Abstract
Environmental stresses, especially heat and drought, severely limit plant growth and negatively affect wheat yield and quality worldwide. Heat shock factors (Hsfs) play a central role in regulating plant responses to various stresses. In this study, the wheat heat shock factor gene TaHsfA2e-5D on chromosome 5D was isolated and functionally characterized, with the goal of investigating its role in responses to heat and drought stresses. Gene expression profiling showed that TaHsfA2e-5D was expressed constitutively in various wheat tissues, most highly in roots at the reproductive stage. The expression of TaHsfA2e-5D was highly up-regulated in wheat seedlings by heat, cold, drought, high salinity, and multiple phytohormones. The TaHsfA2e-5D protein was localized in the nucleus and showed a transcriptional activation activity. Ectopic expression of the TaHsfA2e-5D in yeast exhibited improved thermotolerance. Overexpression of the TaHsfA2e-5D in Arabidopsis results in enhanced tolerance to heat and drought stresses. Furthermore, RT-qPCR analyses revealed that TaHsfA2e-5D functions through increasing the expression of Hsp genes and other stress-related genes, including APX2 and GolS1. Collectively, these results suggest that TaHsfA2e-5D functions as a positive regulator of plants’ responses to heat and drought stresses, which may be of great significance for understanding and improving environmental stress tolerance in crops.
1. Introduction
Wheat (Triticum aestivum L.) is one of the most important staple food crops in the world. Environmental stresses, such as high temperatures and drought, adversely affect wheat viability and yield globally, especially during pollination and grain-filling stages. Plants have developed various morphological, physiological, biochemical and molecular alterations, and adaptation strategies to acquire stress tolerance [1,2]. One of the effective strategies is to transcriptionally regulate the expression of stress-related genes, among which transcription factors play a crucial role by activating and/or inhibiting specific genes [3]. Heat shock factors (Hsfs) are one of the prime transcription factors identified in plants in response to heat stress, which are responsible for activation of heat-responsive genes (for example, Hsp genes) by recognizing conserved heat shock elements (HSEs) in their promoters [4,5]. Hsps rapidly accumulate in response to heat stress, protecting plants against heat stress by re-establishing normal protein conformation, thereby maintaining cellular homeostasis [6]. Hsfs are structurally and functionally conserved throughout eukaryotes, and they consist of a common core structure comprising a DNA-binding domain (DBD) and an oligomerization domain (OD or HR-A/B) [7]. The N-terminal DBD is a typical structure in Hsfs that specifically recognizes HSEs [8]. The adjacent HR-A/B is responsible for Hsf trimerization during transcriptional activation. Hsfs can be divided into three classes (HsfA, HsfB and HsfC) based on the different linkers between HR-A and HR-B [9]. In addition, the nuclear localization signal (NLS), nuclear export signal (NES) and C-terminal transcriptional activation domains (CTAD or AHA) are found in some Hsfs. The conserved DBDs and variable AHAs imply evolutionarily functional conservation and divergence [10].
Hsfs are widespread in plants and play important roles in various plant processes including the regulation of growth and development, signaling networks, responses to abiotic stresses, etc. Several wheat Hsfs have been functionally characterized, mainly focusing on their roles in abiotic stress responses. For instance, overexpression of the seed-preferential TaHsfA2d provided increased heat and salt tolerance in Arabidopsis [11]; additionally, it (renamed as TaHsfA6b) improved thermotolerance in transgenic barley [12]. Arabidopsis overexpressing TaHsfA2-1 or TaHsfA2-10 displayed enhanced thermotolerance by up-regulating the expression of Hsp genes [13,14]. TaHsfA4a conferred cadmium tolerance by increasing the expression levels of metallothionein genes in rice [15]. TaHsfA6f overexpression resulted in improved tolerance to heat, drought and salt stresses in Arabidopsis [16], and as a transcription activator directly regulating TaHSP, TaGAAP and TaRof1 genes, TaHsfA6f exerted a positive effect on thermotolerance in wheat [17]. TaHsfA6e modulates heat and drought tolerance in wheat by regulating the expression of Hsp genes, such as Hsp17, Hsp70 and Hsp90 [18]. Overexpression of TaHsf3 (belonging to class HsfB) enhanced tolerance to extreme temperatures in Arabidopsis by activating Hsp genes, such as Hsp70 [19]. TaHsfC2a can improve thermotolerance through an ABA-mediated regulatory network in wheat [20]. Screening of wheat genome data by different researchers has revealed that 53–82 Hsfs are present in wheat, and the expression of these Hsfs varies greatly across different wheat tissues and in response to various abiotic stresses [21,22,23,24]. This has led us to hypothesize that other uncharacterized wheat Hsfs should also function in abiotic stress responses.
In this study, we identified a Hsf gene on chromosome 5D from the heat-/drought-resistant wheat cultivar TAM107 and designated it as TaHsfA2e-5D. To unravel the biological function of TaHsfA2e-5D, we investigated its expression patterns, subcellular localization, transactivation activity, and stress tolerance in transgenic yeast and Arabidopsis. The findings provide evidence that TaHsfA2e-5D plays a significant role in plant responses to heat and drought stresses, and that TaHsfA2e-5D represents a valuable candidate gene for engineering heat and drought tolerant plants in future.
2. Results
2.1. TaHsfA2e-5D Expression Is Induced by Various Abiotic Stresses and Phytohormones
By digging into published transcriptome sequencing data of wheat (TAM107) seedlings under normal growth conditions and heat stress for 6 h [25], we identified a heat shock factor gene, TaHsfA2e-5D, which was highly up-regulated by heat stress and thus was selected for further functional characterization. Firstly, we investigated the expression profiles of TaHsfA2e-5D in different wheat tissues. As shown in Figure 1A, TaHsfA2e-5D was expressed in all parts of wheat plants examined; the highest expression was noted in the root, stamen and pistil, followed by that in the glume and arista, whereas the lowest expression was found in the leaf, young spike, and stem under normal growth conditions. Next, we explored the expression patterns of TaHsfA2e-5D in response to various abiotic stresses and phytohormones. Under a high-temperature, low-temperature and methyl jasmonate (MeJA) treatments, the expression levels of TaHsfA2e-5D were up-regulated rapidly and peaked at 2, 4 and 1 h, respectively (Figure 1B,E,F). TaHsfA2e-5D expression was induced by PEG6000 or abscisic acid (ABA) at 6 h post treatment, peaked at 12 h, and then decreased sharply (Figure 1C,G). Following NaCl stress, the TaHsfA2e-5D transcript increased gradually from 6 h (Figure 1D). For H2O2 and salicylic acid (SA) treatments, the responses of TaHsfA2e-5D were relatively weak, and its mRNA levels slightly increased after 6 h of treatment (Figure 1H,I). These results implied that TaHsfA2e-5D appears to be involved in different signal transduction pathways and may play an important role in various abiotic stress responses in plants.
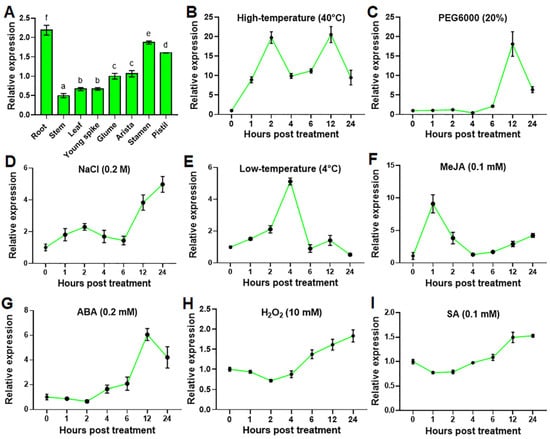
Figure 1.
Expression analyses of TaHsfA2e-5D. (A) Expression levels of TaHsfA2e-5D in different tissues of TAM107 wheat at the booting stage. Different lowercase letters above the bars denote significant differences at the p < 0.05 level. (B–I) Expression patterns of TaHsfA2e-5D during abiotic stresses and phytohormones treatment. Total RNA was extracted from 10-day-old wheat seedlings treated with 40 °C high-temperature (B), 20% PEG6000 (C), 0.2 M NaCl (D), 4 °C low-temperature (E), 0.1 mM MeJA (F), 0.2 mM ABA (G), 10 mM H2O2 (H), and 0.1 mM SA (I), for the indicated time points. The wheat β-actin gene was used as an internal control. All data represent means ± standard deviation (SD) of three replicates.
2.2. Molecular Characterization of TaHsfA2e-5D
The genomic and coding sequence alignment showed that TaHsfA2e-5D (GenBank accession number OM735737) included two exons and contained a 1047-bp open reading frame encoding 348 amino acid residues (Figure 2A and Figure S1). The gene-specific amplicons of TaHsfA2e-5D were observed in four Chinese Spring (CS) nulli-tetrasomic (NT) lines of chromosome 5, but not in lines N5DT5A (nullisomic 5D-tetrasomic 5A) and N5DT5B (nullisomic 5D-tetrasomic 5B), indicating that TaHsfA2e-5D was located on the chromosome 5D (Figure 2B). Phylogenetic analyses indicated that TaHsfA2e-5D and its homologs diverged distinctly between monocot and dicot species (Figure 2C). TaHsfA2e-5D shared the highest similarity with the Aegilops tauschii (the D genome donor species of hexaploid wheat) protein XP_020187328.1, and it was homologous to rice OsHsfA2e (XP_015630486.1) (Figure 2C). In addition, it was found that the TaHsfA2e-5D protein owned a typical DBD domain at the N-terminal, which exhibited significant sequence conservation within Hsf proteins from various species (Figure 2D and Figure S1).
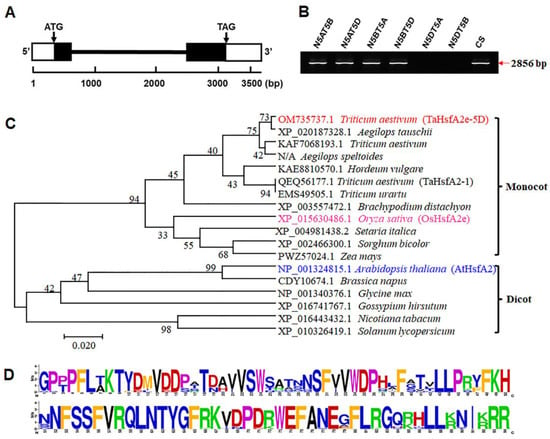
Figure 2.
Molecular characterization of TaHsfA2e-5D. (A) Gene structure of TaHsfA2e-5D. White boxes, black boxes, and black line represent untranslated regions, exons, and intron, respectively. (B) Chromosomal location of TaHsfA2e-5D in CS NT lines. N5AT5B: nullisomic 5A-tetrasomic 5B; N5AT5D: nullisomic 5A-tetrasomic 5D; N5BT5A: nullisomic 5B-tetrasomic 5A; N5BT5D: nullisomic 5B-tetrasomic 5D; N5DT5A: nullisomic 5D-tetrasomic 5A; N5DT5B: nullisomic 5D-tetrasomic 5B; CS: Chinese Spring. (C,D) Phylogenetic relationships (C) and sequences logo (D) of the DBD domain of TaHsfA2e-5D and its homologous proteins from other species.
2.3. Promoter Isolation and Cis-Acting Regulatory Elements Analysis of TaHsfA2e-5D
To understand the underlying mechanism responsible for the regulation of TaHsfA2e-5D expression, we isolated a 2027 bp DNA sequence upstream of the start codon of TaHsfA2e-5D using a PCR strategy. We searched for putative cis-acting regulatory elements in this sequence using PlantCARE (Figure 3). A number of stress-related elements were identified, including drought-responsive element (MBS), dehydration-responsive element (DRE), low-temperature-responsive element (LTR), MeJA-responsive element (CGTCA/TGACG-motif) and ABA-responsive element (ABRE).
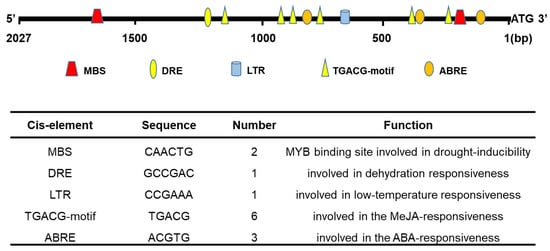
Figure 3.
Putative cis-acting elements in the 5′-flanking region of TaHsfA2e-5D. A region of 2027 bp upstream of the start codon of TaHsfA2e-5D was isolated as its 5′-flanking region and used for the prediction of cis-acting elements. MBS: MYB binding site element; DRE: dehydration-responsive element; LTR: low-temperature-responsive element; ABRE: ABA-responsive element.
2.4. TaHsfA2e-5D Is a Nucleus-localized Transcriptional Activator
Domain analyses revealed that TaHsfA2e-5D possesses NLS and AHA motifs similar to other known HsfAs (Figure S1), indicating that TaHsfA2e-5D is likely to be a nucleus-localized transcriptional activator. In the assessment of subcellular localization of TaHsfA2e-5D through transient transformation in tobacco leaf epidermal cells, the green fluorescence of cells with GFP was distributed throughout the cell, whereas it was only observed in the nucleus of cells with TaHsfA2e-5D-GFP, verifying that TaHsfA2e-5D is a nucleus-localized protein (Figure 4A). The transactivation activity of TaHsfA2e-5D was evaluated with the GAL4 yeast system. As shown in Figure 4B, all transformants grew well on SD/-Trp medium. However, only the yeast harboring pGBKT7-TaHsfA2e-5D and pGBKT7-P53 constructs, but not the yeast carrying pGBKT7-TaHsfA2e-5D CΔ48 and pGBKT7 vectors, grew well on SD/-Trp-His medium, confirming that TaHsfA2e-5D functions as a transcriptional activator and the predicted AHA motif is required for its transactivation activity.
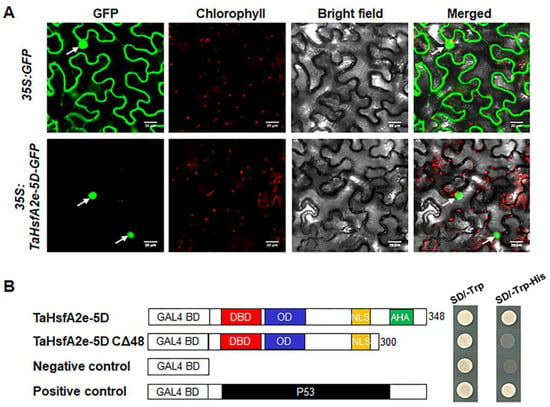
Figure 4.
Subcellular localization and transcriptional activity analyses of TaHsfA2e-5D. (A) Subcellular localization of TaHsfA2e-5D in tobacco leaf epidermal cells. White arrows indicate nuclei. Bars = 20 μm. (B) Transcriptional activation of TaHsfA2e-5D in yeast. Schematic representation of the full-length TaHsfA2e-5D and truncated protein constructs in the pGBKT7 vector. Fusion proteins of the GAL4 DNA-binding domain (BD) and full-length TaHsfA2e-5D or truncated TaHsfA2e-5D with the deletion of 48 C-terminal amino acid residues (CΔ48) were expressed in the yeast strain AH109. The pGBKT7 and pGBKT7-P53 vectors were used as negative and positive controls, respectively.
2.5. Ectopic Expression of TaHsfA2e-5D Improves Thermotolerance in Yeast
The aforementioned results indicated that the expression of TaHsfA2e-5D was remarkably induced by heat stress (Figure 1B). To confirm the function of TaHsfA2e-5D in heat stress responses, we used ectopic expression in yeast to study its effect on thermotolerance. Under normal growth conditions (30 °C), no obvious difference was observed in the growth status between the yeast transformed with pYES2 and that transformed with pYES2-TaHsfA2e-5D, and the growth curves of the two kinds of transformants almost overlapped (Figure 5A,C). However, after heat treatments (40 °C or 50 °C), the growth of both transformants was inhibited, but the pYES2-TaHsfA2e-5D transgenic yeast grew better and had higher survival rates compared with the pYES2 transgenic yeast (Figure 5A,B). Moreover, the OD600 values of pYES2-TaHsfA2e-5D transgenic yeast were obviously higher than those of pYES2 transgenic yeast after 9 h of treatment (Figure 5C). These results demonstrated that ectopic expression of TaHsfA2e-5D improves thermotolerance in yeast.
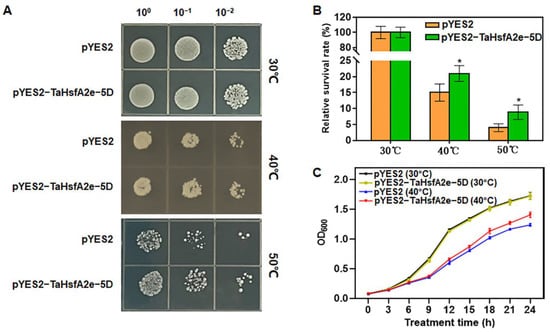
Figure 5.
Thermotolerance assays of transgenic yeast with pYES2 vector alone and pYES2-TaHsfA2e-5D recombinant vector. (A) Growth status of transgenic yeast before (30 °C) and after heat stress (40 °C or 50 °C, 1 h). 100, 10−1 and 10−2 represent the 10-fold serial gradient dilutions. (B) Relative survival rates of yeast harboring pYES2 or pYES2-TaHsfA2e-5D treated with 40 °C or 50 °C for 1 h. (C) Growth curves of transgenic yeast under normal (30 °C) and heat stress (40 °C) conditions. Values are means ± SD of triplicate experiments. * indicates p < 0.05 (Student’s t-test).
2.6. Overexpression of TaHsfA2e-5D Confers Heat and Drought Tolerance in Arabidopsis
To further determine the usefulness of TaHsfA2e-5D in molecular breeding for plant thermotolerance, four single copy TaHsfA2e-5D overexpressing lines (L1, L2, L3 and L4) displaying different transcript levels (Figure S2) were selected to examine the role of TaHsfA2e-5D in thermotolerance in Arabidopsis. As shown in Figure 6A,B, under normal growth conditions, no visible phenotype variation was observed between WT and transgenic lines. However, under heat stress conditions, the TaHsfA2e-5D overexpressing lines were more resistant to heat stress than WT in terms of seedling survival rates (Figure 6). These results proved that overexpression of TaHsfA2e-5D confers thermotolerance in Arabidopsis.
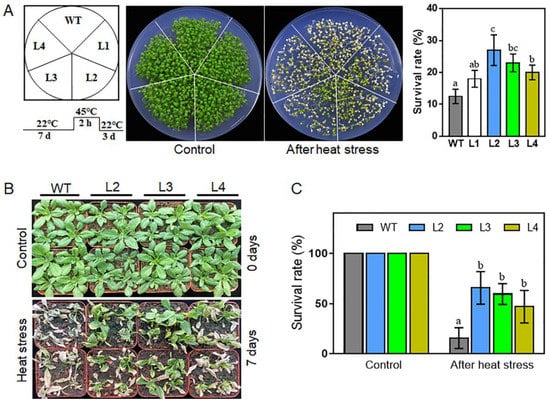
Figure 6.
Thermotolerance assays of WT and TaHsfA2e-5D overexpressing transgenic Arabidopsis lines. (A) Morphology and survival rates of WT and transgenic lines (L1, L2, L3 and L4) grown on MS agar plates after heat treatment. Seven-day-old seedlings were treated at 45 °C for 2 h and recovered at 22 °C for 3 days. (B,C) Comparison of thermotolerance among WT and transgenic lines (L2, L3, and L4) in soil. Phenotype (B) and survival rates (C) of WT and transgenic lines after heat stress at 40/32 °C (day/night) for 7 days. For each experiment, at least 20 plants per line were used; values are means ± SD from three independent measurements. Different lowercase letters above the bars denote significant differences at the p < 0.05 level.
Considering that heat stress and drought stress are closely correlated and that the expression of TaHsfA2e-5D is highly up-regulated by dehydration (PEG6000) stress (Figure 1C), the phenotypes of the transgenic plants under drought stress were investigated. As shown in Figure 7A, after withholding water for 14 days, the WT leaves were severely dehydrated, whereas the three transgenic lines exhibited better growth status than WT. After two days’ recovery, only 25% of the WT plants survived, whereas more than 70% of the transgenic plants survived in lines L2, L3, and L4 (Figure 7B). Additionally, the transgenic lines showed lower rates of water loss than the WT plants (Figure 7C). These data demonstrate that the TaHsfA2e-5D gene also confers drought tolerance in transgenic Arabidopsis.
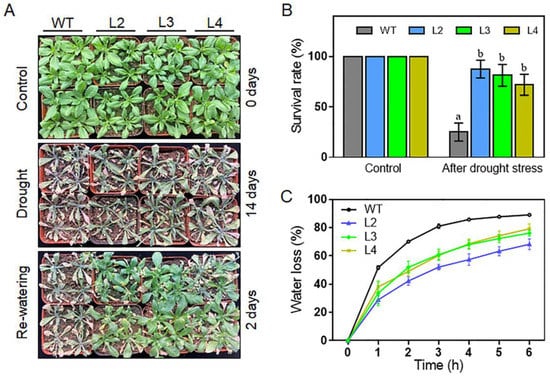
Figure 7.
Comparison of drought tolerance among WT and TaHsfA2e-5D overexpressing transgenic lines. (A) Morphology and (B) survival rates of WT and transgenic lines (L2, L3 and L4) after drought treatment. The 21-day-old seedlings did not receive watering for 14 days until the plants withered. Afterwards, re-watering was applied, and survival rates were measured after re-watering for 2 days under normal conditions. Different lowercase letters above the bars denote significant differences at the p < 0.05 level. (C) Water loss rates of WT and transgenic lines. Values are means ± SD of three replicates.
2.7. Expression Analysis of Stress-Related Genes in TaHsfA2e-5D Transgenic Arabidopsis
To explore the molecular basis of heat and drought resistance conferred by TaHsfA2e-5D in transgenic Arabidopsis, the expression of several stress-responsive genes was examined in WT and transgenic lines by RT-qPCR. Specifically, ten genes positively regulated by AtHsfA2 [26,27], including Hsp17.7-CII, Hsp18.1-CI, Hsp22-ER, Hsp25.3-P, Hsp26.5-P, Hsp70t-2, Hsp90.1, Hsp101, GolS1, and APX2 were selected for analyses. As shown in Figure 8, the expression levels of most genes examined were significantly higher in TaHsfA2e-5D overexpressing lines than in WT, except that there was no significant difference between the WT and transgenic lines for Hsp90.1 and Hsp101 (Figure 8). These results revealed that the enhanced heat and drought tolerance conferred by TaHsfA2e-5D was mediated at least in part by the transcriptional activation of Hsp genes and other stress-related genes.
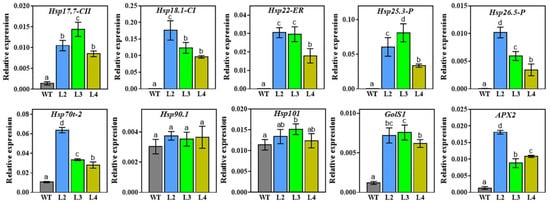
Figure 8.
Expression of stress-related genes in WT and transgenic lines. Total RNAs were isolated from WT and transgenic plants grown for 10 days under normal conditions. Three biological replicates were used for each line. The Arabidopsis Actin2 gene was used as an internal standard. All data represent means ± SD. Different lowercase letters above the bars denote significant differences at the p < 0.05 level. Hsp17.7-CII (AT5G12030), Hsp18.1-CI (At5g59720), Hsp22-ER (At4g10250), Hsp25.3-P (At4g27670), Hsp26.5-P (At1g52560), Hsp70t-2 (At2g32120), Hsp90.1 (At5g52640), Hsp101 (At1g74310), GolS1 (At2g47180), and APX2 (At3g09640).
3. Discussion
High temperatures and drought are increasingly becoming a serious threat to wheat production worldwide. An increase in atmospheric temperature of 1 °C above the wheat’s optimal growing temperature during the reproductive development phase has been presumed to reduce wheat yield by 6–10%, which can greatly threaten food security [28]. It is known that transcription factors play important roles in mediating environmental stress responses by regulating the expression of stress-responsive genes [29]. Among them, Hsfs, which perform a pivotal role in plant adaptation to abiotic stresses, have been extensively studied in various plants [30]. Although 82 non-redundant Hsfs located on 21 chromosomes have been identified in wheat [12], fewer Hsfs have been cloned and characterized because of the complexity of the wheat genome. In this study, we cloned a wheat Hsf gene, TaHsfA2e-5D, which is located on chromosome 5D and the deduced protein features the DBD and OD motifs of HsfAs. Gene expression profiling revealed differential accumulation of TaHsfA2e-5D transcripts in all test tissues (Figure 1A), suggesting that it might play a role in plant growth and development. In spite of high sequence similarities between TaHsfA2e-5D and TaHsfA2-1 (Figure 2C), the expression patterns of these two genes are quite different. TaHsfA2-1 is specifically highly expressed in the mature leaf and lowly expressed in the young root and stamen [14]. Here, it was found that TaHsfA2e-5D transcripts were more abundant in the root and stamen, but less in the leaf (Figure 1A). TaHsfA2-1 was upregulated by heat, drought, H2O2 and SA treatments [14]. In this study, TaHsfA2e-5D was also induced strongly by heat and drought, but weakly by H2O2 and SA treatments (Figure 1). These results suggest that although more than 82 Hsf genes have been found in the wheat genome, further study will be required to elucidate their individual roles.
The sequence of TaHsfA2e-5D is highly similar to its ortholog from diploid progenitor Aegilops tauschii, indicating that TaHsfA2e-5D is highly conserved during polyploidization. Homology analyses showed that TaHsfA2e-5D is homologous to Arabidopsis AtHsfA2 and rice OsHsfA2e (Figure 2C). AtHsfA2 and OsHsfA2e have been reported to be able to respond to various environmental stresses [26,31]. In the present study, TaHsfA2e-5D was induced by various abiotic stresses and phytohormones (Figure 1), suggesting versatile functions of TaHsfA2e-5D, similar to its orthologous genes AtHsfA2 and OsHsfA2e. It was found that TaHsfA2e-5D contained the nuclear localization signal (NLS) sequence (Figure S1), and the subcellular localization assay in tobacco leaf epidermal cells confirmed that TaHsfA2e-5D is a nuclear localized protein (Figure 4A). Similar nuclear localization studies have been reported in AtHsfA2 and OsHsfA2e [26,31]. In addition, TaHsfA2e-5D shows a transcriptional activation activity, and a deletion analysis revealed that the activation domain of TaHsfA2e-5D is localized in the 300–348 region (Figure 4B), which contains an AHA motif (Figure S1) typical of HsfAs [10].
Like other Hsfs, TaHsfA2e-5D was found to confer significant thermotolerance on plants (Figure 5 and Figure 6). Besides, transgenic Arabidopsis lines clearly exhibited better growth status, higher survival rates, and lower water loss rates under drought stress conditions (Figure 7). In recent years, some heat shock factors have been reported to play important roles in other abiotic stresses in addition to heat stress. For example, Arabidopsis plants overexpressing AtHsfA7b exhibited enhanced tolerance against salt and drought stresses [32]. Maize ZmHsf04 conferred increased thermo and salt-stress tolerance in transgenic Arabidopsis [33]. ZmHsf05 improved thermotolerance in Arabidopsis and enhanced drought tolerance in rice [34,35], while overexpression of ZmHsf08 reduced salt and drought tolerance in maize [30]. The recently reported OsHSFA3 played an important role in ABA-mediated drought tolerance in Arabidopsis [36]. TaHsfA6f from wheat could enhance tolerance to heat, drought, and salt stresses of transgenic plants [16,17].
To adapt to heat and drought stresses, plants have developed different response strategies, including modulation of the expression of stress-related genes [37]. Hsps, as key molecular chaperones, assist with the folding, intracellular distribution, and degradation of proteins and thus play central roles in protecting plants from stress damage [6]. Proactive regulation of Hsfs to Hsps is particularly important in abiotic stress responses [37]. In this study, the expression levels of several Hsp genes, such as Hsp17.7-CII, Hsp18.1-CI, Hsp22-ER, Hsp25.3-P, Hsp26.5-P, and Hsp70 were elevated significantly in TaHsfA2e-5D overexpressing Arabidopsis under normal conditions. However, no significant differences in the transcript levels of Hsp90.1 and Hsp101 genes were observed between transgenic and WT plants (Figure 8). This result is not consistent with those of previous studies on AtHsfA2 and OsHsfA2e, in which both genes were directly regulated by AtHsfA2 and OsHsfA2e [26,31]. Beyond Hsp genes, other stress-related genes, APX2 and GolS1, were also up-regulated in TaHsfA2e-5D overexpression plants. The APX2 gene encodes cytosolic ascorbate peroxidase which plays an important role in maintaining the activity of the antioxidant system and protects plants against oxidative damage resulting from adverse stresses [38]. The GolS1 gene encodes galactinol synthase and was previously reported to function primarily in drought, high-salinity, and osmotic stress tolerance [39]. These results indicated that TaHsfA2e-5D positively regulates heat and drought stress responses, at least partly through the boosted expression of stress-responsive genes. Future studies will focus on the elucidation of the stress-responsive regulatory network underlying TaHsfA2e-5D, and the development of heat-/drought-resistance wheat cultivars through genetic manipulation, thereby improving wheat yield under environmental stress conditions.
In conclusion, we cloned and characterized a wheat Hsf gene, TaHsfA2e-5D, which was localized in the nucleus and was demonstrated to function as a transcriptional activator. Overexpression of TaHsfA2e-5D in yeast and Arabidopsis led to increased tolerance to heat and drought stresses. Furthermore, RT-qPCR analyses demonstrated that TaHsfA2e-5D functions through modulating the expression of Hsp genes and other stress-related genes. Taken together, our results suggest that TaHsfA2e-5D could be useful in molecular breeding of wheat or other crops for enhanced stress tolerance, especially during terminal heat and drought stresses.
4. Materials and Methods
4.1. Plant Materials, Growth Conditions, and Stress Treatments
Heat-/drought-resistant wheat cultivar TAM107 used for gene cloning and expression analysis was planted in Hoagland solution. Chinese Spring (CS) nulli-tetrasomic (NT) wheat lines (N5AT5B, N5AT5D, N5BT5A, N5BT5D, N5DT5A and N5DT5B) used for determining chromosomal locations, and tobacco (Nicotiana benthamiana) used for transient transfection were cultivated in soil. Columbia-0 (Arabidopsis thaliana) used for genetic transformation was sown on Murashige and Skoog (MS) agar media or in soil. All the plants were grown in a greenhouse at 22 °C with a 16-h photoperiod.
To explore the tissue-specific expression of TaHsfA2e-5D, we harvested the root, stem, leaf, young spike, glume, arista, stamen and pistil from TAM107 at the booting stage. To study the expression patterns of TaHsfA2e-5D in response to stresses, plants were grown hydroponically in a light chamber with a 16-h photoperiod at 22 °C, and 10-day-old wheat seedlings were treated by submerging the roots in Hoagland solution with 20% PEG 6000 (polyethylene glycol 6000), 0.2 M NaCl, 0.1 mM MeJA, 0.2 mM ABA, 10 mM H2O2, or 0.1 mM SA. For high-temperature and low-temperature stresses, the seedlings were transferred to another growth chamber with a constant temperature of 40 °C or 4 °C and with the same parameters of light and humidity as the previous chamber. For each treatment, five seedlings were separately sampled at 0, 1, 2, 4, 6, 12, and 24 h after treatment. All the samples were stored at −80 °C for RNA extraction.
4.2. Gene Cloning, Chromosomal Location, and Sequence Analysis
Genomic DNA and total RNA were isolated using the CTAB and TRIzol methods (Coolaber, Beijing, China), respectively. First-strand cDNA was synthesized using a Reverse Transcription System (Vazyme, Nanjing, China). Sequence BLAST search was performed through NCBI (https://www.ncbi.nlm.nih.gov/, accessed on 28 February 2022) and IWGSC (http://www.wheatgenome.org/, accessed on 28 February 2022) databases. The genomic and cDNA sequences of TaHsfA2e-5D were obtained from the DNA and cDNA of TAM107 by PCR with gene-specific primers. The chromosomal location of TaHsfA2e-5D was determined by PCR using chromosome specific primers with the DNA of CS NT lines as templates. All PCR reactions were performed using high-fidelity Tks Gflex™ DNA Polymerase (TaKaRa, Dalian, China) according to the manufacturer’s recommended procedures. The PCR products were constructed into the pEASY-Blunt cloning vector (TransGen, Beijing, China) for sequencing. Primer sequence information is presented in Table S1. DNAMAN8 software was used for sequence alignment. Cis-acting regulatory elements were predicted using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 28 February 2022). Phylogenetic tree was constructed using MEGA7 software, and sequence logos were analyzed by WebLogo (http://weblogo.berkeley.edu/, accessed on 28 February 2022) platform.
4.3. Gene Expression Analysis
Gene expression was analyzed by RT-qPCR using SYBR Green Realtime PCR Master Mix (Takara, Dalian, China) on a CFX96 real-time PCR machine (Bio-Rad, Hercules, CA, USA) following the manufacturer’s recommended procedures. The gene expression values were normalized to that of wheat β-actin or Arabidopsis Actin2 using the 2−∆∆CT method [40].
4.4. Subcellular Localization and Transactivation Analysis
The coding sequence (CDS) of TaHsfA2e-5D without a stop codon was inserted into the pCAM35s-GFP vector via an In-Fusion Cloning System (Vazyme, Nanjing, China). pCAM35s-GFP and pCAM35s-TaHsfA2e-5D-GFP were transformed respectively into tobacco leaf epidermal cells using the Agrobacterium infiltration method [41]. Fluorescence signals were visualized and photographed using a laser confocal microscope (Olympus, Tokyo, Japan).
The full-length and truncated TaHsfA2e-5D were respectively introduced into the pGBKT7 vector by the In-Fusion technique. The constructs, including negative control pGBKT7, positive control pGBKT7–P53, pGBKT7-TaHsfA2e-5D and pGBKT7-TaHsfA2e-5D C∆48 (without the C-terminal 48 amino acids) were respectively transformed into yeast strain AH109. Transcriptional activation activities were evaluated according to the growth status of transformants on SD/-Trp and SD/-Trp-His media.
4.5. Transformation and Thermotolerance Assay in Yeast
The CDS of TaHsfA2e-5D was cloned into yeast expression vector pYES2. The pYES2 contains a URA3 selection marker driven by a galactose-inducible (GAL1) promoter. The pYES2-TaHsfA2e-5D and the empty pYES2 control plasmids were transformed into yeast strain INVSc1, respectively. Positive transformants were screened and cultivated at 30 °C in an uracil-deficient synthetic complete (SC-ura) medium with 2% (w/v) glucose. A single positive clone of INVSc1 containing the fused pYES2-TaHsfA2e-5D or the empty pYES2 vector was inoculated in 5 mL of SC-ura liquid medium containing 2% glucose at 30 °C. For heat shock treatment, yeast cells which had been cultured overnight were diluted to an OD600 value of 0.4 into 10 mL induction SC-ura liquid medium (supplemented with 2% galactose). After incubation for approximately 16 h, the yeast cell densities were recalculated and an equal number of yeast cells were re-suspended in 200 μL of sterile water and exposed to normal conditions (30 °C) or heat stress conditions (40 °C or 50 °C) for 1 h. Afterwards, 20 μL of 10-fold serial dilutions were dotted on SC-ura plates, and incubated at 30 °C for 2 days to evaluate the growth status and relative survival rates. Meanwhile, the overnight cultures were resuspended and diluted in SC-ura liquid medium (supplemented with 2% galactose) to obtain an OD600 of 0.08. Furthermore, the dilutions were incubated at normal conditions (30 °C) or heat stress conditions (40 °C), and the OD600 was measured every 3 h to obtain the growth curves.
4.6. Arabidopsis Transformation and Stress Treatments
The CDS of TaHsfA2e-5D was cloned into pSuper1300 vector to generate a TaHsfA2e-5D overexpressing construct. The construct was transformed into wild-type (Columbia-0) Arabidopsis using the Agrobacterium mediated floral-dip method [42]. Single T-DNA insertion and homozygous transgenic lines were screened on MS media containing 25 mg L−1 hygromycin. The expression levels of transgenic plants were analyzed by RT-qPCR.
For heat stress treatment, sterilized seeds of WT and homozygous transgenic lines were planted on MS agar plates. Seven-day-old seedlings of WT and homozygous transgenic lines were placed at 45 °C for 2 h, and the survival rates were calculated after 3-day recovery at 22 °C. Additionally, 7-day-old seedlings were transplanted into nutrient-rich soil for 14 days under normal conditions. The 21-day-old Arabidopsis seedlings (the transgenic lines and the WT plants) were grown at 40/32 °C (day/night) with irrigation every day for 7 days. Then, the phenotypes and survival rates of plants were measured. For drought stress treatment, the 21-day-old Arabidopsis seedlings did not receive watering for 14 days until the plants withered. Afterwards, re-watering was applied, and survival rates were measured after re-watering for 2 days under normal conditions. Water loss rate assay was performed according to a previous study [43]. The above experiments were repeated three times.
4.7. Statistical Analyses
Fisher’s LSD multiple comparisons test following One-Way ANOVA was performed for WT and transgenic Arabidopsis lines overexpressing TaHsfA2e-5D by Prism 7 (GraphPad Software, Version 7.00, San Diego, CA, USA). Differences in relative survival rates between yeast harboring pYES2 and yeast with pYES2-TaHsfA2e-5D were evaluated using a Student’s t test.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23052784/s1.
Author Contributions
Conceptualization, Y.Z. and X.T.; Data curation, H.L., Y.X. and D.M.; Funding acquisition, H.B. and X.T.; Investigation, J.M., J.H., Q.C. and J.Q.; Project administration, H.B.; Supervision, W.L.; Writing—original draft, Y.Z. and X.T.; Writing—review and editing, H.B. and W.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (32072062 and 32001546), the Natural Science Foundation of Hubei Province of China (2019CFB296), the Scientific Research Project of Jingchu University of Technology (QDJ201901) and the Scientific Research Program Guiding Project of Hubei Provincial Department of Education (B2018233).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Anwar, K.; Joshi, R.; Dhankher, O.P.; Singla-Pareek, S.L.; Pareek, A. Elucidating the response of crop plants towards individual, combined and sequentially occurring abiotic stresses. Int. J. Mol. Sci. 2021, 22, 6119. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H. Multifaceted chromatin structure and transcription changes in plant stress response. Int. J. Mol. Sci. 2021, 22, 2013. [Google Scholar] [CrossRef] [PubMed]
- Ohama, N.; Sato, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 2017, 22, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.R.; Goswami, S.; Rai, G.K.; Jain, N.; Singh, P.K.; Mishra, D.; Chaturvedi, K.K.; Kumar, S.; Singh, B.; Singh, G.P.; et al. Protection from terminal heat stress: A trade-off between heat-responsive transcription factors (HSFs) and stress-associated genes (SAGs) under changing environment. Cereal Res. Commun. 2021, 49, 227–234. [Google Scholar] [CrossRef]
- Pirkkala, L.; Nykanen, P.; Sistonen, L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 2001, 15, 1118–1131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pal, S.; Sharma, R. Transcription factors and chaperone proteins play a role in launching a faster response to heat stress and aggregation. Comput. Biol. Chem. 2021, 93, 107534. [Google Scholar] [CrossRef] [PubMed]
- Scharf, K.D.; Berberich, T.; Ebersberger, I.; Nover, L. The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. Biochim. Biophys. Acta 2012, 1819, 104–119. [Google Scholar] [CrossRef]
- Guo, M.; Liu, J.H.; Ma, X.; Luo, D.X.; Gong, Z.H.; Lu, M.H. The Plant Heat Stress Transcription factors (HSFs): Structure, regulation, and function in response to abiotic stresses. Front. Plant Sci. 2016, 7, 114. [Google Scholar] [CrossRef] [Green Version]
- Nover, L.; Bharti, K.; Doring, P.; Mishra, S.K.; Ganguli, A.; Scharf, K.D. Arabidopsis and the heat stress transcription factor world: How many heat stress transcription factors do we need? Cell Stress Chaperones 2001, 6, 177–189. [Google Scholar] [CrossRef]
- Wang, X.; Shi, X.; Chen, S.; Ma, C.; Xu, S. Evolutionary origin, gradual accumulation and functional divergence of heat shock factor gene family with plant evolution. Front. Plant Sci. 2018, 9, 71. [Google Scholar] [CrossRef]
- Chauhan, H.; Khurana, N.; Agarwal, P.; Khurana, J.P.; Khurana, P. A seed preferential heat shock transcription factor from wheat provides abiotic stress tolerance and yield enhancement in transgenic Arabidopsis under heat stress environment. PLoS ONE 2013, 8, e79577. [Google Scholar] [CrossRef] [Green Version]
- Poonia, A.K.; Mishra, S.K.; Sirohi, P.; Chaudhary, R.; Kanwar, M.; Germain, H.; Chauhan, H. Overexpression of wheat transcription factor (TaHsfA6b) provides thermotolerance in barley. Planta 2020, 252, 53. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.L.; Yuan, S.N.; Zhang, H.N.; Zhang, Y.Y.; Zhang, Y.J.; Wang, G.Y.; Li, Y.Q.; Li, G.L. Heat-response patterns of the heat shock transcription factor family in advanced development stages of wheat (Triticum aestivum L.) and thermotolerance-regulation by TaHsfA2-10. BMC Plant Biol. 2020, 20, 364. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Li, G.; Zhang, H.; Zhang, Y.; Zhang, Y.; Duan, S.; Sheteiwy, M.; Zhang, H.; Shao, H.; Guo, X. TaHsfA2-1, a new gene for thermotolerance in wheat seedlings: Characterization and functional roles. J. Plant Physiol. 2020, 246–247, 153135. [Google Scholar] [CrossRef] [PubMed]
- Shim, D.; Hwang, J.U.; Lee, J.; Lee, S.; Choi, Y.; An, G.; Martinoia, E.; Lee, Y. Orthologs of the class A4 heat shock transcription factor HsfA4a confer cadmium tolerance in wheat and rice. Plant Cell 2009, 21, 4031–4043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bi, H.; Zhao, Y.; Li, H.; Liu, W. Wheat heat shock factor TaHsfA6f increases ABA levels and enhances tolerance to multiple abiotic stresses in transgenic plants. Int. J. Mol. Sci. 2020, 21, 3121. [Google Scholar] [CrossRef]
- Xue, G.P.; Drenth, J.; McIntyre, C.L. TaHsfA6f is a transcriptional activator that regulates a suite of heat stress protection genes in wheat (Triticum aestivum L.) including previously unknown Hsf targets. J. Exp. Bot. 2015, 66, 1025–1039. [Google Scholar] [CrossRef] [Green Version]
- Kumar, R.R.; Goswami, S.; Singh, K.; Dubey, K.; Rai, G.K.; Singh, B.; Singh, S.; Grover, M.; Mishra, D.; Kumar, S.; et al. Characterization of novel heat-responsive transcription factor (TaHSFA6e) gene involved in regulation of heat shock proteins (HSPs)-A key member of heat stress-tolerance network of wheat. J. Biotechnol. 2018, 279, 1–12. [Google Scholar] [CrossRef]
- Zhang, S.; Xu, Z.; Li, P.; Yang, L.; Wei, Y.; Chen, M.; Li, L.; Zhang, G.; Ma, Y. Overexpression of TaHSF3 in transgenic Arabidopsis enhances tolerance to extreme temperatures. Plant Mol. Biol. Rep. 2013, 31, 688–697. [Google Scholar] [CrossRef]
- Hu, X.J.; Chen, D.; Lynne, M.C.; Fernanda, D.M.; Zhang, Z.B.; Drenth, J.; Kalaipandian, S.; Chang, H.; Xue, G.P. Heat shock factor C2a serves as a proactive mechanism for heat protection in developing grains in wheat via an ABA-mediated regulatory pathway. Plant Cell Environ. 2018, 41, 79–98. [Google Scholar] [CrossRef]
- Ye, J.; Yang, X.; Hu, G.; Liu, Q.; Li, W.; Zhang, L.; Song, X. Genome-wide investigation of heat shock transcription factor family in wheat (Triticum aestivum L.) and possible roles in anther development. Int. J. Mol. Sci. 2020, 21, 608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duan, S.; Liu, B.; Zhang, Y.; Li, G.; Guo, X. Genome-wide identification and abiotic stress-responsive pattern of heat shock transcription factor family in Triticum aestivum L. BMC Genom. 2019, 20, 257. [Google Scholar] [CrossRef] [PubMed]
- Min, Z.; Shigang, Z.; Rong, L.; Jing, L.; Lu, L.; Chihong, Z.; Zehou, L.; Congpei, L.; Lei, Z.; Levi, Y.; et al. Genome-wide identification, phylogenetic and expression analysis of the heat shock transcription factor family in bread wheat (Triticum aestivum L.). BMC Genom. 2019, 20, 505. [Google Scholar]
- Xue, G.P.; Sadat, S.; Drenth, J.; McIntyre, C.L. The heat shock factor family from Triticum aestivum in response to heat and other major abiotic stresses and their role in regulation of heat shock protein genes. J. Exp. Bot. 2014, 65, 539–557. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Xin, M.; Qin, J.; Peng, H.; Ni, Z.; Yao, Y.; Sun, Q. Temporal transcriptome profiling reveals expression partitioning of homeologous genes contributing to heat and drought acclimation in wheat (Triticum aestivum L.). BMC Plant Biol. 2015, 15, 152. [Google Scholar] [CrossRef] [Green Version]
- Ogawa, D.; Yamaguchi, K.; Nishiuchi, T. High-level overexpression of the Arabidopsis HsfA2 gene confers not only increased themotolerance but also salt/osmotic stress tolerance and enhanced callus growth. J. Exp. Bot. 2007, 58, 3373–3383. [Google Scholar] [CrossRef] [Green Version]
- Nishizawa, A.; Yabuta, Y.; Yoshida, E.; Maruta, T.; Yoshimura, K.; Shigeoka, S. Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J. 2006, 48, 535–547. [Google Scholar] [CrossRef]
- Lesk, C.; Rowhani, P.; Ramankutty, N. Influence of extreme weather disasters on global crop production. Nature 2016, 529, 84–87. [Google Scholar] [CrossRef]
- Hrmova, M.; Hussain, S.S. Plant transcription factors involved in drought and associated stresses. Int. J. Mol. Sci. 2021, 22, 5662. [Google Scholar] [CrossRef]
- Wang, J.; Chen, L.; Long, Y.; Si, W.; Cheng, B.; Jiang, H. A novel heat shock transcription factor (ZmHsf08) negatively regulates salt and drought stress responses in maize. Int. J. Mol. Sci. 2021, 22, 11922. [Google Scholar] [CrossRef]
- Yokotani, N.; Ichikawa, T.; Kondou, Y.; Matsui, M.; Hirochika, H.; Iwabuchi, M.; Oda, K. Expression of rice heat stress transcription factor OsHsfA2e enhances tolerance to environmental stresses in transgenic Arabidopsis. Planta 2008, 227, 957–967. [Google Scholar] [CrossRef] [PubMed]
- Zang, D.; Wang, J.; Zhang, X.; Liu, Z.; Wang, Y. Arabidopsis heat shock transcription factor HSFA7b positively mediates salt stress tolerance by binding to an E-box-like motif to regulate gene expression. J. Exp. Bot. 2019, 70, 5355–5374. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zheng, Q.; Chen, L.; Liang, Y.; Wu, J. Ectopic overexpression of maize heat shock transcription factor gene ZmHsf04 confers increased thermo and salt-stress tolerance in transgenic Arabidopsis. Acta Physiol. Plant. 2018, 40, 9. [Google Scholar] [CrossRef]
- Li, G.; Zhang, H.; Shao, H.; Wang, G.; Zhang, Y.; Zhang, Y.; Zhao, L.; Guo, X.; Sheteiwy, M. ZmHsf05, a new heat shock transcription factor from Zea mays L. improves thermotolerance in Arabidopsis thaliana and rescues thermotolerance defects of the athsfa2 mutant. Plant Sci. 2019, 283, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Si, W.; Liang, Q.; Chen, L.; Song, F.; Chen, Y.; Jiang, H. Ectopic overexpression of maize heat stress transcription factor ZmHsf05 confers drought tolerance in transgenic rice. Genes 2021, 12, 1568. [Google Scholar] [CrossRef]
- Zhu, M.; Zhang, M.; Gao, D.; Zhou, K.; Tang, S.; Zhou, B.; Lv, Y. Rice OsHSFA3 gene improves drought tolerance by modulating polyamine biosynthesis depending on abscisic acid and ROS levels. Int. J. Mol. Sci. 2020, 21, 1857. [Google Scholar] [CrossRef] [Green Version]
- Lamers, J.; van der Meer, T.; Testerink, C. How plants sense and respond to stressful environments. Plant Physiol. 2020, 182, 1624–1635. [Google Scholar] [CrossRef] [Green Version]
- Rossel, J.B.; Walter, P.B.; Hendrickson, L.; Chow, W.S.; Poole, A.; Mullineaux, P.M.; Pogson, B.J. A mutation affecting ASCORBATE PEROXIDASE 2 gene expression reveals a link between responses to high light and drought tolerance. Plant Cell Environ. 2006, 29, 269–281. [Google Scholar] [CrossRef] [Green Version]
- Selvaraj, M.G.; Ishizaki, T.; Valencia, M.; Ogawa, S.; Dedicova, B.; Ogata, T.; Yoshiwara, K.; Maruyama, K.; Kusano, M.; Saito, K.; et al. Overexpression of an Arabidopsis thaliana galactinol synthase gene improves drought tolerance in transgenic rice and increased grain yield in the field. Plant Biotechnol. J. 2017, 15, 1465–1477. [Google Scholar] [CrossRef] [Green Version]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, Y.; Tang, S.; Zhao, Q.; Zhang, Z.; Zhang, H.; Dong, L.; Guo, H.; Xie, Q. An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J. 2010, 61, 893–903. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Tian, X.; Wang, F.; Zhang, L.; Xin, M.; Hu, Z.; Yao, Y.; Ni, Z.; Sun, Q.; Peng, H. Characterization of wheat MYB genes responsive to high temperatures. BMC Plant Biol. 2017, 17, 208. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).