao, B.; Guan, K.-L. The Hippo Pathway in Heart Development, Regeneration, and Diseases. Circ. Res. 2015, 116, 1431–1447. [Google Scholar] [CrossRef]
- D’Amato, G.; Luxán, G.; Mínguez, J.L.D.L.P. Notch signalling in ventricular chamber development & cardiomyopathy. FEBS J. 2016, 283, 4223–4237. [Google Scholar] [CrossRef]
- D’Amato, G.; Luxán, G.; Nieto, G.D.M.; Poveda, B.M.; Torroja, C.; Walter, W.; Bochter, M.S.; Benedito, R.; Cole, S.E.; Martinez, F.; et al. Sequential Notch activation regulates ventricular chamber development. Nat. Cell Biol. 2015, 18, 7–20. [Google Scholar] [CrossRef]
- Samsa, L.; Givens, C.; Tzima, E.; Stainier, D.; Qian, L.; Liu, J. Cardiac contraction activates endocardial Notch signaling to modulate chamber maturation in zebrafish. Development 2015, 142, 4080–4091. [Google Scholar] [CrossRef]
- Yang, Y.; Li, B.; Zhang, X.; Zhao, Q.; Lou, X. The zinc finger protein Zfpm1 modulates ventricular trabeculation through Neuregulin-ErbB signalling. Dev. Biol. 2019, 446, 142–150. [Google Scholar] [CrossRef]
- Zhao, Y.-Y.; Sawyer, D.R.; Baliga, R.R.; Opel, D.J.; Han, X.; Marchionni, M.A.; Kelly, R.A. Neuregulins Promote Survival and Growth of Cardiac Myocytes. J. Biol. Chem. 1998, 273, 10261–10269. [Google Scholar] [CrossRef]
- Hertig, C.M.; Kubalak, S.W.; Wang, Y.; Chien, K.R. Synergistic Roles of Neuregulin-1 and Insulin-like Growth Factor-I in Activation of the Phosphatidylinositol 3-Kinase Pathway and Cardiac Chamber Morphogenesis. J. Biol. Chem. 1999, 274, 37362–37369. [Google Scholar] [CrossRef] [PubMed]
- Lai, D.; Liu, X.; Forrai, A.; Wolstein, O.; Michalicek, J.; Ahmed, I.; Garratt, A.N.; Birchmeier, C.; Zhou, M.; Hartley, L.; et al. Neuregulin 1 Sustains the Gene Regulatory Network in Both Trabecular and Nontrabecular Myocardium. Circ. Res. 2010, 107, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Saga, Y.; Miyagawa-Tomita, S.; Takagi, A.; Kitajima, S.; Miyazaki, J.-I.; Inoue, T. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development 1999, 126, 3437–3447. [Google Scholar] [CrossRef]
- Saga, Y. Mesp1 Expression Is the Earliest Sign of Cardiovascular Development. Trends Cardiovasc. Med. 2000, 10, 345–352. [Google Scholar] [CrossRef]
- Kitajima, S.; Takagi, A.; Inoue, T.; Saga, Y. MesP1 and MesP2 are essential for the development of cardiac mesoderm. Development 2000, 127, 3215–3226. [Google Scholar] [CrossRef] [PubMed]
- Laverriere, A.C.; MacNeill, C.; Mueller, C.; Poelmann, R.E.; Burch, J.B.; Evans, T. GATA-4/5/6, a subfamily of three transcription factors transcribed in developing heart and gut. J. Biol. Chem. 1994, 269, 23177–23184. [Google Scholar] [CrossRef]
- Charron, F.; Nemer, M. GATA transcription factors and cardiac development. Semin. Cell Dev. Biol. 1999, 10, 85–91. [Google Scholar] [CrossRef]
- Belaguli, N.S.; Sepulveda, J.L.; Nigam, V.; Charron, F.; Nemer, M.; Schwartz, R.J. Cardiac Tissue Enriched Factors Serum Response Factor and GATA-4 Are Mutual Coregulators. Mol. Cell. Biol. 2000, 20, 7550–7558. [Google Scholar] [CrossRef]
- Durocher, D.; Charron, F.; Warren, R.; Schwartz, R.J.; Nemer, M. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO J. 1997, 16, 5687–5696. [Google Scholar] [CrossRef]
- Jiang, Y.; Evans, T. TheXenopusGATA-4/5/6 Genes Are Associated with Cardiac Specification and Can Regulate Cardiac-Specific Transcription during Embryogenesis. Dev. Biol. 1996, 174, 258–270. [Google Scholar] [CrossRef][Green Version]
- Harvey, R. NK-2Homeobox Genes and Heart Development. Dev. Biol. 1996, 178, 203–216. [Google Scholar] [CrossRef]
- Biben, C.; Harvey, R.P. Homeodomain factor Nkx2-5 controls left/right asymmetric expression of bHLH gene eHand during murine heart development. Genes Dev. 1997, 11, 1357–1369. [Google Scholar] [CrossRef]
- Lin, Q.; Schwarz, J.; Bucana, C.; Olson, E.N. Control of Mouse Cardiac Morphogenesis and Myogenesis by Transcription Factor MEF2C. Science 1997, 276, 1404–1407. [Google Scholar] [CrossRef]
- Croissant, J.D.; Kim, J.-H.; Eichele, G.; Goering, L.; Lough, J.; Prywes, R.; Schwartz, R.J. Avian Serum Response Factor Expression Restricted Primarily to Muscle Cell Lineages Is Required for α-Actin Gene Transcription. Dev. Biol. 1996, 177, 250–264. [Google Scholar] [CrossRef]
- Kim, S.; Ip, H.S.; Lu, M.M.; Clendenin, C.; Parmacek, M.S. A serum response factor-dependent transcriptional regulatory program identifies distinct smooth muscle cell sublineages. Mol. Cell. Biol. 1997, 17, 2266–2278. [Google Scholar] [CrossRef]
- Ocaña, O.H.; Coskun, H.; Minguillón, C.; Murawala, P.; Tanaka, E.M.; Galceran, J.; Muñoz-Chápuli, R.; Nieto, M.A. A right-handed signalling pathway drives heart looping in vertebrates. Nature 2017, 549, 86–90. [Google Scholar] [CrossRef]
- Ammirabile, G.; Tessari, A.; Pignataro, V.; Szumska, D.; Sardo, F.S.; Benes, J.; Balistreri, M.; Bhattacharya, S.; Sedmera, D.; Campione, M. Pitx2 confers left morphological, molecular, and functional identity to the sinus venosus myocardium. Cardiovasc. Res. 2011, 93, 291–301. [Google Scholar] [CrossRef]
- Campione, M.; Ros, M.; Icardo, J.M.; Piedra, E.; Christoffels, V.M.; Schweickert, A.; Blum, M.; Franco, D.; Moorman, A.F. Pitx2 Expression Defines a Left Cardiac Lineage of Cells: Evidence for Atrial and Ventricular Molecular Isomerism in the iv/iv Mice. Dev. Biol. 2001, 231, 252–264. [Google Scholar] [CrossRef]
- Campione, M.; Acosta, L.; Martinez, S.; Icardo, J.; Aranega, A.; Franco, D. Pitx2 and Cardiac Development: A Molecular Link between Left/Right Signaling and Congenital Heart Disease. Cold Spring Harb. Symp. Quant. Biol. 2002, 67, 89–96. [Google Scholar] [CrossRef]
- Franco, D.; Campione, M.; Kelly, R.; Zammit, P.S.; Buckingham, M.; Lamers, W.H.; Moorman, A.F.M. Multiple transcriptional domains, with distinct left and right components, in the atrial chambers of the developing heart. Circ. Res. 2000, 87, 984–991. [Google Scholar] [CrossRef]
- Campione, M.; Franco, D. Current Perspectives in Cardiac Laterality. J. Cardiovasc. Dev. Dis. 2016, 3, 34. [Google Scholar] [CrossRef]
- Xu, H.; Morishima, M.; Wylie, J.N.; Schwartz, R.J.; Bruneau, B.G.; Lindsay, E.A.; Baldini, A. Tbx1has a dual role in the morphogenesis of the cardiac outflow tract. Development 2004, 131, 3217–3227. [Google Scholar] [CrossRef]
- Zhang, Z.; Huynh, T.; Baldini, A. Mesodermal expression ofTbx1is necessary and sufficient for pharyngeal arch and cardiac outflow tract development. Development 2006, 133, 3587–3595. [Google Scholar] [CrossRef]
- Greulich, F.; Rudat, C.; Kispert, A. Mechanisms of T-box gene function in the developing heart. Cardiovasc. Res. 2011, 91, 212–222. [Google Scholar] [CrossRef] [PubMed]
- Aanhaanen, W.T.; Brons, J.F.; Domínguez, J.N.; Rana, M.S.; Norden, J.; Airik, R.; Wakker, V.; Vries, C.D.G.-D.; Brown, N.A.; Kispert, A.; et al. The Tbx2 + Primary Myocardium of the Atrioventricular Canal Forms the Atrioventricular Node and the Base of the Left Ventricle. Circ. Res. 2009, 104, 1267–1274. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, J.K.; Ohgi, M.; Koshiba-Takeuchi, K.; Shiratori, H.; Sakaki, I.; Ogura, K.; Saijoh, Y.; Ogura, T. Tbx5specifies the left/right ventricles and ventricular septum position during cardiogenesis. Development 2003, 130, 5953–5964. [Google Scholar] [CrossRef] [PubMed]
- Moskowitz, I.P.G.; Pizard, A.; Patel, V.V.; Bruneau, B.; Kim, J.B.; Kupershmidt, S.; Roden, D.; Berul, C.I.; Seidman, C.E.; Seidman, J.G. The T-Box transcription factor Tbx5 is required for the patterning and maturation of the murine cardiac conduction system. Development 2004, 131, 4107–4116. [Google Scholar] [CrossRef]
- Bruneau, B.G.; Logan, M.; Davis, N.; Levi, T.; Tabin, C.J.; Seidman, J.G.; Seidman, C.E. Chamber-Specific Cardiac Expression of Tbx5 and Heart Defects in Holt–Oram Syndrome. Dev. Biol. 1999, 211, 100–108. [Google Scholar] [CrossRef]
- Bruneau, B.; Nemer, G.; Schmitt, J.P.; Charron, F.; Robitaille, L.; Caron, S.; Conner, D.A.; Gessler, M.; Nemer, M.; Seidman, C.E.; et al. A Murine Model of Holt-Oram Syndrome Defines Roles of the T-Box Transcription Factor Tbx5 in Cardiogenesis and Disease. Cell 2001, 106, 709–721. [Google Scholar] [CrossRef]
- Greulich, F.; Farin, H.F.; Schuster-Gossler, K.; Kispert, A. Tbx18 function in epicardial development. Cardiovasc. Res. 2012, 96, 476–483. [Google Scholar] [CrossRef]
- Wu, S.-P.; Dong, X.-R.; Regan, J.N.; Su, C.; Majesky, M.W. Tbx18 regulates development of the epicardium and coronary vessels. Dev. Biol. 2013, 383, 307–320. [Google Scholar] [CrossRef]
- Firulli, A.B.; McFadden, D.G.; Lin, Q.; Srivastava, D.; Olson, E.N. Heart and extra-embryonic mesodermal defects in mouse embryos lacking the bHLH transcription factor Hand1. Nat. Genet. 1998, 18, 266–270. [Google Scholar] [CrossRef]
- Riley, P.; Anaon-Cartwight, L.; Cross, J.C. The Hand1 bHLH transcription factor is essential for placentation and cardiac morphogenesis. Nat. Genet. 1998, 18, 271–275. [Google Scholar] [CrossRef]
- Srivastava, D.; Cserjesi, P.; Olson, E.N. A Subclass of bHLH Proteins Required for Cardiac Morphogenesis. Science 1995, 270, 1995–1999. [Google Scholar] [CrossRef]
- Thomas, T.; Yamagishi, H.; Overbeek, P.A.; Olson, E.N.; Srivastava, D. The bHLH Factors, dHAND and eHAND, Specify Pulmonary and Systemic Cardiac Ventricles Independent of Left–Right Sidedness. Dev. Biol. 1998, 196, 228–236. [Google Scholar] [CrossRef]
- Pereira, F.; Qiu, Y.; Zhou, G.; Tsai, M.-J.; Tsai, S.Y. The orphan nuclear receptor COUP-TFII is required for angiogenesis and heart development. Genes Dev. 1999, 13, 1037–1049. [Google Scholar] [CrossRef]
- Fischer, A.; Schumacher, N.; Maier, M.; Sendtner, M.; Gessler, M. The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev. 2004, 18, 901–911. [Google Scholar] [CrossRef]
- Nakagawaab, O.; Nakagawaab, M.; Richardson, J.A.; Olson, E.N.; Srivastava, D. HRT1, HRT2, and HRT3: A New Subclass of bHLH Transcription Factors Marking Specific Cardiac, Somitic, and Pharyngeal Arch Segments. Dev. Biol. 1999, 216, 72–84. [Google Scholar] [CrossRef]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef]
- Hombach, S.; Kretz, M. Non-coding RNAs: Classification, Biology and Functioning. Adv. Exp. Med. Biol. 2016, 937, 3–17. [Google Scholar] [CrossRef]
- Ounzain, S.; Crippa, S.; Pedrazzini, T. Small and long non-coding RNAs in cardiac homeostasis and regeneration. Biochim. et Biophys. Acta 2013, 1833, 923–933. [Google Scholar] [CrossRef]
- Chen, J.; Wang, D.-Z. microRNAs in cardiovascular development. J. Mol. Cell. Cardiol. 2012, 52, 949–957. [Google Scholar] [CrossRef]
- Beermann, J.; Piccoli, M.-T.; Viereck, J.; Thum, T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol. Rev. 2016, 96, 1297–1325. [Google Scholar] [CrossRef]
- Poller, W.; Dimmeler, S.; Heymans, S.; Zeller, T.; Haas, J.; Karakas, M.; Leistner, D.M.; Jakob, P.; Nakagawa, S.; Blankenberg, S.; et al. Non-coding RNAs in cardiovascular diseases: Diagnostic and therapeutic perspectives. Eur. Heart J. 2018, 39, 2704–2716. [Google Scholar] [CrossRef] [PubMed]
- Dykes, I.M.; Emanueli, C. Transcriptional and Post-transcriptional Gene Regulation by Long Non-coding RNA. Genom. Proteom. Bioinform. 2017, 15, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Shi, Z.-M.; Chang, Y.-N.; Hu, Z.-M.; Qi, H.-X.; Hong, W. The ways of action of long non-coding RNAs in cytoplasm and nucleus. Gene 2014, 547, 1–9. [Google Scholar] [CrossRef]
- Wojciechowska, A.; Braniewska, A.; Kozar-Kamińska, K. MicroRNA in cardiovascular biology and disease. Adv. Clin. Exp. Med. 2017, 26, 868–874. [Google Scholar] [CrossRef]
- Barwari, T.; Joshi, A.; Mayr, M. MicroRNAs in Cardiovascular Disease. J. Am. Coll. Cardiol. 2016, 68, 2577–2584. [Google Scholar] [CrossRef]
- Wong, L.L.; Wang, J.; Liew, O.W.; Richards, A.M.; Chen, Y.-T. MicroRNA and Heart Failure. Int. J. Mol. Sci. 2016, 17, 502. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Lin, S.; Gregory, R.I. MicroRNA biogenesis pathways in cancer. Nat. Cancer 2015, 15, 321–333. [Google Scholar] [CrossRef]
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018, 19, 143–157. [Google Scholar] [CrossRef]
- Bhan, A.; Soleimani, M.; Mandal, S.S. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017, 77, 3965–3981. [Google Scholar] [CrossRef]
- Dahariya, S.; Paddibhatla, I.; Kumar, S.; Raghuwanshi, S.; Pallepati, A.; Gutti, R.K. Long non-coding RNA: Classification, biogenesis and functions in blood cells. Mol. Immunol. 2019, 112, 82–92. [Google Scholar] [CrossRef]
- Yuan, Z.; Huang, W. New Developments in Exosomal lncRNAs in Cardiovascular Diseases. Front. Cardiovasc. Med. 2021, 8, 709169. [Google Scholar] [CrossRef]
- Zheng, D.; Huo, M.; Li, B.; Wang, W.; Piao, H.; Wang, Y.; Zhu, Z.; Li, D.; Wang, T.; Liu, K. The Role of Exosomes and Exosomal MicroRNA in Cardiovascular Disease. Front. Cell Dev. Biol. 2021, 8, 616161. [Google Scholar] [CrossRef]
- Barron, M.; Gao, M.; Lough, J. Requirement for BMP and FGF signaling during cardiogenic induction in non-precardiac mesoderm is specific, transient, and cooperative. Dev. Dyn. 2000, 218, 383–393. [Google Scholar] [CrossRef]
- Lough, J.; Barron, M.; Brogley, M.; Sugi, Y.; Bolender, D.L.; Zhu, X. Combined BMP-2 and FGF-4, but Neither Factor Alone, Induces Cardiogenesis in Non-Precardiac Embryonic Mesoderm. Dev. Biol. 1996, 178, 198–202. [Google Scholar] [CrossRef][Green Version]
- Schultheiss, T.M.; Burch, J.B.; Lassar, A.B. A role for bone morphogenetic proteins in the induction of cardiac myogenesis. Genes Dev. 1997, 11, 451–462. [Google Scholar] [CrossRef]
- Zhang, H.; Bradley, A. Mice deficient for BMP2 are nonviable and have defects in amnion/chorion and cardiac development. Development 1996, 122, 2977–2986. [Google Scholar] [CrossRef]
- Sugi, Y.; Yamamura, H.; Okagawa, H.; Markwald, R.R. Bone morphogenetic protein-2 can mediate myocardial regulation of atrioventricular cushion mesenchymal cell formation in mice. Dev. Biol. 2004, 269, 505–518. [Google Scholar] [CrossRef]
- Luna-Zurita, L.; Prados, B.; Grego-Bessa, J.; Luxán, G.; del Monte, G.; Benguría, A.; Adams, R.H.; Pérez-Pomares, J.M.; de la Pompa, J.L. Integration of a Notch-dependent mesenchymal gene program and Bmp2-driven cell invasiveness regulates murine cardiac valve formation. J. Clin. Investig. 2010, 120, 3493–3507. [Google Scholar] [CrossRef]
- Winnier, G.; Blessing, M.; Labosky, P.A.; Hogan, B.L. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 1995, 9, 2105–2116. [Google Scholar] [CrossRef] [PubMed]
- Waldo, K.L.; Kumiski, D.H.; Wallis, K.T.; Stadt, H.A.; Hutson, M.R.; Platt, D.H.; Kirby, M.L. Conotruncal myocardium arises from a secondary heart field. Development 2001, 128, 3179–3188. [Google Scholar] [CrossRef]
- Bernardo, A.S.; Faial, T.; Gardner, L.; Niakan, K.K.; Ortmann, D.; Senner, C.E.; Callery, E.M.; Trotter, M.W.; Hemberger, M.; Smith, J.C.; et al. BRACHYURY and CDX2 Mediate BMP-Induced Differentiation of Human and Mouse Pluripotent Stem Cells into Embryonic and Extraembryonic Lineages. Cell Stem Cell 2011, 9, 144–155. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Sanchez, C.; Franco, D.; Bonet, F.; Garcia-Lopez, V.; Aranega, A.; Martinez, F.B. Negative Fgf8-Bmp2 feed-back is regulated by miR-130 during early cardiac specification. Dev. Biol. 2015, 406, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, C.A.; Eisenberg, L.M. WNT11 promotes cardiac tissue formation of early mesoderm. Dev. Dyn. 1999, 216, 45–58. [Google Scholar] [CrossRef]
- Ueno, S.; Weidinger, G.; Osugi, T.; Kohn, A.D.; Golob, J.L.; Pabon, L.; Reinecke, H.; Moon, R.T.; Murry, C.E. Biphasic role for Wnt/beta-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proc. Natl. Acad. Sci. USA 2007, 104, 9685–9690. [Google Scholar] [CrossRef]
- Afouda, B.A.; Hoppler, S. Different requirements for GATA factors in cardiogenesis are mediated by non-canonical Wnt signaling. Dev. Dyn. 2011, 240, 649–662. [Google Scholar] [CrossRef]
- Samuel, L.J.; Latinkić, B.V. Early Activation of FGF and Nodal Pathways Mediates Cardiac Specification Independently of Wnt/β-Catenin Signaling. PLoS ONE 2009, 4, e7650. [Google Scholar] [CrossRef]
- Klaus-Bergmann, A.; Saga, Y.; Taketo, M.M.; Tzahor, E.; Birchmeier, W. Distinct roles of Wnt/beta-catenin and Bmp signaling during early cardiogenesis. Proc. Natl. Acad. Sci. USA 2007, 104, 18531–18536. [Google Scholar] [CrossRef]
- Gessert, S.; Kühl, M. The Multiple Phases and Faces of Wnt Signaling During Cardiac Differentiation and Development. Circ. Res. 2010, 107, 186–199. [Google Scholar] [CrossRef]
- Onizuka, T.; Yuasa, S.; Kusumoto, D.; Shimoji, K.; Egashira, T.; Ohno, Y.; Kageyama, T.; Tanaka, T.; Hattori, F.; Fujita, J.; et al. Wnt2 accelerates cardiac myocyte differentiation from ES-cell derived mesodermal cells via non-canonical pathway. J. Mol. Cell. Cardiol. 2011, 52, 650–659. [Google Scholar] [CrossRef]
- Medley, T.L.; Furtado, M.; Lam, N.T.; Idrizi, R.; Williams, D.; Verma, P.J.; Costa, M.; Kaye, D.M. Effect of Oxygen on Cardiac Differentiation in Mouse iPS Cells: Role of Hypoxia Inducible Factor-1 and Wnt/Beta-Catenin Signaling. PLoS ONE 2013, 8, e80280. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, J.; Cui, J.; Sun, M.; Du, W.; Chen, T.; Ming, X.; Zhang, L.; Tian, J.; Li, J.; et al. MiR218 Modulates Wnt Signaling in Mouse Cardiac Stem Cells by Promoting Proliferation and Inhibiting Differentiation through a Positive Feedback Loop. Sci. Rep. 2016, 6, 20968. [Google Scholar] [CrossRef]
- Wang, D.; Liu, C.; Wang, Y.; Wang, W.; Wang, K.; Wu, X.; Li, Z.; Zhao, C.; Li, L.; Peng, L. Impact of miR-26b on cardiomyocyte differentiation in P19 cells through regulating canonical/non-canonical Wnt signalling. Cell Prolif. 2017, 50, e12371. [Google Scholar] [CrossRef]
- Liu, X.; Yang, L.; Wang, H.; Xu, G.; Zhu, S.; Li, M.; Hu, X.; Zhu, J.; Zhu, C.; Xu, J.; et al. Effects of miR-19b knockdown on the cardiac differentiation of P19 mouse embryonic carcinoma cells. Mol. Med. Rep. 2014, 11, 2504–2512. [Google Scholar] [CrossRef]
- Qin, D.-N.; Qian, L.; Hu, D.-L.; Yu, Z.-B.; Han, S.-P.; Zhu, C.; Wang, X.; Hu, X. Effects of miR-19b Overexpression on Proliferation, Differentiation, Apoptosis and Wnt/β-Catenin Signaling Pathway in P19 Cell Model of Cardiac Differentiation In Vitro. Cell Biophys. 2013, 66, 709–722. [Google Scholar] [CrossRef]
- Lu, T.-Y.; Lin, B.; Li, Y.; Arora, A.; Han, L.; Cui, C.; Coronnello, C.; Sheng, Y.; Benos, P.V.; Yang, L. Overexpression of microRNA-1 promotes cardiomyocyte commitment from human cardiovascular progenitors via suppressing WNT and FGF signaling pathways. J. Mol. Cell. Cardiol. 2013, 63, 146–154. [Google Scholar] [CrossRef]
- Xu, J.; Gruber, P.J.; Chien, K.R. SMAD4 Is Essential for Human Cardiac Mesodermal Precursor Cell Formation. Stem Cells 2018, 37, 216–225. [Google Scholar] [CrossRef]
- López-Sánchez, C.; Climent, V.; Schoenwolf, G.; Álvarez, I.; García-Martínez, V. Induction of cardiogenesis by Hensen’s node and fibroblast growth factors. Cell Tissue Res. 2002, 309, 237–249. [Google Scholar] [CrossRef]
- Coppola, A.; Romito, A.; Borel, C.; Gehrig, C.; Gagnebin, M.; Falconnet, E.; Izzo, A.; Altucci, L.; Banfi, S.; Antonarakis, S.; et al. Cardiomyogenesis is controlled by the miR-99a/let-7c cluster and epigenetic modifications. Stem Cell Res. 2014, 12, 323–337. [Google Scholar] [CrossRef]
- Chen, X.; Wang, L.; Huang, R.; Qiu, H.; Wang, P.; Wu, D.; Zhu, Y.; Ming, J.; Wang, Y.; Wang, J.; et al. Dgcr8 deletion in the primitive heart uncovered novel microRNA regulating the balance of cardiac-vascular gene program. Protein Cell 2019, 10, 327–346. [Google Scholar] [CrossRef]
- Shen, X.; Soibam, B.; Benham, A.; Xu, X.; Chopra, M.; Peng, X.; Yu, W.; Bao, W.; Liang, R.; Azares, A.; et al. miR-322/-503 cluster is expressed in the earliest cardiac progenitor cells and drives cardiomyocyte specification. Proc. Natl. Acad. Sci. USA 2016, 113, 9551–9556. [Google Scholar] [CrossRef]
- Yao, C.-X.; Wei, Q.-X.; Zhang, Y.-Y.; Wang, W.-P.; Xue, L.-X.; Yang, F.; Zhang, S.-F.; Xiong, C.-J.; Li, W.-Y.; Wei, Z.-R.; et al. miR-200b targets GATA-4 during cell growth and differentiation. RNA Biol. 2013, 10, 465–480. [Google Scholar] [CrossRef]
- Han, M.; Yang, Z.; Sayed, D.; He, M.; Gao, S.; Lin, L.; Yoon, S.; Abdellatif, M. GATA4 expression is primarily regulated via a miR-26b-dependent post-transcriptional mechanism during cardiac hypertrophy. Cardiovasc. Res. 2012, 93, 645–654. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, S.; Guo, C.; Li, J.; Sang, W. Downregulation of miR-200c protects cardiomyocytes from hypoxia-induced apoptosis by targeting GATA-4. Int. J. Mol. Med. 2017, 39, 1589–1596. [Google Scholar] [CrossRef]
- Kay, M.; Soltani, B.M.; Aghdaei, F.H.; Ansari, H.; Baharvand, H. Hsa-miR-335 regulates cardiac mesoderm and progenitor cell differentiation. Stem Cell Res. Ther. 2019, 10, 191. [Google Scholar] [CrossRef]
- Qian, L.; Wythe, J.D.; Liu, J.; Cartry, J.; Vogler, G.; Mohapatra, B.; Otway, R.T.; Huang, Y.; King, I.N.; Maillet, M.; et al. Tinman/Nkx2-5 acts via miR-1 and upstream of Cdc42 to regulate heart function across species. J. Cell Biol. 2011, 193, 1181–1196. [Google Scholar] [CrossRef] [PubMed]
- Hoelscher, S.C.; Stich, T.; Diehm, A.; Lahm, H.; Dreßen, M.; Zhang, Z.; Neb, I.; Aherrahrou, Z.; Erdmann, J.; Schunkert, H.; et al. miR-128a Acts as a Regulator in Cardiac Development by Modulating Differentiation of Cardiac Progenitor Cell Populations. Int. J. Mol. Sci. 2020, 21, 1158. [Google Scholar] [CrossRef] [PubMed]
- Arasaratnam, D.; Bell, K.M.; Sim, C.B.; Koutsis, K.; Anderson, D.J.; Qian, E.L.; Stanley, E.G.; Elefanty, A.G.; Cheung, M.M.; Oshlack, A.; et al. The role of cardiac transcription factor NKX2-5 in regulating the human cardiac miRNAome. Sci. Rep. 2019, 9, 15928. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Wu, D.; Wang, C.; Guo, Z.; Li, B.; Zuo, Z. Hexabromocyclododecane exposure induces cardiac hypertrophy and arrhythmia by inhibiting miR-1 expression via up-regulation of the homeobox gene Nkx2.5. J. Hazard. Mater. 2015, 302, 304–313. [Google Scholar] [CrossRef]
- Chinchilla, A.; Lozano, E.; Daimi, H.; Esteban, F.J.; Crist, C.; Aranega, A.E.; Franco, D. MicroRNA profiling during mouse ventricular maturation: A role for miR-27 modulating Mef2c expression. Cardiovasc. Res. 2010, 89, 98–108. [Google Scholar] [CrossRef]
- Chen, H.-P.; Wen, J.; Tan, S.-R.; Kang, L.-M.; Zhu, G.-C. MiR-199a-3p inhibition facilitates cardiomyocyte differentiation of embryonic stem cell through promotion of MEF2C. J. Cell. Physiol. 2019, 234, 23315–23325. [Google Scholar] [CrossRef]
- Xu, M.; Chen, X.; Chen, D.; Yu, B.; Li, M.; He, J.; Huang, Z. MicroRNA-499-5p regulates skeletal myofiber specification via NFATc1/MEF2C pathway and Thrap1/MEF2C axis. Life Sci. 2018, 215, 236–245. [Google Scholar] [CrossRef]
- Shi, Y.; Mao, X.; Cai, M.; Hu, S.; Lai, X.; Chen, S.; Jia, X.; Wang, J.; Lai, S. miR-194-5p negatively regulates the proliferation and differentiation of rabbit skeletal muscle satellite cells. Mol. Cell. Biochem. 2020, 476, 425–433. [Google Scholar] [CrossRef]
- Cheng, X.; Du, J.; Shen, L.; Tan, Z.; Jiang, D.; Jiang, A.; Li, Q.; Tang, G.; Jiang, Y.; Wang, J.; et al. MiR-204-5p regulates C2C12 myoblast differentiation by targeting MEF2C and ERRγ. Biomed. Pharmacother. 2018, 101, 528–535. [Google Scholar] [CrossRef]
- Gagan, J.; Dey, B.K.; Layer, R.; Yan, Z.; Dutta, A. Notch3 and Mef2c Proteins Are Mutually Antagonistic via Mkp1 Protein and miR-1/206 MicroRNAs in Differentiating Myoblasts. J. Biol. Chem. 2012, 287, 40360–40370. [Google Scholar] [CrossRef]
- Tang, C.-M.; Liu, F.-Z.; Zhu, J.-N.; Fu, Y.-H.; Lin, Q.-X.; Deng, C.-Y.; Hu, Z.-Q.; Yang, H.; Zheng, X.-L.; Cheng, J.-D.; et al. Myocyte-specific enhancer factor 2C: A novel target gene of miR-214-3p in suppressing angiotensin II-induced cardiomyocyte hypertrophy. Sci. Rep. 2016, 6, 36146. [Google Scholar] [CrossRef]
- Zhang, R.; Sui, L.; Hong, X.; Yang, M.; Li, W. MiR-448 promotes vascular smooth muscle cell proliferation and migration in through directly targeting MEF2C. Environ. Sci. Pollut. Res. 2017, 24, 22294–22300. [Google Scholar] [CrossRef]
- Klattenhoff, C.A.; Scheuermann, J.C.; Surface, L.E.; Bradley, R.K.; Fields, P.A.; Steinhauser, M.L.; Ding, H.; Butty, V.L.; Torrey, L.; Haas, S.; et al. Braveheart, a Long Noncoding RNA Required for Cardiovascular Lineage Commitment. Cell 2013, 152, 570–583. [Google Scholar] [CrossRef]
- Hou, J.; Long, H.; Zhou, C.; Zheng, S.; Wu, H.; Guo, T.; Wu, Q.; Zhong, T.; Wang, T. Long noncoding RNA Braveheart promotes cardiogenic differentiation of mesenchymal stem cells in vitro. Stem Cell Res. Ther. 2017, 8, 4. [Google Scholar] [CrossRef]
- García-Padilla, C.; Domínguez, J.N.; Aránega, A.E.; Franco, D. Differential chamber-specific expression and regulation of long non-coding RNAs during cardiac development. Biochim. Biophys. Acta 2019, 1862, 194435. [Google Scholar] [CrossRef]
- Yin, A.; Feng, M.; Cheng, Z.; Zhang, Q.; Li, H.; Xu, J.; Zhang, H.; Li, Y.; Qian, L. Altered DNA Methylation of Long Noncoding RNA uc.167 Inhibits Cell Differentiation in Heart Development. BioMed Res. Int. 2018, 2018, 4658024. [Google Scholar] [CrossRef]
- Liu, H.; Hu, Y.; Yin, J.; Yan, X.; Chen, W.; Wang, X.; Han, S.; Yu, Z.; Li, M. Effects of long non-coding RNA uc.245 on cardiomyocyte-like differentiation in P19 cells via FOG2. Gene 2019, 694, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Shen, Y.; Ruan, Z.; Li, X.; Chen, Y.; Yuan, W.; Ding, X.; Zhu, L.; Qian, L. LncRNA-uc.167 influences cell proliferation, apoptosis and differentiation of P19 cells by regulating Mef2c. Gene 2016, 590, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-H.; Chang, M.-W.; Pandey, P.R.; Tsitsipatis, D.; Yang, X.; Martindale, J.L.; Munk, R.; De, S.; Abdelmohsen, K.; Gorospe, M. Interaction of OIP5-AS1 with MEF2C mRNA promotes myogenic gene expression. Nucleic Acids Res. 2020, 48, 12943–12956. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.-J.; Lee, K.-H.; Son, Y.; Nam, A.-R.; Moon, E.-H.; Pyun, J.-H.; Park, J.; Kang, J.-S.; Lee, Y.J.; Cho, J.-Y. Spatiotemporal expression of long noncoding RNA Moshe modulates heart cell lineage commitment. RNA Biol. 2021, 18, 640–654. [Google Scholar] [CrossRef]
- Grote, P.; Wittler, L.; Hendrix, D.; Koch, F.; Währisch, S.; Beisaw, A.; Macura, K.; Bläss, G.; Kellis, M.; Werber, M.; et al. The Tissue-Specific lncRNA Fendrr Is an Essential Regulator of Heart and Body Wall Development in the Mouse. Dev. Cell 2013, 24, 206–214. [Google Scholar] [CrossRef]
- Zhu, H. Forkhead box transcription factors in embryonic heart development and congenital heart disease. Life Sci. 2016, 144, 194–201. [Google Scholar] [CrossRef]
- Watanabe, M.; Whitman, M. FAST-1 is a key maternal effector of mesoderm inducers in the early Xenopus embryo. Development 1999, 126, 5621–5634. [Google Scholar] [CrossRef]
- Weisberg, E.; Winnier, G.E.; Chen, X.; Farnsworth, C.L.; Hogan, B.L.; Whitman, M. A mouse homologue of FAST-1 transduces TGFβ superfamily signals and is expressed during early embryogenesis. Mech. Dev. 1998, 79, 17–27. [Google Scholar] [CrossRef]
- von Both, I.; Silvestri, C.; Erdemir, T.; Lickert, H.; Walls, J.R.; Henkelman, R.M.; Rossant, J.; Harvey, R.P.; Attisano, L.; Wrana, J.L. Foxh1 is essential for development of the anterior heart field. Dev. Cell 2004, 7, 331–345. [Google Scholar] [CrossRef]
- Huang, C.-X.; Chen, N.; Wu, X.-J.; He, Y.; Huang, C.-H.; Liu, H.; Wang, W.-M.; Wang, H.-L. Zebrafish let-7b acts downstream of hypoxia-inducible factor-1α to assist in hypoxia-mediated cell proliferation and cell cycle regulation. Life Sci. 2017, 171, 21–29. [Google Scholar] [CrossRef]
- Heigwer, J.; Kutzner, J.; Haeussler, M.; Burkhalter, M.D.; Draebing, T.; Juergensen, L.; Katus, H.A.; Philipp, M.; Westhoff, J.H.; Hassel, D. miR-103/107 regulates left-right asymmetry in zebrafish by modulating Kupffer’s vesicle development and ciliogenesis. Biochem. Biophys. Res. Commun. 2020, 527, 432–439. [Google Scholar] [CrossRef]
- Fischer, P.; Chen, H.; Pacho, F.; Rieder, D.; Kimmel, R.A.; Meyer, D. FoxH1 represses miR-430 during early embryonic development of zebrafish via non-canonical regulation. BMC Biol. 2019, 17, 61. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, L.; Meng, Y.; Huang, L. Benzyl isothiocyanate inhibits breast cancer cell tumorigenesis via repression of the FoxH1-Mediated Wnt/β-catenin pathway. Int. J. Clin. Exp. Med. 2015, 8, 17601–17611. [Google Scholar]
- Vincent, S.D.; Buckingham, M. Chapter One-How to Make a Heart: The Origin and Regulation of Cardiac Progenitor Cells. Curr. Top. Dev. Biol. 2010, 90, 1–41. [Google Scholar]
- Evans, S.M.; Yelon, D.; Conlon, F.L.; Kirby, M.L. Myocardial Lineage Development. Circ. Res. 2010, 107, 1428–1444. [Google Scholar] [CrossRef]
- Ren, J.; Miao, D.; Li, Y.; Gao, R. Spotlight on Isl1: A Key Player in Cardiovascular Development and Diseases. Front. Cell Dev. Biol. 2021, 9, 793605. [Google Scholar] [CrossRef]
- Wang, J.; Greene, S.B.; Bonilla-Claudio, M.; Tao, Y.; Zhang, J.; Huang, Z.; Black, B.L.; Wang, F.; Martin, J.F. Bmp-signaling regulates myocardial differentiation from cardiac progenitors through a micro RNA-mediated mechanism. Dev. Cell 2010, 19, 903–912. [Google Scholar] [CrossRef]
- Zlabinger, K.; Spannbauer, A.; Traxler, D.; Gugerell, A.; Lukovic, D.; Winkler, J.; Mester-Tonczar, J.; Podesser, B.; Gyöngyösi, M. MiR-21, MiR-29a, GATA4, and MEF2c Expression Changes in Endothelin-1 and Angiotensin II Cardiac Hypertrophy Stimulated Isl-1+Sca-1+c-kit+ Porcine Cardiac Progenitor Cells In Vitro. Cells 2019, 8, 1416. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, X.; Li, F.; Ji, Q. MiR-128-3p accelerates cardiovascular calcification and insulin resistance through ISL1-dependent Wnt pathway in type 2 diabetes mellitus rats. J. Cell. Physiol. 2018, 234, 4997–5010. [Google Scholar] [CrossRef]
- Steimle, J.D.; Moskowitz, I.P. Chapter Seven—TBX5: A Key Regulator of Heart Development. Curr. Top. Dev. Biol. 2017, 122, 195–221. [Google Scholar]
- D’Aurizio, R.; Russo, F.; Chiavacci, E.; Baumgart, M.; Groth, M.; D’Onofrio, M.; Arisi, I.; Rainaldi, G.; Pitto, L.; Pellegrini, M. Discovering miRNA Regulatory Networks in Holt–Oram Syndrome Using a Zebrafish Model. Front. Bioeng. Biotechnol. 2016, 4, 60. [Google Scholar] [CrossRef][Green Version]
- Chiavacci, E.; Daurizio, R.; Guzzolino, E.; Russo, F.; Baumgart, M.; Groth, M.; Mariani, L.; D’Onofrio, M.; Arisi, I.; Pellegrini, M.; et al. MicroRNA 19a replacement partially res-cues fin and cardiac defects in zebrafish model of Holt Oram syndrome. Sci. Rep. 2015, 5, 18240. [Google Scholar] [CrossRef]
- Boon, R.A.; Iekushi, K.; Lechner, S.; Seeger, T.; Fischer, A.; Heydt, S.; Kaluza, D.; Tréguer, K.; Carmona, G.; Bonauer, A.; et al. MicroRNA-34a regulates cardiac ageing and function. Nature 2013, 495, 107–110. [Google Scholar] [CrossRef]
- Fish, J.E.; Wythe, J.D.; Xiao, T.; Bruneau, B.G.; Stainier, D.Y.R.; Srivastava, D.; Woo, S. A Slit/miR-218/Robo regulatory loop is required during heart tube formation in zebrafish. Development 2011, 138, 1409–1419. [Google Scholar] [CrossRef]
- Qu, X.; Jia, H.; Garrity, D.M.; Tompkins, K.; Batts, L.; Appel, B.; Zhong, T.P.; Baldwin, H.S. ndrg4 is required for normal myocyte proliferation during early cardiac development in zebrafish. Dev. Biol. 2008, 317, 486–496. [Google Scholar] [CrossRef]
- Chiavacci, E.; Dolfi, L.; Verduci, L.; Meghini, F.; Gestri, G.; Evangelista, M.; Wilson, S.W.; Cremisi, F.; Pitto, L. MicroRNA 218 Mediates the Effects of Tbx5a Over-Expression on Zebrafish Heart Development. PLoS ONE 2012, 7, e50536. [Google Scholar] [CrossRef]
- Alzein, M.; Lozano-Velasco, E.; Hernández-Torres, F.; García-Padilla, C.; Domínguez, J.N.; Aránega, A.; Franco, D. Differential Spatio-Temporal Regulation of T-Box Gene Expression by microRNAs during Cardiac Development. J. Cardiovasc. Dev. Dis. 2021, 8, 56. [Google Scholar] [CrossRef]
- Ma, J.; Chen, S.; Hao, L.; Sheng, W.; Chen, W.; Ma, X.; Zhang, B.; Ma, D.; Huang, G. Hypermethylation-mediated down-regulation of lncRNA TBX5-AS1:2 in Tetralogy of Fallot inhibits cell proliferation by reducingTBX5expression. J. Cell. Mol. Med. 2020, 24, 6472–6484. [Google Scholar] [CrossRef]
- Hori, Y.; Tanimoto, Y.; Takahashi, S.; Furukawa, T.; Koshiba-Takeuchi, K.; Takeuchi, J.K. Important cardiac transcription factor genes are accompanied by bidirectional long non-coding RNAs. BMC Genom. 2018, 19, 967. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.H.; Nadadur, R.D.; Hilvering, C.; Bianchi, V.; Werner, M.; Mazurek, S.R.; Gadek, M.; Shen, K.M.; Goldman, J.A.; Tyan, L.; et al. Transcription-factor-dependent enhancer transcription defines a gene regulatory network for cardiac rhythm. eLife 2017, 6, e31683. [Google Scholar] [CrossRef] [PubMed]
- Shiratori, H.; Hamada, H. TGFβ signaling in establishing left–right asymmetry. Semin. Cell Dev. Biol. 2014, 32, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Hamada, H.; Meno, C.; Watanabe, D.; Saijoh, Y. Establishment of vertebrate left–right asymmetry. Nat. Rev. Genet. 2002, 3, 103–113. [Google Scholar] [CrossRef]
- Franco, D. The Role of Pitx2 during Cardiac Development Linking Left–Right Signaling and Congenital Heart Diseases. Trends Cardiovasc. Med. 2003, 13, 157–163. [Google Scholar] [CrossRef]
- Grimes, D.T.; Burdine, R.D. Left–Right Patterning: Breaking Symmetry to Asymmetric Morphogenesis. Trends Genet. 2017, 33, 616–628. [Google Scholar] [CrossRef]
- Franco, D.; Sedmera, D.; Lozano-Velasco, E. Multiple Roles of Pitx2 in Cardiac Development and Disease. J. Cardiovasc. Dev. Dis. 2017, 4, 16. [Google Scholar] [CrossRef]
- Franco, D.; Christoffels, V.M.; Campione, M. Homeobox transcription factor Pitx2: The rise of an asymmetry gene in cardiogenesis and arrhythmogenesis. Trends Cardiovasc. Med. 2014, 24, 23–31. [Google Scholar] [CrossRef]
- Gage, P.; Suh, H.; Camper, S. Dosage requirement of Pitx2 for development of multiple organs. Development 1999, 126, 4643–4651. [Google Scholar] [CrossRef]
- Lu, M.-F.; Pressman, C.L.; Dyer, R.; Johnson, R.L.; Martin, J.F. Function of Rieger syndrome gene in left–right asymmetry and craniofacial development. Nature 1999, 401, 276–278. [Google Scholar] [CrossRef]
- Barroso-Deljesus, A.; Lucena-Aguilar, G.; Sanchez, L.; Ligero, G.; Gutierrez-Aranda, I.; Menendez, P. The Nodal inhibitor Lefty is negatively modulated by the microRNA miR-302 in human embryonic stem cells. FASEB J. 2011, 25, 1497–1508. [Google Scholar] [CrossRef]
- Rosa, A.; Spagnoli, F.M.; Brivanlou, A.H. The miR-430/427/302 Family Controls Mesendodermal Fate Specification via Species-Specific Target Selection. Dev. Cell 2009, 16, 517–527. [Google Scholar] [CrossRef]
- Rosa, A.; Papaioannou, M.D.; Krzyspiak, J.E.; Brivanlou, A.H. miR-373 is regulated by TGFβ signaling and promotes mesendoderm differentiation in human Embryonic Stem Cells. Dev. Biol. 2014, 391, 81–88. [Google Scholar] [CrossRef]
- Choi, W.-Y.; Giraldez, A.J.; Schier, A.F. Target Protectors Reveal Dampening and Balancing of Nodal Agonist and Antagonist by miR-430. Science 2007, 318, 271–274. [Google Scholar] [CrossRef]
- Ma, H.; Lin, Y.; Zhao, Z.-A.; Lu, X.; Yu, Y.; Zhang, X.; Wang, Q.; Li, L. MicroRNA-127 Promotes Mesendoderm Differentiation of Mouse Embryonic Stem Cells by Targeting Left-Right Determination Factor 2. J. Biol. Chem. 2016, 291, 12126–12135. [Google Scholar] [CrossRef]
- Yang, F.; Qi, J. miR-430a regulates the development of left–right asymmetry by targeting sqt in the teleost. Gene 2020, 745, 144628. [Google Scholar] [CrossRef]
- Li, M.; Hu, X.; Zhu, J.; Zhu, C.; Zhu, S.; Liu, X.; Xu, J.; Han, S.; Yu, Z. Overexpression of miR-19b Impairs Cardiac Development in Zebrafish by Targeting ctnnb1. Cell. Physiol. Biochem. 2014, 33, 1988–2002. [Google Scholar] [CrossRef]
- Ryan, A.K.; Blumberg, B.; Rodriguez-Esteban, C.; Yonei-Tamura, S.; Tamura, K.; Tsukui, T.; De La Peña, J.; Sabbagh, W.; Greenwald, J.; Choe, S.; et al. Pitx2 determines left–right asymmetry of internal organs in vertebrates. Nature 1998, 394, 545–551. [Google Scholar] [CrossRef]
- Piedra, M.E.; Icardo, J.M.; Albajar, M.; Rodriguez-Rey, J.C.; Ros, M.A. Pitx2 participates in the late phase of the pathway controlling left-right asymmetry. Cell 1998, 94, 319–324. [Google Scholar] [CrossRef]
- Logan, M.; Pagán-Westphal, S.M.; Smith, D.M.; Paganessi, L.; Tabin, C.J. The Transcription Factor Pitx2 Mediates Situs-Specific Morphogenesis in Response to Left-Right Asymmetric Signals. Cell 1998, 94, 307–317. [Google Scholar] [CrossRef]
- Campione, M.; Steinbeisser, H.; Schweickert, A.; Deissler, K.; van Bebber, F.; Lowe, L.A.; Nowotschin, S.; Viebahn, C.; Haffter, P.; Kuehn, M.R.; et al. The homeobox gene Pitx2: Mediator of asymmetric left-right signaling in vertebrate heart and gut looping. Development 1999, 126, 1225–1234. [Google Scholar] [CrossRef]
- Bisgrove, B.W.; Essner, J.J.; Yost, H.J. Multiple pathways in the midline regulate concordant brain, heart and gut left-right asymmetry. Development 2000, 127, 3567–3579. [Google Scholar] [CrossRef]
- Bisgrove, B.W.; Yost, H.J. Classification of left-right patterning defects in zebrafish, mice, and humans. Am. J. Med Genet. 2001, 101, 315–323. [Google Scholar] [CrossRef]
- Branford, W.W.; Essner, J.; Yost, H. Regulation of Gut and Heart Left–Right Asymmetry by Context-Dependent Interactions between Xenopus Lefty and BMP4 Signaling. Dev. Biol. 2000, 223, 291–306. [Google Scholar] [CrossRef]
- Chinchilla, A.; Daimi, H.; Lozano-Velasco, E.; Dominguez, J.N.; Caballero, R.; Delpo, E.; Tamargo, J.; Cinca, J.; Hove-Madsenet, L.; Aranega, A.E.; et al. PITX2 insufficiency leads to atrial electrical and structural remodeling linked to arrhythmogenesis. Circ. Cardiovasc. Genet. 2011, 4, 269–279. [Google Scholar] [CrossRef]
- Wang, J.; Klysik, E.; Sood, S.; Johnson, R.L.; Wehrens, X.H.T.; Martin, J.F. Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc. Natl. Acad. Sci. USA 2010, 107, 9753–9758. [Google Scholar] [CrossRef]
- Kirchhof, P.; Kahr, P.C.; Kaese, S.; Piccini, I.; Vokshi, I.; Scheld, H.-H.; Rotering, H.; Fortmueller, L.; Laakmann, S.; Verheule, S.; et al. PITX2c Is Expressed in the Adult Left Atrium, and Reducing Pitx2c Expression Promotes Atrial Fibrillation Inducibility and Complex Changes in Gene Expression. Circ. Cardiovasc. Genet. 2011, 4, 123–133. [Google Scholar] [CrossRef]
- Lozano-Velasco, E.; Hernández-Torres, F.; Daimi, H.; Serra, S.A.; Herraiz, A.; Hove-Madsen, L.; Aránega, A.; Franco, D. Pitx2 impairs calcium handling in a dose-dependent manner by modulating Wnt signalling. Cardiovasc. Res. 2016, 109, 55–66. [Google Scholar] [CrossRef]
- Lozano-Velasco, E.; Wangensteen, R.; Quesada, A.; Garcia-Padilla, C.; Osorio, J.A.; Ruiz-Torres, M.D.; Aranega, A.; Franco, D. Hyperthyroidism, but not hypertension, impairs PITX2 expression leading to Wnt-microRNA-ion channel remodeling. PLoS ONE 2017, 12, e0188473. [Google Scholar] [CrossRef]
- Franco, D.; Aranega, A.; Dominguez, J.N. Non-coding RNAs and Atrial Fibrillation. In Non-coding RNAs in Cardiovascular Diseases; Springer: Berlin/Heidelberg, Germany, 2020; Volume 1229, pp. 311–325. [Google Scholar] [CrossRef]
- Petkova, M.; Atkinson, A.J.; Yanni, J.; Stuart, L.; Aminu, A.J.; Ivanova, A.D.; Pustovit, K.B.; Geragthy, C.; Feather, A.; Li, N.; et al. Identification of Key Small Non-Coding MicroRNAs Controlling Pacemaker Mechanisms in the Human Sinus Node. J. Am. Heart Assoc. 2020, 9, e016590. [Google Scholar] [CrossRef]
- García-Padilla, C.; Aránega, A.; Franco, D. The role of long non-coding RNAs in cardiac development and disease. AIMS Genet. 2018, 5, 124–140. [Google Scholar] [CrossRef] [PubMed]
- Ocaña, O.H.; Córcoles, R.; Fabra, Á.; Moreno-Bueno, G.; Acloque, H.; Vega, S.; Barrallo-Gimeno, A.; Cano, A.; Nieto, M.A. Metastatic Colonization Requires the Re-pression of the Epithelial-Mesenchymal Transition Inducer Prrx1. Cancer Cell. 2012, 22, 709–724. [Google Scholar] [CrossRef] [PubMed]
- Fazilaty, H.; Rago, L.; Youssef, K.K.; Ocaña, O.H.; Garcia-Asencio, F.; Arcas, A.; Galceran, J.; Nieto, M.A. A gene regulatory network to control EMT programs in development and disease. Nat. Commun. 2019, 10, 5115. [Google Scholar] [CrossRef] [PubMed]
- Rago, L.; Castroviejo, N.; Fazilaty, H.; Garcia-Asencio, F.; Ocaña, O.H.; Galceran, J.; Nieto, M.A. MicroRNAs Establish the Right-Handed Dominance of the Heart Laterality Pathway in Vertebrates. Dev. Cell 2019, 51, 446–459. [Google Scholar] [CrossRef]
- Welsh, I.C.; Kwak, H.; Chen, F.L.; Werner, M.; Shopland, L.S.; Danko, C.G.; Lis, J.T.; Zhang, M.; Martin, J.F.; Kurpios, N.A. Chromatin Architecture of the Pitx2 Locus Requires CTCF- and Pitx2-Dependent Asymmetry that Mirrors Embryonic Gut Laterality. Cell Rep. 2015, 13, 337–349. [Google Scholar] [CrossRef]
- Gore-Panter, S.R.; Hsu, J.; Barnard, J.; Moravec, C.S.; Van Wagoner, D.R.; Chung, M.K.; Smith, J.D. PANCR, the PITX2 Adjacent Noncoding RNA, Is Expressed in Human Left Atria and Regulates PITX2c Expression. Circ. Arrhythmia Electrophysiol. 2016, 9, e003197. [Google Scholar] [CrossRef]
- Cao, Y.; Duca, S.; Cao, J. Epicardium in Heart Development. Cold Spring Harb. Perspect. Biol. 2019, 12, a037192. [Google Scholar] [CrossRef]
- Quijada, P.; Trembley, M.A.; Small, E.M. The Role of the Epicardium During Heart Development and Repair. Circ. Res. 2020, 126, 377–394. [Google Scholar] [CrossRef]
- Pomares, J.M.P.; De La Pompa, J.L. Signaling During Epicardium and Coronary Vessel Development. Circ. Res. 2011, 109, 1429–1442. [Google Scholar] [CrossRef]
- Dueñas, A.; Aranega, A.E.; Franco, D. More than Just a Simple Cardiac Envelope; Cellular Contributions of the Epicardium. Front. Cell Dev. Biol. 2017, 5, 44. [Google Scholar] [CrossRef]
- Sridurongrit, S.; Larsson, J.; Schwartz, R.; Ruiz-Lozano, P.; Kaartinen, V. Signaling via the Tgf-β type I receptor Alk5 in heart development. Dev. Biol. 2008, 322, 208–218. [Google Scholar] [CrossRef]
- Azhar, M.; Schultz, J.E.J.; Grupp, I.; Dorn, G.W.; Meneton, P.; Molin, D.G.; Groot, A.C.G.-D.; Doetschman, T. Transforming growth factor beta in cardiovascular development and function. Cytokine Growth Factor Rev. 2003, 14, 391–407. [Google Scholar] [CrossRef]
- Allison, P.; Espiritu, D.; Barnett, J.V.; Camenisch, T.D. Type III TGFβ receptor and Src direct hyaluronan-mediated invasive cell motility. Cell. Signal. 2014, 27, 453–459. [Google Scholar] [CrossRef]
- Austin, A.F.; Compton, L.A.; Love, J.D.; Brown, C.B.; Barnett, J.V. Primary and immortalized mouse epicardial cells undergo differentiation in response to TGFβ. Dev. Dyn. 2008, 237, 366–376. [Google Scholar] [CrossRef]
- Clark, C.R.; Robinson, J.Y.; Sanchez, N.S.; Townsend, T.A.; Arrieta, J.A.; Merryman, W.D.; Trykall, D.Z.; Olivey, H.E.; Hong, C.C.; Barnett, J.V. Common pathways regulate Type III TGFβ receptor-dependent cell invasion in epicardial and endocardial cells. Cell. Signal. 2016, 28, 688–698. [Google Scholar] [CrossRef]
- Bax, N.A.M.; Van Oorschot, A.A.M.; Maas, S.; Braun, J.; Van Tuyn, J.; De Vries, A.A.F.; Groot, A.C.G.-D.; Goumans, M.-J. In vitro epithelial-to-mesenchymal transformation in human adult epicardial cells is regulated by TGFβ-signaling and WT1. Basic Res. Cardiol. 2011, 106, 829–847. [Google Scholar] [CrossRef]
- Hill, C.R.; Sanchez, N.S.; Love, J.D.; Arrieta, J.A.; Hong, C.C.; Brown, C.B.; Austin, A.F.; Barnett, J.V. BMP2 signals loss of epithelial character in epicardial cells but requires the Type III TGFβ receptor to promote invasion. Cell. Signal. 2012, 24, 1012–1022. [Google Scholar] [CrossRef][Green Version]
- Morabito, C.J.; Dettman, R.; Kattan, J.; Collier, J.M.; Bristow, J. Positive and Negative Regulation of Epicardial–Mesenchymal Transformation during Avian Heart Development. Dev. Biol. 2001, 234, 204–215. [Google Scholar] [CrossRef]
- Sánchez, N.S.; Barnett, J.V. TGFβ and BMP-2 regulate epicardial cell invasion via TGFβR3 activation of the Par6/Smurf1/RhoA pathway. Cell. Signal. 2012, 24, 539–548. [Google Scholar] [CrossRef]
- Brønnum, H.; Andersen, D.C.; Schneider, M.; Sandberg, M.B.; Eskildsen, T.; Nielsen, S.B.; Kalluri, R.; Sheikh, S.P. miR-21 Promotes Fibrogenic Epithelial-to-Mesenchymal Transition of Epicardial Mesothelial Cells Involving Programmed Cell Death 4 and Sprouty-1. PLoS ONE 2013, 8, e56280. [Google Scholar] [CrossRef]
- Pontemezzo, E.; Foglio, E.; Vernucci, E.; Magenta, A.; D’Agostino, M.; Sileno, S.; Astanina, E.; Bussolino, F.; Pellegrini, L.; Germani, A.; et al. miR-200c-3p Regulates Epitelial-to-Mesenchymal Transition in Epicardial Mesothelial Cells by Targeting Epicardial Follistatin-Related Protein 1. Int. J. Mol. Sci. 2021, 22, 4971. [Google Scholar] [CrossRef]
- Tandon, P.; Miteva, Y.V.; Kuchenbrod, L.M.; Cristea, I.M.; Conlon, F.L. Tcf21 regulates the specification and maturation of proepicardial cells. Development 2013, 140, 2409–2421. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Lin, S.; Wang, S.; Chen, X. The Role of Transcription Factor 21 in Epicardial Cell Differentiation and the Development of Coronary Heart Disease. Front Cell Dev. Biol. 2020, 8, 457. [Google Scholar] [CrossRef] [PubMed]
- Seeger, T.; Xu, Q.-F.; Muhly-Reinholz, M.; Fischer, A.; Kremp, E.-M.; Zeiher, A.M.; Dimmeler, S. Inhibition of let-7 augments the recruitment of epicardial cells and improves cardiac function after myocardial infarction. J. Mol. Cell. Cardiol. 2016, 94, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.M.; Dykeman, C.A.; King, B.L.; Yin, V.P. Modulation of TNFα Activity by the microRNA Let-7 Coordi-nates Zebrafish Heart Regeneration. iScience 2019, 15, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.L.; Haas, U.; Diaz, R.; Leeper, N.J.; Kundu, R.K.; Patlolla, B.; Assimes, T.L.; Kaiser, F.J.; Perisic, L.; Hedin, U.; et al. Coronary Heart Disease-Associated Variation in TCF21 Disrupts a miR-224 Binding Site and miRNA-Mediated Regulation. PLoS Genet. 2014, 10, e1004263. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Gao, X.; Luo, H.; Huang, Q.; Su, D.; Tan, X.; Lu, C. TCF21 rs12190287 Polymorphisms Are Associated with Ventricu-lar Septal Defects in a Chinese Population. Genet. Test. Mol. Biomark. 2017, 21, 312–315. [Google Scholar] [CrossRef]
- Dechamethakun, S.; Ikeda, S.; Arai, T.; Sato, N.; Sawabe, M.; Muramatsu, M. Associations between the CDKN2A/B, ADTRP and PDGFD Polymorphisms and the Development of Coronary Atherosclerosis in Japanese Patients. J. Atheroscler. Thromb. 2014, 21, 680–690. [Google Scholar] [CrossRef]
- Fujimaki, T.; Oguri, M.; Horibe, H.; Kato, K.; Matsuoka, R.; Abe, S.; Tokoro, F.; Arai, M.; Noda, T.; Watanabe, S.; et al. Association of a transcription factor 21 gene polymorphism with hypertension. Biomed. Rep. 2015, 3, 118–122. [Google Scholar] [CrossRef]
- Wirtwein, M.; Melander, O.; Sjőgren, M.; Hoffmann, M.; Narkiewicz, K.; Gruchala, M.; Sobiczewski, W. Relationship between selected DNA polymorphisms and coronary artery disease complications. Int. J. Cardiol. 2016, 228, 814–820. [Google Scholar] [CrossRef]
- Zhao, Q.; Wirka, R.; Nguyen, T.; Nagao, M.; Cheng, P.; Miller, C.L.; Kim, J.B.; Pjanic, M.; Quertermous, T. TCF21 and AP-1 interact through epigenetic modifi-cations to regulate coronary artery disease gene expression. Genome Med. 2019, 11, 23. [Google Scholar] [CrossRef]
- Nagao, M.; Lyu, Q.; Zhao, Q.; Wirka, R.C.; Bagga, J.; Nguyen, T.; Cheng, P.; Kim, J.; Pjanic, M.; Miano, J.M.; et al. Coronary Disease-Associated Gene TCF21 Inhibits Smooth Muscle Cell Differentiation by Blocking the Myocardin-Serum Response Factor Pathway. Circ. Res. 2020, 126, 517–529. [Google Scholar] [CrossRef]
- Bastami, M.; Ghaderian, S.M.; Omrani, M.D.; Mirfakhraie, R.; Vakili, H.; Parsa, S.A.; Nariman-Saleh-Fam, Z.; Masotti, A. MiRNA-Related Polymorphisms in miR-146a and TCF21 Are Associated with Increased Susceptibility to Coronary Artery Disease in an Iranian Population. Genet. Test. Mol. Biomark. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Aminu, A.J.; Petkova, M.; Chen, W.; Yin, Z.; Kuzmin, V.S.; Atkinson, A.J.; Dobrzynski, H. MiR-486-3p and MiR-938—Important Inhibitors of Pacemaking Ion Channels and/or Markers of Immune Cells. Appl. Sci. 2021, 11, 11366. [Google Scholar] [CrossRef]
- Yanni, J.; D’Souza, A.; Wang, Y.; Li, N.; Hansen, B.J.; Zakharkin, S.O.; Smith, M.; Hayward, C.; Whitson, B.A.; Mohler, P.J.; et al. Silencing miR-370-3p rescues funny current and sinus node function in heart failure. Sci. Rep. 2020, 10, 11279. [Google Scholar] [CrossRef]
- Carmona, R.; Guadix, J.A.; Cano, E.; Ruiz-Villalba, A.; Portillo-Sánchez, V.; Pérez-Pomares, J.M.; Muñoz-Chápuli, R. The embryonic epicardium: An essential element of cardiac development. J. Cell. Mol. Med. 2010, 14, 2066–2072. [Google Scholar] [CrossRef]
- Martínez-Estrada, O.M.; Lettice, A.L.; Essafi, A.; Guadix, J.A.; Slight, J.; Velecela, V.; Hall, E.; Reichmann, J.; Devenney, P.S.; Hohenstein, P.; et al. Wt1 is required for cardiovascular progenitor cell formation through transcriptional control of Snail and E-cadherin. Nat. Genet. 2009, 42, 89–93. [Google Scholar] [CrossRef]
- von Gise, A.; Zhou, B.; Honor, L.B.; Ma, Q.; Petryk, A.; Pu, W.T. WT1 regulates epicardial epithelial to mesenchymal transition through β-catenin and retinoic acid signaling pathways. Dev. Biol. 2011, 356, 421–431. [Google Scholar] [CrossRef]
- Bakker, M.L.; Christoffels, V.M.; Moorman, A.F.M. The Cardiac Pacemaker and Conduction System Develops From Embryonic Myocardium that Retains Its Primitive Phenotype. J. Cardiovasc. Pharmacol. 2010, 56, 6–15. [Google Scholar] [CrossRef]
- Lv, L.; Chen, G.; Zhou, J.; Li, J.; Gong, J. WT1-AS promotes cell apoptosis in hepatocellular carcinoma through down-regulating of WT1. J. Exp. Clin. Cancer Res. 2015, 34, 119. [Google Scholar] [CrossRef]
- Zhang, Y.; Na, R.; Wang, X. LncRNA WT1-AS up-regulates p53 to inhibit the proliferation of cervical squamous carcino-ma cells. BMC Cancer 2019, 19, 1052. [Google Scholar] [CrossRef]
- Wang, Q.; Ge, X.; Zhang, J.; Chen, L. Effect of lncRNA WT1-AS regulating WT1 on oxidative stress injury and apoptosis of neurons in Alzheimer’s disease via inhibition of the miR-375/SIX4 axis. Aging 2020, 12, 23974–23995. [Google Scholar] [CrossRef]
- Christoffels, V.M.; Smits, G.J.; Kispert, A.; Moorman, A.F.M. Development of the Pacemaker Tissues of the Heart. Circ. Res. 2010, 106, 240–254. [Google Scholar] [CrossRef]
- Hoogaars, W.M.H.; Barnett, P.; Moorman, A.F.M.; Christoffels, V.M. Cardiovascular development: Towards biomedical applicability. Cell. Mol. Life Sci. 2007, 64, 646. [Google Scholar] [CrossRef]
- Singh, R.; Hoogaars, W.M.; Barnett, P.; Grieskamp, T.; Rana, M.S.; Buermans, H.; Farin, H.F.; Petry, M.; Heallen, T.; Martin, J.F.; et al. Tbx2 and Tbx3 induce atrioventricular myocardial development and endocardial cushion formation. Cell. Mol. Life Sci. 2012, 69, 1377–1389. [Google Scholar] [CrossRef]
- Wang, J.; Bai, Y.; Li, N.; Ye, W.; Zhang, M.; Greene, S.B.; Tao, Y.; Chen, Y.; Wehrens, X.H.; Martin, J.F. Pitx2-microRNA pathway that delimits sinoatrial node development and inhibits predisposition to atrial fibrillation. Proc. Natl. Acad. Sci. USA 2014, 111, 9181–9186. [Google Scholar] [CrossRef]
- Benzoni, P.; Nava, L.; Giannetti, F.; Guerini, G.; Gualdoni, A.; Bazzini, C.; Milanesi, R.; Bucchi, A.; Baruscotti, M.; Barbuti, A. Dual role of miR-1 in the development and function of sinoatrial cells. J. Mol. Cell. Cardiol. 2021, 157, 104–112. [Google Scholar] [CrossRef]
- D’Souza, A.; Bucchi, A.; Johnsen, A.B.; Logantha, S.J.; Monfredi, O.; Yanni, J.; Prehar, S.; Hart, G.; Cartwright, E.; Wisløff, U.; et al. Exercise training reduces resting heart rate via downregulation of the funny channel HCN4. Nat. Commun. 2014, 5, 3775. [Google Scholar] [CrossRef]
- Zhao, H.; Wang, F.; Zhang, W.; Yang, M.; Tang, Y.; Wang, X.; Zhao, Q.; Huang, C. Overexpression of TBX3 in human induced pluripotent stem cells (hiPSCs) increases their differentiation into cardiac pacemaker-like cells. Biomed. Pharmacother. 2020, 130, 110612. [Google Scholar] [CrossRef]
- Jiang, K.; Ren, C.; Nair, V. MicroRNA-137 represses Klf4 and Tbx3 during differentiation of mouse embryonic stem cells. Stem Cell Res. 2013, 11, 1299–1313. [Google Scholar] [CrossRef]
- Han, J.; Yuan, P.; Yang, H.; Zhang, J.; Soh, B.S.; Li, P.; Lim, S.L.; Cao, S.; Tay, J.; Orlov, Y.; et al. Tbx3 improves the germ-line competency of induced pluripotent stem cells. Nature 2010, 463, 1096–1100. [Google Scholar] [CrossRef]
- Russell, R.; Ilg, M.; Lin, Q.; Wu, G.; Lechel, A.; Bergmann, W.; Eiseler, T.; Linta, L.; Kumar, P.P.; Klingenstein, M.; et al. A Dynamic Role of TBX3 in the Pluripotency Circuitry. Stem Cell Rep. 2015, 5, 1155–1170. [Google Scholar] [CrossRef] [PubMed]
- Niwa, H.; Ogawa, K.; Shimosato, D.; Adachi, K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature 2009, 460, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Liu, M.; Zhao, C.; Chen, C.; Kong, Q.; Cai, Z.; Li, D. MiR-1/133 attenuates cardiomyocyte apoptosis and electrical remodeling in mice with viral myocarditis. Cardiol. J. 2020, 27, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, J.; Huo, C.; Ding, N.; Li, J.; Xiao, J.; Lin, X.; Cai, B.; Zhang, Y.; Xu, J. Dynamic Organization of lncRNA and Circular RNA Regulators Collectively Controlled Cardiac Differentiation in Humans. EBioMedicine 2017, 24, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Salguero-Jiménez, A.; Grego-Bessa, J.; D’Amato, G.; Jiménez-Borreguero, L.J.; De La Pompa, J.L. Myocardial Notch1-Rbpj deletion does not affect NOTCH signaling, heart development or function. PLoS ONE 2018, 13, e0203100. [Google Scholar] [CrossRef]
- MacGrogan, D.; Münch, J.; De La Pompa, J.L. Notch and interacting signalling pathways in cardiac development, disease, and regeneration. Nat. Rev. Cardiol. 2018, 15, 685–704. [Google Scholar] [CrossRef] [PubMed]
- Luxán, G.; D’Amato, G.; MacGrogan, D.; De La Pompa, J.L. Endocardial Notch Signaling in Cardiac Development and Disease. Circ. Res. 2016, 118, e1–e18. [Google Scholar] [CrossRef]
- Gassmann, M.; Casagranda, F.; Orioli, D.; Simon, H.; Lai, C.; Klein, R.; Lemke, G. Aberrant neural and cardiac development in mice lacking the ErbB4 neuregulin receptor. Nature 1995, 378, 390–394. [Google Scholar] [CrossRef]
- Lee, K.-F.; Simon, H.; Chen, H.; Bates, B.; Hung, M.-C.; Hauser, C. Requirement for neuregulin receptor erbB2 in neural and cardiac development. Nature 1995, 378, 394–398. [Google Scholar] [CrossRef]
- Rupert, C.; Coulombe, K.L. The Roles of Neuregulin-1 in Cardiac Development, Homeostasis, and Disease. Biomark. Insights 2015, 10s1, BMI-S20061. [Google Scholar] [CrossRef]
- Kirabo, A.; Ryzhov, S.; Gupte, M.; Sengsayadeth, S.; Gumina, R.J.; Sawyer, D.B.; Galindo, C.L. Neuregulin-1β induces proliferation, survival and paracrine signaling in normal human cardiac ventricular fibroblasts. J. Mol. Cell. Cardiol. 2017, 105, 59–69. [Google Scholar] [CrossRef]
- Sun, M.; Yan, X.; Bian, Y.; Caggiano, A.O.; Morgan, J.P. Improving murine embryonic stem cell differentiation into cardiomyocytes with neuregulin-1: Differential expression of microRNA. Am. J. Physiol. Physiol. 2011, 301, C21–C30. [Google Scholar] [CrossRef]
- von Gise, A.; Lin, Z.; Schlegelmilch, K.; Honor, L.B.; Pan, G.M.; Buck, J.N.; Ma, Q.; Ishiwata, T.; Zhou, B.; Camargo, F.D.; et al. YAP1, the nuclear target of Hippo signaling, stimulates heart growth through cardiomyocyte proliferation but not hypertrophy. Proc. Natl. Acad. Sci. USA 2012, 109, 2394–2399. [Google Scholar] [CrossRef]
- Xiao, Y.; Leach, J.; Wang, J.; Martin, J.F. Hippo/Yap Signaling in Cardiac Development and Regeneration. Curr. Treat. Opt. Cardiovasc. Med. 2016, 18, 38. [Google Scholar] [CrossRef]
- Moya, I.M.; Halder, G. Hippo–YAP/TAZ signalling in organ regeneration and regenerative medicine. Nat. Rev. Mol. Cell Biol. 2018, 20, 211–226. [Google Scholar] [CrossRef]
- Mia, M.M.; Singh, M.K. The Hippo Signaling Pathway in Cardiac Development and Diseases. Front. Cell Dev. Biol. 2019, 7, 211. [Google Scholar] [CrossRef]
- Xie, Y.; Wang, Q.; Gao, N.; Wu, F.; Lan, F.; Zhang, F.; Jin, L.; Huang, Z.; Ge, J.; Wang, H.; et al. MircroRNA-10b Promotes Human Embryonic Stem Cell-Derived Cardiomyocyte Proliferation via Novel Target Gene LATS1. Mol. Ther. Nucleic Acids 2019, 19, 437–445. [Google Scholar] [CrossRef]
- Wu, K.-H.; Xiao, Q.-R.; Yang, Y.; Xu, J.-L.; Zhang, F.; Liu, C.-M.; Zhang, Z.-M.; Lu, Y.-Q.; Huang, N.-P. MicroRNA-34a modulates the Notch signaling pathway in mice with congenital heart disease and its role in heart development. J. Mol. Cell. Cardiol. 2017, 114, 300–308. [Google Scholar] [CrossRef]
- Han, C.; Song, Y.; Lian, C. MiR-769 Inhibits Colorectal Cancer Cell Proliferation and Invasion by Targeting HEY1. Med. Sci. Monit. 2018, 24, 9232–9239. [Google Scholar] [CrossRef]
- Wei, B.; Liu, Y.; Guan, H. MicroRNA-145-5p attenuates high glucose-induced apoptosis by targeting the Notch signaling pathway in podocytes. Exp. Ther. Med. 2020, 19, 1915–1924. [Google Scholar] [CrossRef]
- Lina, S.; Lihong, Q.; Di, Y.; Bo, Y.; Xiaolin, L.; Jing, M. microRNA-146a and Hey2 form a mutual negative feedback loop to regulate the inflammatory response in chronic apical periodontitis. J. Cell. Biochem. 2018, 120, 645–657. [Google Scholar] [CrossRef]
- Wu, D.-C.; Zhang, M.-F.; Su, S.-G.; Fang, H.-Y.; Wang, X.-H.; He, D.; Xie, Y.-Y.; Liu, X.-H. HEY2, a target of miR-137, indicates poor outcomes and promotes cell proliferation and migration in hepatocellular carcinoma. Oncotarget 2016, 7, 38052–38063. [Google Scholar] [CrossRef]
- Bao, Y.; Lu, Y.; Feng, W.; Yu, H.; Tao, Y.; Shi, Q.; Chen, W.; Wang, X. COUP-TFII promotes epithelial-mesenchymal transition by inhibiting miR-34a expression in colorectal cancer. Int. J. Oncol. 2019, 54, 1337–1344. [Google Scholar] [CrossRef] [PubMed]
- Jeong, B.-C.; Kang, I.-H.; Hwang, Y.-C.; Kim, S.-H.; Koh, J.-T. MicroRNA-194 reciprocally stimulates osteogenesis and inhibits adipogenesis via regulating COUP-TFII expression. Cell Death Dis. 2014, 5, e1532. [Google Scholar] [CrossRef] [PubMed]
- Kang, I.-H.; Jeong, B.-C.; Hur, S.-W.; Choi, H.; Choi, S.-H.; Ryu, J.-H.; Hwang, Y.-C.; Koh, J.-T. MicroRNA-302a Stimulates Osteoblastic Differentiation by Repressing COUP-TFII Expression. J. Cell. Physiol. 2014, 230, 911–921. [Google Scholar] [CrossRef] [PubMed]
- Lu, F.; Langenbacher, A.; Chen, J.-N. Tbx20 drives cardiac progenitor formation and cardiomyocyte proliferation in zebrafish. Dev. Biol. 2016, 421, 139–148. [Google Scholar] [CrossRef]
- Gupta, M.K.; Rao, T.N. Hearty miR-363 controls HAND1 in cardiac cell specification. Stem Cell Res. Ther. 2014, 5, 89. [Google Scholar] [CrossRef]
- Wagh, V.; Pomorski, A.; Wilschut, K.J.; Piombo, S.; Bernstein, H.S. MicroRNA-363 negatively regulates the left ventricular determining transcription factor HAND1 in human embryonic stem cell-derived cardiomyocytes. Stem Cell Res. Ther. 2014, 5, 75. [Google Scholar] [CrossRef]
- Cao, L.; Kong, L.-P.; Yu, Z.-B.; Han, S.-P.; Bai, Y.-F.; Zhu, J.; Hu, X.; Zhu, C.; Zhu, S.; Guo, X.-R. microRNA expression profiling of the developing mouse heart. Int. J. Mol. Med. 2012, 30, 1095–1104. [Google Scholar] [CrossRef]
- Zhao, Y.; Samal, E.; Srivastava, D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 2005, 436, 214–220. [Google Scholar] [CrossRef]
- Vo, N.K.; Dalton, R.P.; Liu, N.; Olson, E.N.; Goodman, R.H. Affinity purification of microRNA-133a with the cardiac transcription factor, Hand2. Proc. Natl. Acad. Sci. USA 2010, 107, 19231–19236. [Google Scholar] [CrossRef]
- Liu, L.; Yuan, Y.; He, X.; Xia, X.; Mo, X. MicroRNA-1 upregulation promotes myocardiocyte proliferation and suppresses apoptosis during heart development. Mol. Med. Rep. 2017, 15, 2837–2842. [Google Scholar] [CrossRef][Green Version]
- Lavallée, G.; Andelfinger, G.; Nadeau, M.; Lefebvre, C.; Nemer, G.; Horb, M.E.; Nemer, M. The Kruppel-like transcription factor KLF13 is a novel regulator of heart development. EMBO J. 2006, 25, 5201–5213. [Google Scholar] [CrossRef]
- Gu, M.; Wang, J.; Wang, Y.; Xu, Y.; Zhang, Y.; Wu, W.; Liao, S. MiR-147b inhibits cell viability and promotes apoptosis of rat H9c2 cardiomyocytes via down-regulating KLF13 expression. Acta Biochim. Biophys. Sin. 2018, 50, 288–297. [Google Scholar] [CrossRef]
- Bayoumi, A.S.; Park, K.-M.; Wang, Y.; Teoh, J.-P.; Aonuma, T.; Tang, Y.; Su, H.; Weintraub, N.L.; Kim, I.-M. A carvedilol-responsive microRNA, miR-125b-5p protects the heart from acute myocardial infarction by repressing pro-apoptotic bak1 and klf13 in cardiomyocytes. J. Mol. Cell. Cardiol. 2017, 114, 72–82. [Google Scholar] [CrossRef]
- Mikhailov, A.T.; Torrado, M. Myocardial transcription factors in diastolic dysfunction: Clues for model systems and disease. Heart Fail. Rev. 2016, 21, 783–794. [Google Scholar] [CrossRef]
- Kong, L.; Hu, N.; Du, X.; Wang, W.; Chen, H.; Li, W.; Wei, S.; Zhuang, H.; Li, X.; Li, C. Upregulation of miR-483-3p contributes to endothelial progenitor cells dysfunction in deep vein thrombosis patients via SRF. J. Transl. Med. 2016, 14, 23. [Google Scholar] [CrossRef]
- Xu, D.; Guo, Y.; Liu, T.; Li, S.; Sun, Y. miR-22 contributes to endosulfan-induced endothelial dysfunction by targeting SRF in HUVECs. Toxicol. Lett. 2017, 269, 33–40. [Google Scholar] [CrossRef]
- You, G.; Zu, B.; Wang, B.; Fu, Q.; Li, F. Identification of miRNA–mRNA–TFs Regulatory Network and Crucial Pathways Involved in Tetralogy of Fallot. Front. Genet. 2020, 11, 552. [Google Scholar] [CrossRef]
- Wei, X.; Hou, X.; Li, J.; Liu, Y. miRNA-181a/b Regulates Phenotypes of Vessel Smooth Muscle Cells Through Serum Response Factor. DNA Cell Biol. 2017, 36, 127–135. [Google Scholar] [CrossRef]
- Bolte, C.; Zhang, Y.; Wang, I.-C.; Kalin, T.V.; Molkentin, J.D.; Kalinichenko, V.V. Expression of Foxm1 Transcription Factor in Cardiomyocytes Is Required for Myocardial Development. PLoS ONE 2011, 6, e22217. [Google Scholar] [CrossRef]
- Chen, F.; Kook, H.; Milewski, R.; Gitler, A.D.; Lu, M.M.; Li, J.; Nazarian, R.; Schnepp, R.; Jen, K.-Y.; Biben, C.; et al. Hop Is an Unusual Homeobox Gene that Modulates Cardiac Development. Cell 2002, 110, 713–723. [Google Scholar] [CrossRef]
- Hadji, F.; Boulanger, M.-C.; Guay, S.-P.; Gaudreault, N.; Amellah, S.; Mkannez, G.; Bouchareb, R.; Marchand, J.T.; Nsaibia, M.J.; Guauque-Olarte, S.; et al. Altered DNA Methylation of Long Noncoding RNA H19 in Calcific Aortic Valve Disease Promotes Mineralization by Silencing NOTCH1. Circulation 2016, 134, 1848–1862. [Google Scholar] [CrossRef]
- Anderson, K.M.; Anderson, D.M.; McAnally, J.R.; Shelton, J.M.; Bassel-Duby, R.; Olson, E.N. Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature 2016, 539, 433–436. [Google Scholar] [CrossRef]
- Han, X.; Zhang, J.; Liu, Y.; Fan, X.; Ai, S.; Luo, Y.; Li, X.; Jin, H.; Luo, S.; Zheng, H.; et al. The lncRNA Hand2os1/Uph locus orchestrates heart development through regulation of precise expression of Hand2. Development 2019, 146, dev176198. [Google Scholar] [CrossRef]
- Anderson, D.M.; Anderson, K.M.; Nelson, B.R.; McAnally, J.R.; Bezprozvannaya, S.; Shelton, J.M.; Bassel-Duby, R.; Olson, E.N. A myocardin-adjacent lncRNA balances SRF-dependent gene transcription in the heart. Genes Dev. 2021, 35, 835–840. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, W.; Lin, M.; Wu, W.; Jiang, P.; Tou, E.; Xue, M.; Richards, A.; Jourd’Heuil, D.; Asif, A.; et al. MYOSLID Is a Novel Serum Response Factor–Dependent Long Noncoding RNA That Amplifies the Vascular Smooth Muscle Differentiation Program. Arter. Thromb. Vasc. Biol. 2016, 36, 2088–2099. [Google Scholar] [CrossRef]
- Liu, J.; Li, Y.; Lin, B.; Sheng, Y.; Yang, L. HBL1 Is a Human Long Noncoding RNA that Modulates Cardiomyocyte Development from Pluripotent Stem Cells by Counteracting MIR1. Dev. Cell 2017, 42, 333–348.e5. [Google Scholar] [CrossRef]
- Xiao, X.; Zhou, T.; Guo, S.; Guo, C.; Zhang, Q.; Dong, N.; Wang, Y. LncRNA MALAT1 sponges miR-204 to promote osteoblast differentiation of human aortic valve interstitial cells through up-regulating Smad4. Int. J. Cardiol. 2017, 243, 404–412. [Google Scholar] [CrossRef]
- Leisegang, M.S.; Bibli, S.-I.; Günther, S.; Pflüger-Müller, B.; Oo, J.A.; Höper, C.; Seredinski, S.; Yekelchyk, M.; Schmitz-Rixen, T.; Schürmann, C.; et al. Pleiotropic effects of laminar flow and statins depend on the Krüppel-like factor-induced lncRNA MANTIS. Eur. Heart J. 2019, 40, 2523–2533. [Google Scholar] [CrossRef] [PubMed]
- Shyu, K.; Wang, B.; Fang, W.; Pan, C.; Lin, C. Hyperbaric oxygen-induced long non-coding RNA MALAT1 exosomes suppress MicroRNA-92a expression in a rat model of acute myocardial infarction. J. Cell. Mol. Med. 2020, 24, 12945–12954. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Wang, J.; Morikawa, Y.; Bonilla-Claudio, M.; Klysik, E.; Martin, J.F. Bmp signaling represses Vegfa to promote outflow tract cushion development. Development 2013, 140, 3395–3402. [Google Scholar] [CrossRef] [PubMed]
- Stottmann, R.W.; Choi, M.; Mishina, Y.; Meyers, E.N.; Klingensmith, J. BMP receptor IA is required in mammalian neural crest cells for development of the cardiac outflow tract and ventricular myocardium. Development 2004, 131, 2205–2218. [Google Scholar] [CrossRef]
- Cai, C.-L.; Liang, X.; Shi, Y.; Chu, P.-H.; Pfaff, S.L.; Chen, J.; Evans, S. Isl1 Identifies a Cardiac Progenitor Population that Proliferates Prior to Differentiation and Contributes a Majority of Cells to the Heart. Dev. Cell 2003, 5, 877–889. [Google Scholar] [CrossRef]
- Engleka, K.A.; Manderfield, L.J.; Brust, R.D.; Li, L.; Cohen, A.; Dymecki, S.M.; Epstein, J.A. Islet1Derivatives in the Heart Are of Both Neural Crest and Second Heart Field Origin. Circ. Res. 2012, 110, 922–926. [Google Scholar] [CrossRef]
- Liu, W.; Selever, J.; Wang, D.; Lu, M.-F.; Moses, K.A.; Schwartz, R.J.; Martin, J.F. Bmp4 signaling is required for outflow-tract septation and branchial-arch artery remodeling. Proc. Natl. Acad. Sci. USA 2004, 101, 4489–4494. [Google Scholar] [CrossRef]
- Solloway, M.J.; Robertson, E.J. Early embryonic lethality in Bmp5; Bmp7 double mutant mice suggests functional redundancy within the 60A subgroup. Development 1999, 126, 1753–1768. [Google Scholar] [CrossRef]
- Dobaczewski, M.; Chen, W.; Frangogiannis, N.G. Transforming growth factor (TGF)-β signaling in cardiac remodeling. J. Mol. Cell. Cardiol. 2011, 51, 600–606. [Google Scholar] [CrossRef]
- Hurlstone, A.; Haramis, A.-P.G.; Wienholds, E.; Begthel, H.; Korving, J.; Van Eeden, F.; Cuppen, E.; Zivkovic, D.; Plasterk, R.H.A.; Clevers, H. The Wnt/β-catenin pathway regulates cardiac valve formation. Nature 2003, 425, 633–637. [Google Scholar] [CrossRef]
- Jarrett, M.J.; Houk, A.K.; McCuistion, P.E.; Weyant, M.J.; Reece, T.B.; Meng, X.; Fullerton, D.A. Wnt Signaling Mediates Pro-Fibrogenic Activity in Human Aortic Valve Interstitial Cells. Ann. Thorac. Surg. 2021, 112, 519–525. [Google Scholar] [CrossRef]
- Cai, X.; Zhang, W.; Hu, J.; Zhang, L.; Sultana, N.; Wu, B.; Cai, W.; Zhou, B.; Cai, C.-L. Tbx20 acts upstream of Wnt signaling to regulate endocardial cushion formation and valve remodeling during mouse cardiogenesis. Development 2013, 140, 3176–3187. [Google Scholar] [CrossRef]
- Kim, Y.-S.; Kim, M.-J.; Koo, T.-H.; Kim, J.-D.; Koun, S.; Ham, H.J.; Lee, Y.M.; Rhee, M.; Yeo, S.-Y.; Huh, T.-L. Histone deacetylase is required for the activation of Wnt/β-catenin signaling crucial for heart valve formation in zebrafish embryos. Biochem. Biophys. Res. Commun. 2012, 423, 140–146. [Google Scholar] [CrossRef]
- Alfieri, C.M.; Cheek, J.; Chakraborty, S.; Yutzey, K.E. Wnt signaling in heart valve development and osteogenic gene induction. Dev. Biol. 2010, 338, 127–135. [Google Scholar] [CrossRef]
- Verhoeven, M.C.; Haase, C.; Christoffels, V.M.; Weidinger, G.; Bakkers, J. Wnt signaling regulates atrioventricular canal formation upstream of BMP and Tbx2. Birth Defects Res. Part A: Clin. Mol. Teratol. 2011, 91, 435–440. [Google Scholar] [CrossRef]
- Bosada, F.M.; Devasthali, V.; Jones, K.A.; Stankunas, K. Wnt/β-catenin signaling enables developmental transitions during valvulogenesis. Development 2016, 143, 1041–1054. [Google Scholar] [CrossRef]
- Fang, M.; Wang, C.; Zheng, C.; Luo, J.; Hou, S.; Liu, K.; Li, X. Mir-29b promotes human aortic valve interstitial cell calcification via inhibiting TGF-β3 through activation of wnt3/β-catenin/Smad3 signaling. J. Cell. Biochem. 2017, 119, 5175–5185. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, Z.; Zhang, L.; Wang, J.; Zhang, M.; Zhu, B. miR-27a protects human mitral valve interstitial cell from TNF-α-induced inflammatory injury via up-regulation of NELL-1. Braz. J. Med Biol. Res. 2018, 51. [Google Scholar] [CrossRef]
- Lv, X.; Li, J.; Hu, Y.; Wang, S.; Yang, C.; Li, C.; Zhong, G. Overexpression of miR-27b-3p Targeting Wnt3a Regulates the Signaling Pathway of Wnt/β-Catenin and Attenuates Atrial Fibrosis in Rats with Atrial Fibrillation. Oxid. Med. Cell. Longev. 2019, 2019, 1–13. [Google Scholar] [CrossRef]
- Yang, M.; Kong, D.; Chen, J. Inhibition of miR-148b ameliorates myocardial ischemia/reperfusion injury via regulation of Wnt/β-catenin signaling pathway. J. Cell. Physiol. 2019, 234, 17757–17766. [Google Scholar] [CrossRef]
- Nakajima, Y.; Miyazono, K.; Kato, M.; Takase, M.; Yamagishi, T.; Nakamura, H. Extracellular Fibrillar Structure of Latent TGFβ Binding Protein-1: Role in TGFβ-dependent Endothelial-Mesenchymal Transformation during Endocardial Cushion Tissue Formation in Mouse Embryonic Heart. J. Cell Biol. 1997, 136, 193–204. [Google Scholar] [CrossRef]
- Brown, C.B.; Boyer, A.S.; Runyan, R.B.; Barnett, J.V. Requirement of Type III TGF-β Receptor for Endocardial Cell Transformation in the Heart. Science 1999, 283, 2080–2082. [Google Scholar] [CrossRef]
- Todorovic, V.; Finnegan, E.; Freyer, L.; Zilberberg, L.; Ota, M.; Rifkin, D.B. Long form of latent TGF-β binding protein 1 (Ltbp1L) regulates cardiac valve development. Dev. Dyn. 2010, 240, 176–187. [Google Scholar] [CrossRef]
- Ten Dijke, P.; Egorova, A.D.; Goumans, M.-J.T.H.; Poelmann, R.E.; Hierck, B.P. TGF-β Signaling in Endothelial-to-Mesenchymal Transition: The Role of Shear Stress and Primary Cilia. Sci. Signal. 2012, 5, pt2. [Google Scholar] [CrossRef]
- Wang, J.; Sridurongrit, S.; Dudas, M.; Thomas, P.; Nagy, A.; Schneider, M.; Epstein, J.A.; Kaartinen, V. Atrioventricular cushion transformation is mediated by ALK2 in the developing mouse heart. Dev. Biol. 2005, 286, 299–310. [Google Scholar] [CrossRef]
- Ramsdell, A.F.; Markwald, R.R. Induction of Endocardial Cushion Tissue in the Avian Heart is Regulated, in Part, by TGFβ-3-Mediated Autocrine Signaling. Dev. Biol. 1997, 188, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Townsend, T.A.; Robinson, J.Y.; How, T.; DeLaughter, D.M.; Blobe, G.C.; Barnett, J.V. Endocardial cell epithelial-mesenchymal transformation requires Type III TGFβ receptor interaction with GIPC. Cell. Signal. 2012, 24, 247–256. [Google Scholar] [CrossRef]
- Lagendijk, A.K.; Goumans, M.J.; Burkhard, S.B.; Bakkers, J. MicroRNA-23 Restricts Cardiac Valve Formation by Inhibiting Has2 and Extracellular Hyaluronic Acid Production. Circ. Res. 2011, 109, 649–657. [Google Scholar] [CrossRef]
- Bonet, F.; Dueñas, Á.; López-Sánchez, C.; García-Martínez, V.; Aránega, A.E.; Franco, D. MiR-23b and miR-199a impair epithelial-to-mesenchymal transition during atrioventricular endocardial cushion formation. Dev. Dyn. 2015, 244, 1259–1275. [Google Scholar] [CrossRef]
- Koenig, S.N.; Bosse, K.; Majumdar, U.; Bonachea, E.M.; Radtke, F.; Garg, V. Endothelial Notch1 Is Required for Proper Development of the Semilunar Valves and Cardiac Outflow Tract. J. Am. Heart Assoc. 2016, 5, e003075. [Google Scholar] [CrossRef]
- Yang, J.-H.; Wylie-Sears, J.; Bischoff, J. Opposing actions of Notch1 and VEGF in post-natal cardiac valve endothelial cells. Biochem. Biophys. Res. Commun. 2008, 374, 512–516. [Google Scholar] [CrossRef] [PubMed]
- Nigam, V.; Srivastava, D. Notch1 represses osteogenic pathways in aortic valve cells. J. Mol. Cell. Cardiol. 2009, 47, 828–834. [Google Scholar] [CrossRef] [PubMed]
- Bosse, K.; Hans, C.P.; Zhao, N.; Koenig, S.N.; Huang, N.; Guggilam, A.; LaHaye, S.; Tao, G.; Lucchesi, P.A.; Lincoln, J.; et al. Endothelial nitric oxide signaling regulates Notch1 in aortic valve disease. J. Mol. Cell. Cardiol. 2013, 60, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Timmerman, L.A.; Grego-Bessa, J.; Raya, A.; Bertrán, E.; Pérez-Pomares, J.M.; Díez, J.; Aranda, S.; Palomo, S.; McCormick, F.; Izpisúa-Belmonte, J.C.; et al. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 2003, 18, 99–115. [Google Scholar] [CrossRef]
- Torregrosa-Carrión, R.; Zurita, L.L.; García-Marqués, F.; D’Amato, G.; Piñeiro-Sabarís, R.; Bonzón-Kulichenko, E.; Vázquez, J.; de la Pompa, J.L. NOTCH Activation Promotes Valve Formation by Regulating the Endocardial Secretome. Mol. Cell. Proteom. 2019, 18, 1782–1795. [Google Scholar] [CrossRef]
- Toshima, T.; Watanabe, T.; Narumi, T.; Otaki, Y.; Shishido, T.; Aono, T.; Goto, J.; Watanabe, K.; Sugai, T.; Takahashi, T.; et al. Therapeutic inhibition of microRNA-34a ameliorates aortic valve calcification via modulation of Notch1-Runx2 signaling. Cardiovasc. Res. 2020, 116, 983–994. [Google Scholar] [CrossRef]
- Raddatz, A.M.; Roest, M.J.V.; Merryman, W.D. Notch1 suppression by microRNA-34a: A new mechanism of calcific aortic valve disease. Cardiovasc. Res. 2019, 116, 871–873. [Google Scholar] [CrossRef]
- Wang, L.; Tang, R.; Zhang, Y.; Chen, S.; Guo, Y.; Wang, X.; Liu, Z.; Liu, H.; Zhang, X.; Liu, B.C. PTH-induced EndMT via miR-29a-5p/GSAP/Notch1 pathway contributed to valvular calcification in rats with CKD. Cell Prolif. 2021, 54, e13018. [Google Scholar] [CrossRef]
- Dai, Y.; Yan, T.; Gao, Y. Silence of miR-32-5p promotes endothelial cell viability by targeting KLF2 and serves as a diagnostic biomarker of acute myocardial infarction. Diagn. Pathol. 2020, 15, 19. [Google Scholar] [CrossRef]
- Natarelli, L.; Schober, A. Janus-Faced Role of Krüppel-Like Factor 2–Dependent Regulation of MicroRNAs in Endothelial Proliferation. Arter. Thromb. Vasc. Biol. 2014, 34, 1605–1606. [Google Scholar] [CrossRef][Green Version]
- Liu, H.; Li, G.; Zhao, W.; Hu, Y. Inhibition of MiR-92a may protect endothelial cells after acute myocardial infarction in rats: Role of KLF2/4. Med. Sci. Monit. 2016, 22, 2451–2462. [Google Scholar] [CrossRef]
- Demolli, S.; Doebele, C.; Doddaballapur, A.; Lang, V.; Fisslthaler, B.; Chavakis, E.; Vinciguerra, M.; Sciacca, S.; Henschler, R.; Hecker, M.; et al. MicroRNA-30 mediates anti-inflammatory effects of shear stress and KLF2 via repression of angiopoietin 2. J. Mol. Cell. Cardiol. 2015, 88, 111–119. [Google Scholar] [CrossRef]
- Hergenreider, E.; Heydt, S.; Tréguer, K.; Boettger, T.; Horrevoets, A.J.G.; Zeiher, A.M.; Scheffer, M.P.; Frangakis, A.S.; Yin, X.; Mayr, M.; et al. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat. Cell Biol. 2012, 14, 249–256. [Google Scholar] [CrossRef]
- Sindi, H.A.; Russomanno, G.; Satta, S.; Salam, V.A.; Jo, K.B.; Qazi-Chaudhry, B.; Ainscough, A.J.; Szulcek, R.; Bogaard, H.J.; Morgan, C.C.; et al. Therapeutic potential of KLF2-induced exosomal microRNAs in pulmonary hypertension. Nat. Commun. 2020, 11, 1185. [Google Scholar] [CrossRef]
- Cui, S.; Liu, Z.; Tao, B.; Fan, S.; Pu, Y.; Meng, X.; Li, D.; Xia, H.; Xu, L. MiR-145 attenuates cardiac fibrosis through the AKT/GSK-3β/β-catenin signaling pathway by directly targeting SOX9 in fibroblasts. J. Cell Physiol. 2020, 122, 209–221. [Google Scholar] [CrossRef]
- Li, J.; Cao, Y.; Ma, X.-J.; Wang, H.-J.; Zhang, J.; Luo, X.; Chen, W.; Wu, Y.; Meng, Y.; Yuan, Y.; et al. Roles of miR-1-1 and miR-181c in ventricular septal defects. Int. J. Cardiol. 2013, 168, 1441–1446. [Google Scholar] [CrossRef]
- Cheng, N.; Li, L.; Wu, Y.; Wang, M.; Yang, M.; Wei, S.; Wang, R. microRNA-30e up-regulation alleviates myocardial ischemia-reperfusion injury and promotes ventricular remodeling via SOX9 repression. Mol. Immunol. 2020, 130, 96–103. [Google Scholar] [CrossRef]
- Zhang, Y.; Tian, C.; Liu, X.; Zhang, H. Identification of Genetic Biomarkers for Diagnosis of Myocardial Infarction Compared with Angina Patients. Cardiovasc. Ther. 2020, 2020, 8535314. [Google Scholar] [CrossRef]
- Saadat, S.; Noureddini, M.; Mahjoubin-Tehran, M.; Nazemi, S.; Shojaie, L.; Aschner, M.; Maleki, B.; Abbasi-Kolli, M.; Rajabi Moghadam, H.; Alani, B.; et al. Pivot-al Role of TGF-β/Smad Signaling in Cardiac Fibrosis: Non-coding RNAs as Effectual Players. Front. Cardiovasc. Med. 2021, 7, 256. [Google Scholar] [CrossRef]
- Milano, G.; Biemmi, V.; Lazzarini, E.; Balbi, C.; Ciullo, A.; Bolis, S.; Ameri, P.; Di Silvestre, D.; Mauri, P.; Barile, L.; et al. Intravenous administration of cardiac progenitor cell-derived exosomes protects against doxorubicin/trastuzumab-induced cardiac toxicity. Cardiovasc. Res. 2020, 116, 383–392. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, Q.; Feng, Y.; Chen, X.; Yang, L.; Xu, M.; Wang, X.; Li, W.; Niu, X.; Gao, D. MicroRNA-26a Protects the Heart Against Hypertension-Induced Myocardial Fibrosis. J. Am. Heart Assoc. 2020, 9, e017970. [Google Scholar] [CrossRef]
- Dong, D.; Zhang, Y.; Reece, E.A.; Wang, L.; Harman, C.R.; Yang, P. microRNA expression profiling and functional annotation analysis of their targets modulated by oxidative stress during embryonic heart development in diabetic mice. Reprod. Toxicol. 2016, 65, 365–374. [Google Scholar] [CrossRef]
- Baldini, A.; Fulcoli, F.; Illingworth, E. Chapter Eight—Tbx1: Transcriptional and Developmental Functions. Curr. Top. Dev. Biol. 2017, 122, 223–243. [Google Scholar] [CrossRef]
- Pan, Y.; Wang, Z.-G.; Liu, X.-Y.; Zhao, H.; Zhou, N.; Zheng, G.-F.; Qiu, X.-B.; Li, R.-G.; Yuan, F.; Shi, H.-Y.; et al. A Novel TBX1 Loss-of-Function Mutation Associated with Congenital Heart Disease. Pediatr. Cardiol. 2015, 36, 1400–1410. [Google Scholar] [CrossRef]
- Cao, M.-L.; Zhu, B.-L.; Sun, Y.-Y.; Qiu, G.-R.; Fu, W.-N.; Jiang, H.-K. MicroRNA-144 Regulates Cardiomyocyte Proliferation and Apoptosis by Targeting TBX1 through the JAK2/STAT1 Pathway. Cytogenet. Genome Res. 2019, 159, 190–200. [Google Scholar] [CrossRef]
- Aravalli, R.N.; Greten, T.F. FoxC1: Novel Regulator of Inflammation-Induced Metastasis in Hepatocellular Carcinoma. Gastroenterology 2015, 149, 861–863. [Google Scholar] [CrossRef] [PubMed]
- Prasitsak, T.; Nandar, M.; Okuhara, S.; Ichinose, S.; Ota, M.S.; Iseki, S. Foxc1is required for early stage telencephalic vascular development. Dev. Dyn. 2015, 244, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Lambers, E.; Arnone, B.; Fatima, A.; Qin, G.; Wasserstrom, J.A.; Kume, T. Foxc1 Regulates Early Cardiomyogenesis and Functional Properties of Embryonic Stem Cell Derived Cardiomyocytes. Stem Cells 2016, 34, 1487–1500. [Google Scholar] [CrossRef] [PubMed]
- Henn, D.; Abu-Halima, M.; Wermke, D.; Falkner, F.; Thomas, B.; Köpple, C.; Ludwig, N.; Schulte, M.; Brockmann, M.A.; Kim, Y.-J.; et al. MicroRNA-regulated pathways of flow-stimulated angiogenesis and vascular remodeling in vivo. J. Transl. Med. 2019, 17, 22. [Google Scholar] [CrossRef]
- Mellor, R.H.; Brice, G.; Stanton, A.W.; French, J.; Smith, A.; Jeffery, S.; Levick, J.R.; Burnand, K.G.; Mortimer, P.S. Mutations in FOXC2 Are Strongly Associated With Primary Valve Failure in Veins of the Lower Limb. Circulation 2007, 115, 1912–1920. [Google Scholar] [CrossRef]
- Zeng, N.; Huang, Y.-Q.; Yan, Y.-M.; Hu, Z.-Q.; Zhang, Z.; Feng, J.-X.; Guo, J.-S.; Zhu, J.-N.; Fu, Y.-H.; Wang, X.-P.; et al. Diverging targets mediate the pathological roleof miR-199a-5p and miR-199a-3p by promoting cardiac hypertrophy and fibrosis. Mol. Ther. Nucleic Acids 2021, 26, 1035–1050. [Google Scholar] [CrossRef]
- Cao, Y.; Cao, Z.; Wang, W.; Jie, X.; Li, L. MicroRNA-199a-5p regulates FOXC2 to control human vascular smooth muscle cell phenotypic switch. Mol. Med. Rep. 2021, 24, 627. [Google Scholar] [CrossRef]
- Du, B.; Cawthorn, W.P.; Su, A.; Doucette, C.R.; Yao, Y.; Hemati, N.; Kampert, S.; McCoin, C.; Broome, D.T.; Rosen, C.J.; et al. The Transcription Factor Paired-Related Homeobox 1 (Prrx1) Inhibits Adipogenesis by Activating Transforming Growth Factor-β (TGFβ) Signaling. J. Biol. Chem. 2013, 288, 3036–3047. [Google Scholar] [CrossRef]
- Zhong, X.; Yu, X.; Wen, X.; Chen, L.; Gu, N. Activation of the LINC00242/miR-141/FOXC1 axis underpins the development of gastric cancer. Cancer Cell Int. 2020, 20, 272. [Google Scholar] [CrossRef]
- Jiang, Y.; Mo, H.; Luo, J.; Zhao, S.; Liang, S.; Zhang, M.; Yuan, J. HOTAIR Is a Potential Novel Biomarker in Patients with Congenital Heart Diseases. BioMed Res. Int. 2018, 2018, 850657. [Google Scholar] [CrossRef]
- Moorman, A.F.M.; Christoffels, V.M. Cardiac Chamber Formation: Development, Genes, and Evolution. Physiol. Rev. 2003, 83, 1223–1267. [Google Scholar] [CrossRef]
- Kelly, R.G.; Buckingham, M.E.; Moorman, A.F. Heart Fields and Cardiac Morphogenesis. Cold Spring Harb. Perspect. Med. 2014, 4, a015750. [Google Scholar] [CrossRef]
- Kelly, R.G.; Buckingham, M.E. The anterior heart-forming field: Voyage to the arterial pole of the heart. Trends Genet. 2002, 18, 210–216. [Google Scholar] [CrossRef]
- Erhardt, S.; Zheng, M.; Zhao, X.; Le, T.; Findley, T.; Wang, J. The Cardiac Neural Crest Cells in Heart Development and Congenital Heart Defects. J. Cardiovasc. Dev. Dis. 2021, 8, 89. [Google Scholar] [CrossRef]
- Saretzki, E.; Pankratz, F.; Engert, B.; Grundmann, S.; Bode, C.; Moser, M.; Zhou, Q.; Esser, J.S. Bone morphogenetic protein 4 regulates microRNAs miR-494 and miR-126–5p in control of endothelial cell function in angiogenesis. Thromb. Haemost. 2017, 117, 734–749. [Google Scholar] [CrossRef]
- Yanagawa, B.; Lovren, F.; Pan, Y.; Garg, V.; Quan, A.; Tang, G.; Singh, K.K.; Shukla, P.C.; Kalra, N.P.; Peterson, M.D.; et al. miRNA-141 is a novel regulator of BMP-2–mediated calcification in aortic stenosis. J. Thorac. Cardiovasc. Surg. 2012, 144, 256–262.e2. [Google Scholar] [CrossRef]
- Balderman, J.A.F.; Lee, H.; Mahoney, C.E.; Handy, D.E.; White, K.; Annis, S.; Lebeche, D.; Hajjar, R.J.; Loscalzo, J.; Leopold, J.A. Bone Morphogenetic Protein-2 Decreases MicroRNA-30b and MicroRNA-30c to Promote Vascular Smooth Muscle Cell Calcification. J. Am. Heart Assoc. 2012, 1, e003905. [Google Scholar] [CrossRef]
- Song, R.; Fullerton, D.A.; Ao, L.; Zhao, K.-S.; Meng, X. An epigenetic regulatory loop controls pro-osteogenic activation by TGF-β1 or bone morphogenetic protein 2 in human aortic valve interstitial cells. J. Biol. Chem. 2017, 292, 8657–8666. [Google Scholar] [CrossRef]
- Yousefi, F.; Shabaninejad, Z.; Vakili, S.; Derakhshan, M.; Movahedpour, A.; Dabiri, H.; Ghasemi, Y.; Mahjoubin-Tehran, M.; Nikoozadeh, A.; Savardashtaki, A.; et al. TGF-β and WNT signaling pathways in cardiac fibrosis: Non-coding RNAs come into focus. Cell Commun. Signal. 2020, 18, 87. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, Y.; Zhu, H.; Hu, J.; Xie, Z. MiR-34a/miR-93 target c-Ski to modulate the proliferaton of rat cardiac fibroblasts and extracellular matrix deposition in vivo and in vitro. Cell. Signal. 2018, 46, 145–153. [Google Scholar] [CrossRef]
- He, X.; Zhang, K.; Gao, X.; Li, L.; Tan, H.; Chen, J.; Zhou, Y. Rapid atrial pacing induces myocardial fibrosis by down-regulating Smad7 via microRNA-21 in rabbit. Heart Vessel. 2016, 31, 1696–1708. [Google Scholar] [CrossRef]
- Zhang, D.; Cui, Y.; Li, B.; Luo, X.; Li, B.; Tang, Y. MiR-155 regulates high glucose-induced cardiac fibrosis via the TGF-β signaling pathway. Mol. BioSyst. 2016, 13, 215–224. [Google Scholar] [CrossRef]
- Wang, Y.; Cai, H.; Li, H.; Gao, Z.; Song, K. Atrial overexpression of microRNA-27b attenuates angiotensin II-induced atrial fibrosis and fibrillation by targeting ALK5. Hum. Cell 2018, 31, 251–260. [Google Scholar] [CrossRef]
- Hong, Y.; Cao, H.; Wang, Q.; Ye, J.; Sui, L.; Feng, J.; Cai, X.; Song, H.; Zhang, X.; Chen, X. MiR-22 may Suppress Fibrogenesis by Targeting TGFβR I in Cardiac Fibroblasts. Cell. Physiol. Biochem. 2016, 40, 1345–1353. [Google Scholar] [CrossRef]
- Yu, R.-B.; Li, K.; Wang, G.; Gao, G.-M.; Du, J.-X. MiR-23 enhances cardiac fibroblast proliferation and suppresses fibroblast apoptosis via targeting TGF-β1 in atrial fibrillation. Eur. Rev. Med Pharmacol. Sci. 2019, 23, 4419–4424. [Google Scholar]
- Xu, J.; Wu, H.; Chen, S.; Qi, B.; Zhou, G.; Cai, L.; Zhao, L.; Wei, Y.; Liu, S. Micro RNA -30c suppresses the pro-fibrogenic effects of cardiac fibroblasts induced by TGF -β1 and prevents atrial fibrosis by targeting TGF β RII. J. Cell. Mol. Med. 2018, 22, 3045–3057. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.J.Y.; Zhuang, J.Z.J.; Lin, Y.L.Y.; Wang, X.W.X.; Chen, J.C.J.; Han, F.H.F. Long noncoding RNA SNHG6 contributes to ventricular septal defect formation via negative regulation of miR-101 and activation of Wnt/β-catenin pathway. Die Pharm. -Int. J. Pharm. Sci. 2019, 74, 23–28. [Google Scholar] [CrossRef]
- Zhao, X.; Wei, X.; Wang, X.; Qi, G. Long non-coding RNA NORAD regulates angiogenesis of human umbilical vein endothelial cells via miR-590-3p under hypoxic conditions. Mol. Med. Rep. 2020, 21, 2560–2570. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Luo, M.; Gao, Z.; Han, X.; Yan, Z.; Xie, S.; Zhao, H.; Sun, H. TUG1 knockdown suppresses cardiac fibrosis after myocardial infarction. Mamm. Genome 2021, 32, 435–442. [Google Scholar] [CrossRef] [PubMed]
- García-Padilla, C.; Domínguez, J.N.; Lodde, V.; Munk, R.; Abdelmohsen, K.; Gorospe, M.; Jiménez-Sábado, V.; Ginel, A.; Hove-Madsen, L.; Aránega, A.E.; et al. Identification of atrial-enriched lncRNA Walras linked to cardiomyocyte cytoarchitecture and atrial fibrillation. FASEB J. 2021, 36, e22051. [Google Scholar] [CrossRef]
- Das, A.; Shyamal, S.; Sinha, T.; Mishra, S.S.; Panda, A.C. Identification of Potential circRNA-microRNA-mRNA Regulatory Network in Skeletal Muscle. Front. Mol. Biosci. 2021, 8, 762185. [Google Scholar] [CrossRef]
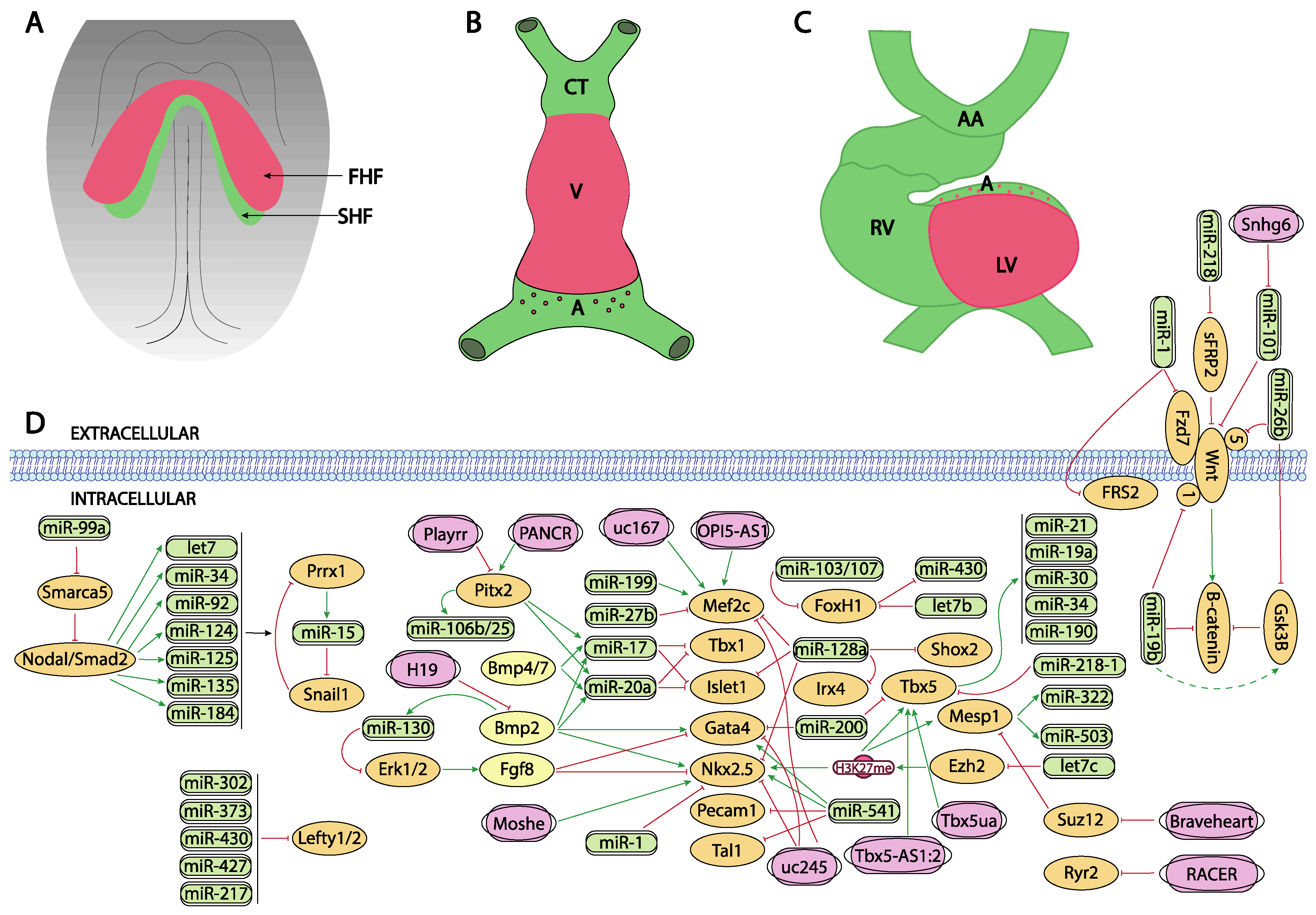

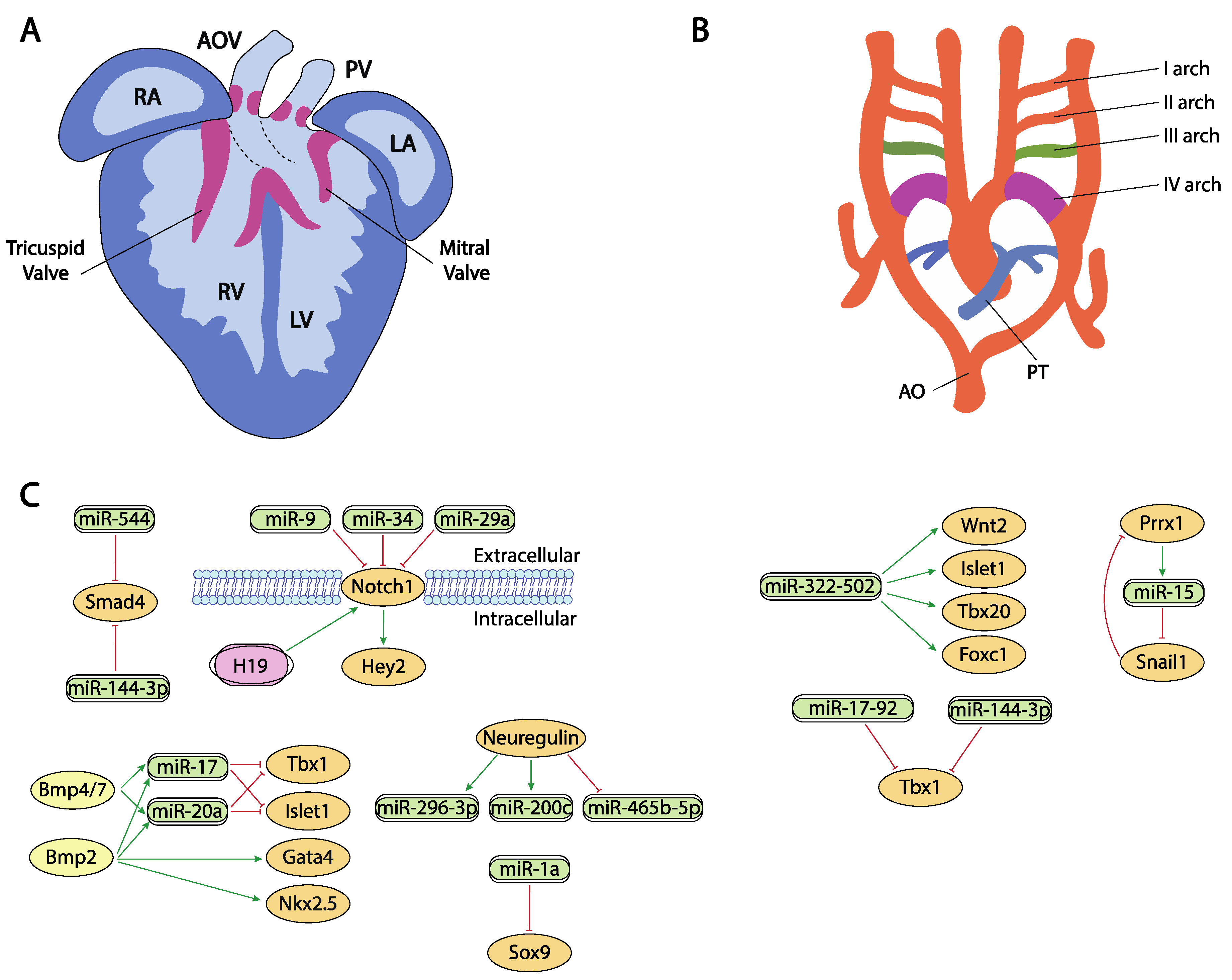
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lozano-Velasco, E.; Garcia-Padilla, C.; del Mar Muñoz-Gallardo, M.; Martinez-Amaro, F.J.; Caño-Carrillo, S.; Castillo-Casas, J.M.; Sanchez-Fernandez, C.; Aranega, A.E.; Franco, D. Post-Transcriptional Regulation of Molecular Determinants during Cardiogenesis. Int. J. Mol. Sci. 2022, 23, 2839. https://doi.org/10.3390/ijms23052839
Lozano-Velasco E, Garcia-Padilla C, del Mar Muñoz-Gallardo M, Martinez-Amaro FJ, Caño-Carrillo S, Castillo-Casas JM, Sanchez-Fernandez C, Aranega AE, Franco D. Post-Transcriptional Regulation of Molecular Determinants during Cardiogenesis. International Journal of Molecular Sciences. 2022; 23(5):2839. https://doi.org/10.3390/ijms23052839
Chicago/Turabian StyleLozano-Velasco, Estefania, Carlos Garcia-Padilla, Maria del Mar Muñoz-Gallardo, Francisco Jose Martinez-Amaro, Sheila Caño-Carrillo, Juan Manuel Castillo-Casas, Cristina Sanchez-Fernandez, Amelia E. Aranega, and Diego Franco. 2022. "Post-Transcriptional Regulation of Molecular Determinants during Cardiogenesis" International Journal of Molecular Sciences 23, no. 5: 2839. https://doi.org/10.3390/ijms23052839
APA StyleLozano-Velasco, E., Garcia-Padilla, C., del Mar Muñoz-Gallardo, M., Martinez-Amaro, F. J., Caño-Carrillo, S., Castillo-Casas, J. M., Sanchez-Fernandez, C., Aranega, A. E., & Franco, D. (2022). Post-Transcriptional Regulation of Molecular Determinants during Cardiogenesis. International Journal of Molecular Sciences, 23(5), 2839. https://doi.org/10.3390/ijms23052839







