Changes in Metabotropic Glutamate Receptor Gene Expression in Rat Brain in a Lithium–Pilocarpine Model of Temporal Lobe Epilepsy
Abstract
1. Introduction
2. Results
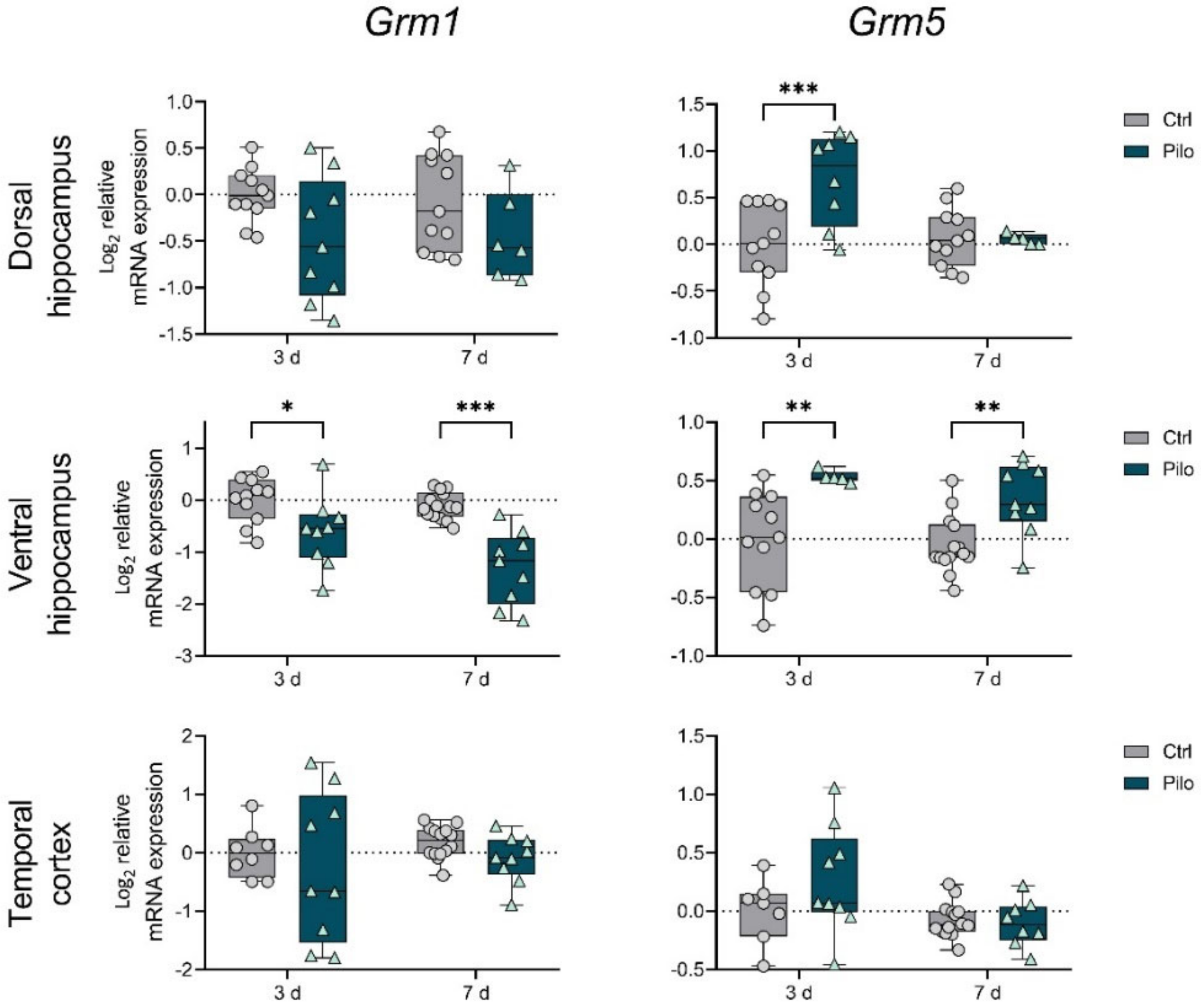
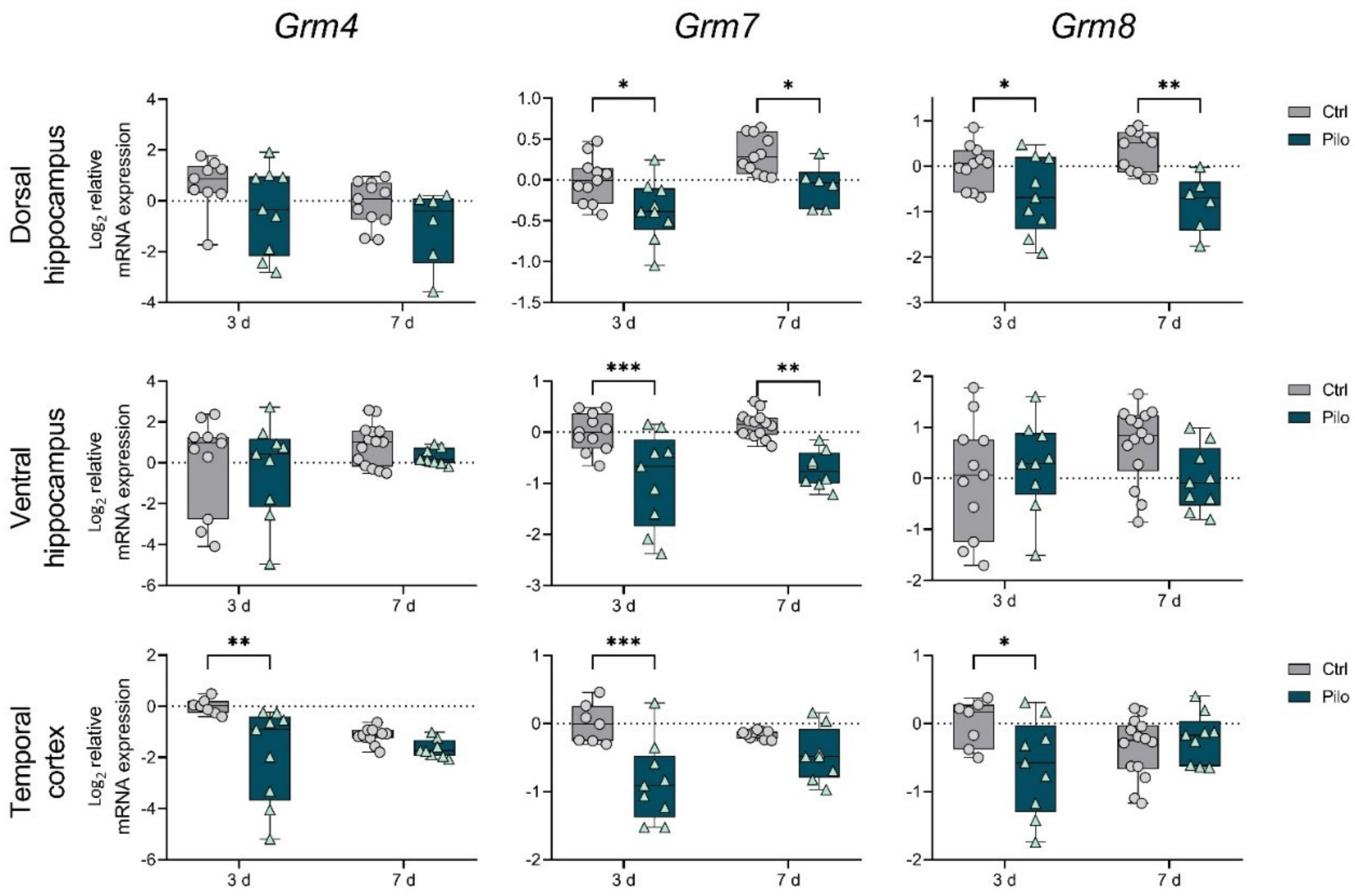
2.1. Changes in the Gene Expression of mGluRs in the Latent Phase of the Model
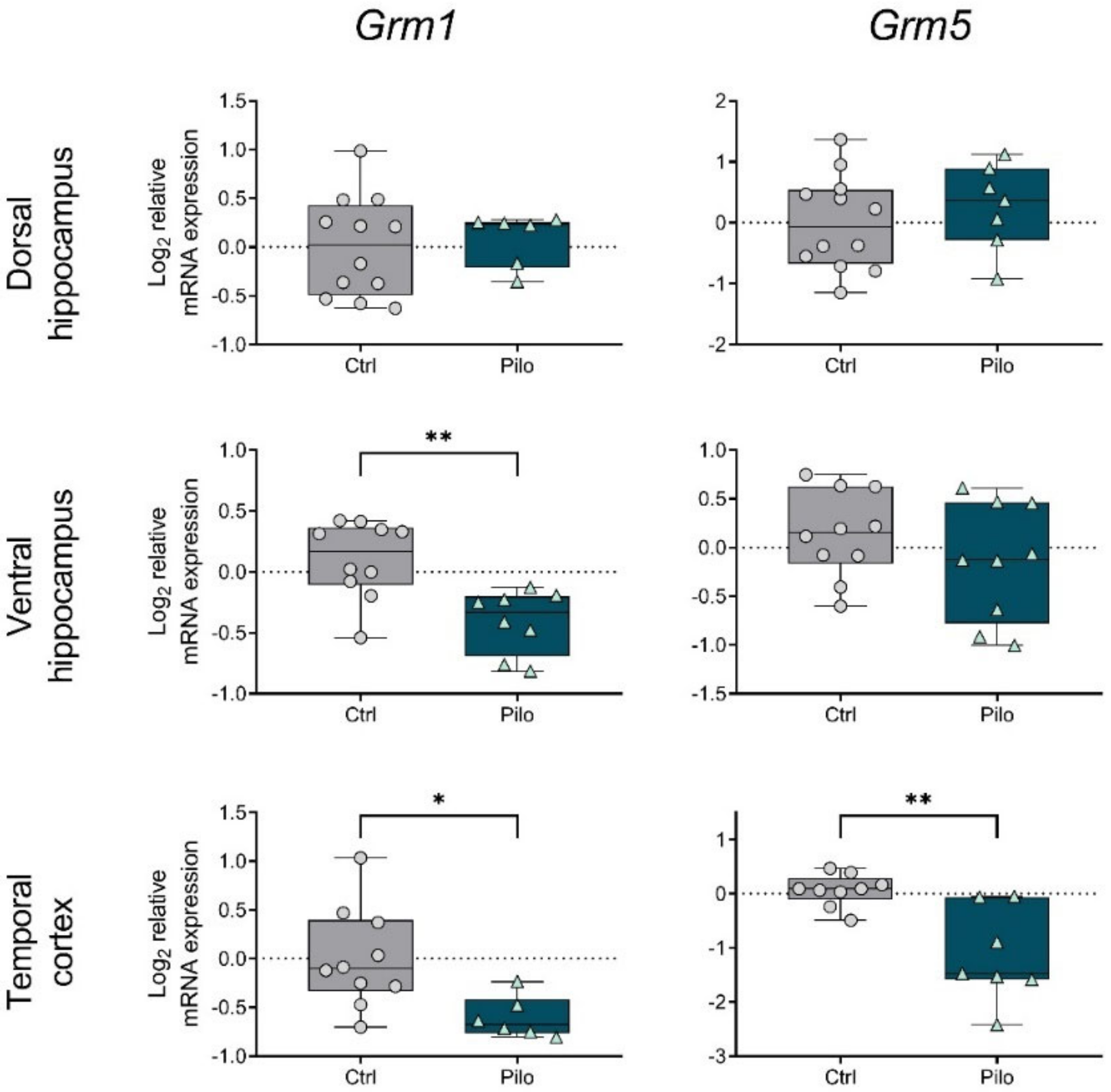
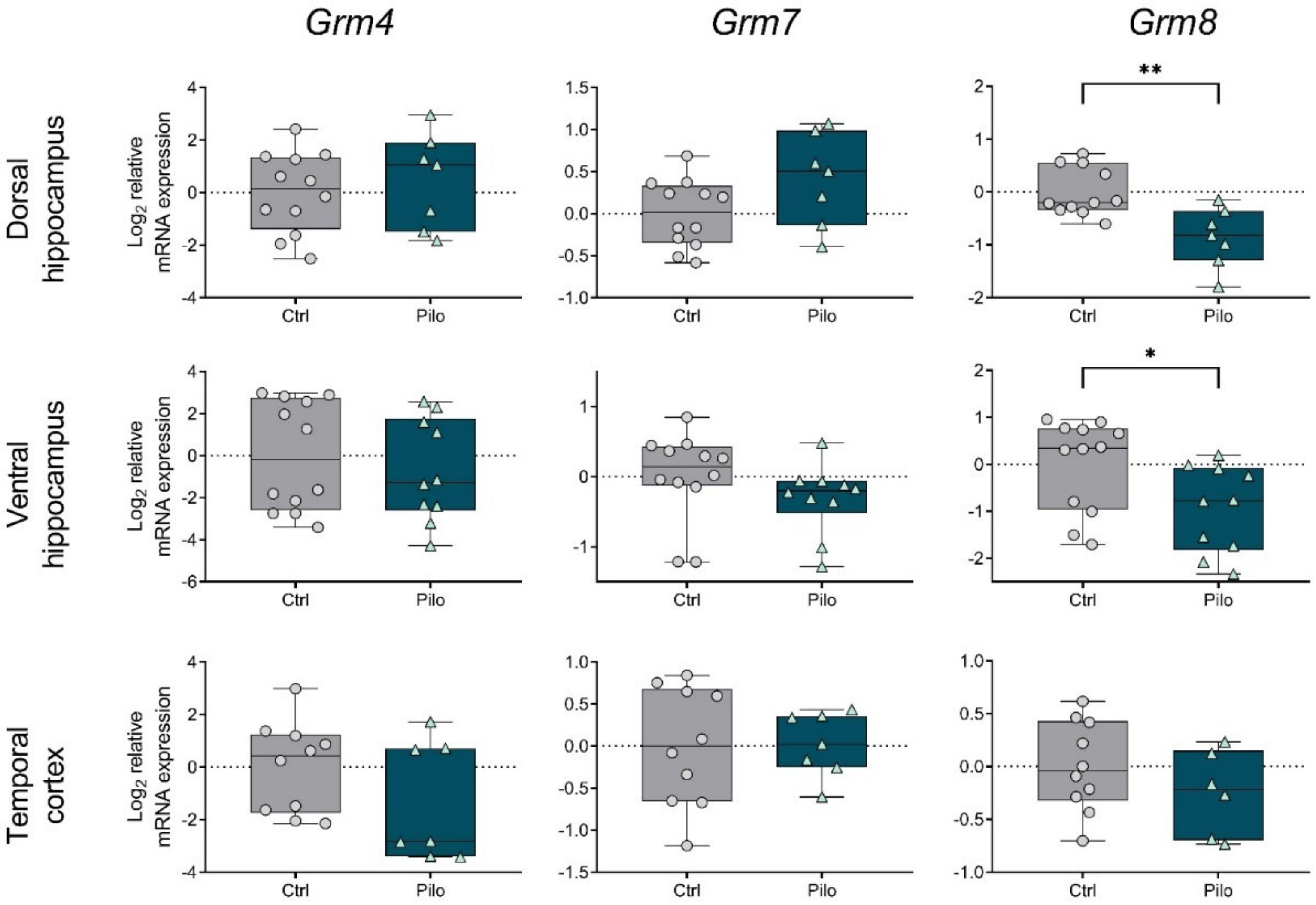
2.2. Changes in the Gene Expression of mGluRs in the Chronic Phase of the Model
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Lithium–Pilocarpine Model
4.3. Spontaneous Recurrent Seizure Registration
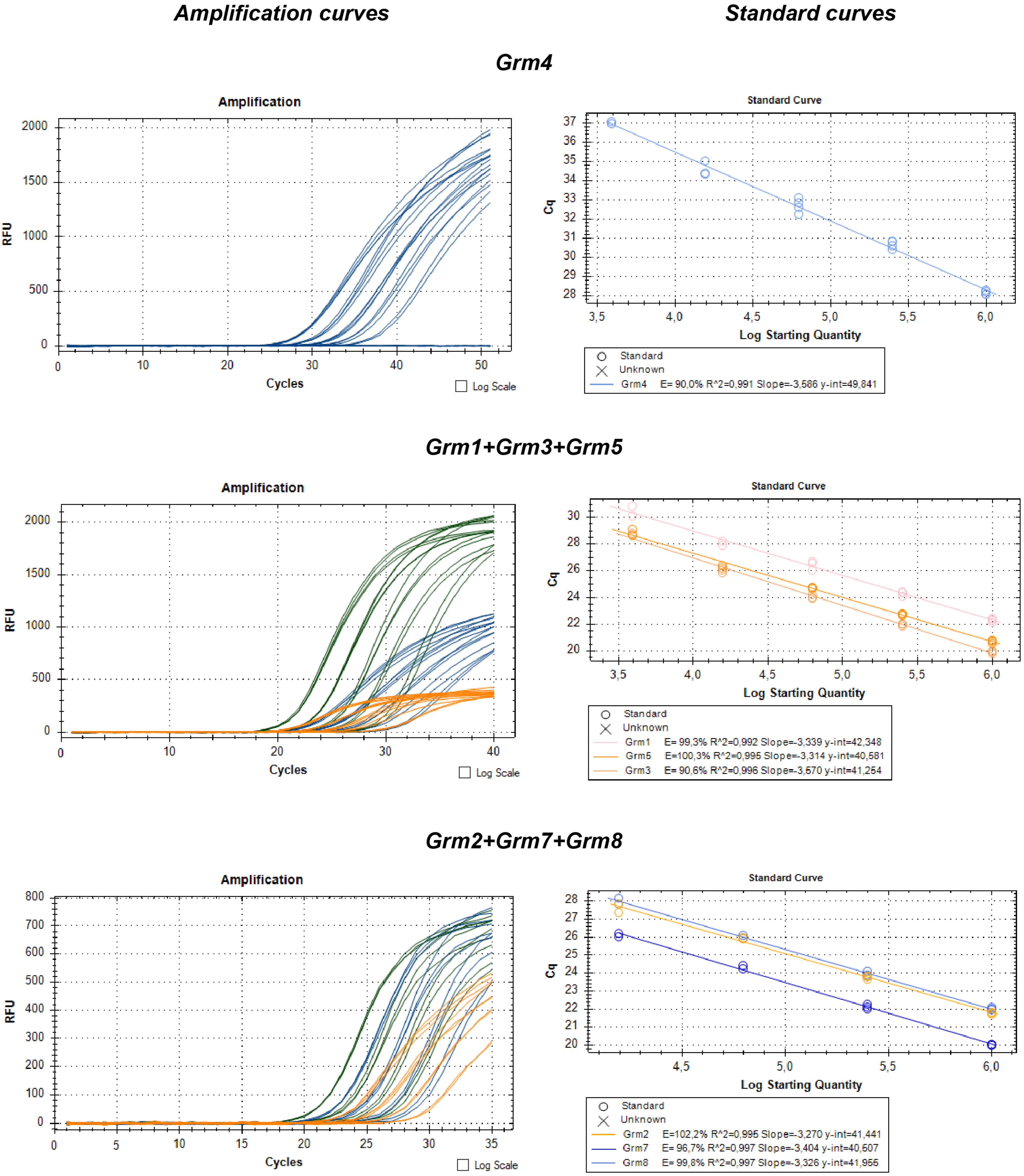
4.4. Reverse Transcription Followed by Quantitative Real-Time PCR (RT-qPCR)
4.5. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Gene Symbol RefSeq Accession Number | Nucleotide Sequences (Forward, Reverse, TaqMan Probe) | Final Primer and Probe Concentrations (nM) | Reference |
|---|---|---|---|
| Grm1 NM_001114330.1 | GGGAATGCCAAGAAGAGGCAG CTGCAGTGTGGGGGTTTTCAA FAM-GCCCAGCAGCCAGTGTCCGTCGGC-BHQ1 | 400 200 | This work |
| Grm2 NM_001105711.1 | TCCAGTGATTATCGGGTGCAG AACTTGGGTGCAAAGAGGCA FAM-TGCGTGTCCGTCAGCCTCAGTGGCT-BHQ1 | 200 100 | This work |
| Grm3 NM_001105712.1 | CAGGAGTTGACGGTGCGAG GCCTGTCCTTCAGATAAGGGAG ROX-TCGGTGACGGGCTCTTTCAGCCCAA-BHQ2 | 200 200 | This work |
| Grm4 NM_022666.1 | GGCAGTGCGAGCAGCTAAGG CCGGTCACTCCTACCAACCG FAM-CTCCCTGAGCTCCCCCGGAGCAGC-BHQ1 | 200 150 | This work |
| Grm5 NM_017012.1 | ATGCATGTAGGAGACGGCAA TTTCCGTTGGAGCTTAGGGTTT HEX-CGTCCGCTGCCAGCAGATCCAGCA-BHQ2 | 400 200 | [57] (primers); this work (probe) |
| Grm7 NM_031040.1 | CCAGACAACAAACACAACCAACC GCGTTCCCTTCTGTGTCTTCTTC HEX-TGCAGTGGGGCAAAGGAGTCCGAG-BHQ2 | 200 100 | [58] (primers); this work (probe) |
| Grm8 NM_022202.1 | TCATCGGGCACTGGACAAAT CACGGTTTTCTTCCTCTCCCC ROX-TGTCTGCAGCCTGCCGTGCAAGCCC-BHQ2 | 300 100 | This work |
| Actb NM_031144 | TGTCACCAACTGGGACGATA GGGGTGTTGAAGGTCTCAAA FAM-CGTGTGGCCCCTGAGGAGCAC-BHQ1 | 200 200 | [59] (primers); [54] (probe) |
| Gapdh NM_017008 | TGCACCACCAACTGCTTAG GGATGCAGGGATGATGTTC R6G-ATCACGCCACAGCTTTCCAGAGGG-BHQ2 | 200 100 | [60] |
| B2m NM_012512 | TGCCATTCAGAAAACTCCCC GAGGAAGTTGGGCTTCCCATT ROX-ATTCAAGTGTACTCTCGCCATCCACCG-BHQ1 | 200 100 | [61] |
| Rpl13a NM_173340 | GGATCCCTCCACCCTATGACA CTGGTACTTCCACCCGACCTC FAM-CTGCCCTCAAGGTTGTGCGGCT-BHQ1 | 200 100 | [62] (primers); [54] (probe) |
| Sdha NM_130428 | AGACGTTTGACAGGGGAATG TCATCAATCCGCACCTTGTA R6G-ACCTGGTGGAGACGCTGGAGCT-BHQ2 | 200 100 | [63] (primers); [54] (probe) |
| Ppia NM_017101 | AGGATTCATGTGCCAGGGTG CTCAGTCTTGGCAGTGCAGA ROX-CACGCCATAATGGCACTGGTGGCA-BHQ1 | 200 100 | [36] |
| Hprt1 NM_012583 | TCCTCAGACCGCTTTTCCCGC TCATCATCACTAATCACGACGCTGG FAM-CCGACCGGTTCTGTCATGTCGACCCT-BHQ1 | 200 100 | [64] (primers); [54] (probe) |
| Pgk1 NM_053291 | ATGCAAAGACTGGCCAAGCTAC AGCCACAGCCTCAGCATATTTC R6G-TGCTGGCTGGATGGGCTTGGA-BHQ2 | 200 100 | [65] (primers); [54] (probe) |
| Ywhaz NM_013011 | GATGAAGCCATTGCTGAACTTG GTCTCCTTGGGTATCCGATGTC ROX-TGAAGAGTCGTACAAAGACAGCACGC-BHQ1 | 200 100 | [65] (primers); [54] (probe) |
References
- Banerjee, P.N.; Filippi, D.; Allen Hauser, W. The descriptive epidemiology of epilepsy—A review. Epilepsy Res. 2009, 85, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Janmohamed, M.; Brodie, M.J.; Kwan, P. Pharmacoresistance—Epidemiology, mechanisms, and impact on epilepsy treatment. Neuropharmacology 2020, 168, 107790. [Google Scholar] [CrossRef] [PubMed]
- Löscher, W. The holy grail of epilepsy prevention: Preclinical approaches to antiepileptogenic treatments. Neuropharmacology 2020, 167, 107605. [Google Scholar] [CrossRef] [PubMed]
- Scharfman, H.E. The neurobiology of epilepsy. Curr. Neurol. Neurosci. Rep. 2007, 7, 348–354. [Google Scholar] [CrossRef]
- Lasoñ, W.; Chlebicka, M.; Rejdak, K. Research advances in basic mechanisms of seizures and antiepileptic drug action. Pharmacol. Rep. 2013, 65, 787–801. [Google Scholar] [CrossRef]
- Celli, R.; Santolini, I.; Van Luijtelaar, G.; Ngomba, R.T.; Bruno, V.; Nicoletti, F. Targeting metabotropic glutamate receptors in the treatment of epilepsy: Rationale and current status. Expert Opin. Ther. Targets 2019, 23, 341–351. [Google Scholar] [CrossRef]
- Anwyl, R. Metabotropic glutamate receptor-dependent long-term potentiation. Neuropharmacology 2009, 56, 735–740. [Google Scholar] [CrossRef]
- Nicoletti, F.; Bockaert, J.; Collingridge, G.L.; Conn, P.J.; Ferraguti, F.; Schoepp, D.D.; Wroblewski, J.T.; Pin, J.P. Metabotropic glutamate receptors: From the workbench to the bedside. Neuropharmacology 2011, 60, 1017–1041. [Google Scholar] [CrossRef]
- Tang, F.; Bradford, H.; Ling, E.-A. Metabotropic Glutamate Receptors in the Control of Neuronal Activity and as Targets for Development of Anti-Epileptogenic Drugs. Curr. Med. Chem. 2009, 16, 2189–2204. [Google Scholar] [CrossRef]
- Niswender, C.M.; Conn, P.J. Metabotropic Glutamate Receptors: Physiology, Pharmacology, and Disease. Annu. Rev. Pharmacol. Toxicol. 2010, 50, 295. [Google Scholar] [CrossRef]
- Di Menna, L.; Joffe, M.E.; Iacovelli, L.; Orlando, R.; Lindsley, C.W.; Mairesse, J.; Gressèns, P.; Cannella, M.; Caraci, F.; Copani, A.; et al. Functional partnership between mGlu3 and mGlu5 metabotropic glutamate receptors in the central nervous system. Neuropharmacology 2018, 128, 301–313. [Google Scholar] [CrossRef] [PubMed]
- Aronica, E.; van Vliet, E.A.; Mayboroda, O.A.; Troost, D.; da Silva, F.H.; Gorter, J.A. Upregulation of metabotropic glutamate receptor subtype mGluR3 and mGluR5 in reactive astrocytes in a rat model of mesial temporal lobe epilepsy. Eur. J. Neurosci. 2000, 12, 2333–2344. [Google Scholar] [CrossRef] [PubMed]
- Blümcke, I.; Becker, A.J.; Klein, C.; Scheiwe, C.; Lie, A.A.; Beck, H.; Waha, A.; Friedl, M.G.; Kuhn, R.; Emson, P.; et al. Temporal Lobe Epilepsy Associated Up-Regulation of Metabotropic Glutamate Receptors: Correlated Changes in mGluR1 mRNA and Protein Expression in Experimental Animals and Human Patients. J. Neuropathol. Exp. Neurol. 2000, 59, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.-R.; Lee, W.-L.; Yeo, T.T. Expression of the group I metabotropic glutamate receptor in the hippocampus of patients with mesial temporal lobe epilepsy. J. Neurocytol. 2002, 30, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Sanabria, E.R.; Otalora, L.F.P.; Arshadmansab, M.F.; Herrera, B.; Francisco, S.; Ermolinsky, B.S. Impaired expression and function of group II metabotropic glutamate receptors in pilocarpine-treated chronically epileptic rats. Brain Res. 2008, 1240, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Curia, G.; Longo, D.; Biagini, G.; Jones, R.S.G.; Avoli, M. The pilocarpine model of temporal lobe epilepsy. J. Neurosci. Methods 2008, 172, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Lévesque, M.; Biagini, G.; de Curtis, M.; Gnatkovsky, V.; Pitsch, J.; Wang, S.; Avoli, M. The pilocarpine model of mesial temporal lobe epilepsy: Over one decade later, with more rodent species and new investigative approaches. Neurosci. Biobehav. Rev. 2021, 130, 274–291. [Google Scholar] [CrossRef]
- Bartolomei, F.; Khalil, M.; Wendling, F.; Sontheimer, A.; Regis, J.; Ranjeva, J.-P.; Guye, M.; Chauvel, P. Entorhinal Cortex Involvement in Human Mesial Temporal Lobe Epilepsy: An Electrophysiologic and Volumetric Study. Epilepsia 2005, 46, 677–687. [Google Scholar] [CrossRef]
- Mathern, G.W.; Adelson, P.D.; Cahan, L.D.; Leite, J.P. Hippocampal neuron damage in human epilepsy: Meyer’s hypothesis revisited. Prog. Brain Res. 2002, 135, 237–251. [Google Scholar] [CrossRef]
- Mathern, G.W.; Kuhlman, P.A.; Mendoza, D.; Pretorius, J.K. Human fascia dentata anatomy and hippocampal neuron densities differ depending on the epileptic syndrome and age at first seizure. J. Neuropathol. Exp. Neurol. 1997, 56, 199–212. [Google Scholar] [CrossRef][Green Version]
- Racine, R.J. Modification of seizure activity by electrical stimulation. II. Motor seizure. Electroencephalogr. Clin. Neurophysiol. 1972, 32, 281–294. [Google Scholar] [CrossRef]
- Clifford, D.B.; Olney, J.W.; Maniotis, A.; Collins, R.C.; Zorumski, C.F. The functional anatomy and pathology of lithium-pilocarpine and high-dose pilocarpine seizures. Neuroscience 1987, 23, 953–968. [Google Scholar] [CrossRef]
- Nagao, T.; Alonso, A.; Avoli, M. Epileptiform activity induced by pilocarpine in the rat hippocampal-entorhinal slice preparation. Neuroscience 1996, 72, 399–408. [Google Scholar] [CrossRef]
- Smolders, I.; Khan, G.M.; Manil, J.; Ebinger, G.; Michotte, Y. NMDA receptor-mediated pilocarpine-induced seizures: Characterization in freely moving rats by microdialysis. Br. J. Pharmacol. 1997, 121, 1171–1179. [Google Scholar] [CrossRef] [PubMed]
- Inostroza, M.; Cid, E.; Menendez de la Prida, L.; Sandi, C. Different emotional disturbances in two experimental models of temporal Lobe Epilepsy in rats. PLoS ONE 2012, 7, e038959. [Google Scholar] [CrossRef] [PubMed]
- Smolensky, I.V.; Zubareva, O.E.; Kalemenev, S.V.; Lavrentyeva, V.V.; Dyomina, A.V.; Karepanov, A.A.; Zaitsev, A. V Impairments in cognitive functions and emotional and social behaviors in a rat lithium-pilocarpine model of temporal lobe epilepsy. Behav. Brain Res. 2019, 372, 112044. [Google Scholar] [CrossRef]
- De Lanerolle, N.C.; Kim, J.H.; Williamson, A.; Spencer, S.S.; Zaveri, H.P.; Eid, T.; Spencer, D.D. A retrospective analysis of hippocampal pathology in human temporal lobe epilepsy: Evidence for distinctive patient subcategories. Epilepsia 2003, 44, 677–687. [Google Scholar] [CrossRef]
- Das, A.; Wallace, G.C.; Holmes, C.; McDowell, M.L.; Smith, J.A.; Marshall, J.D.; Bonilha, L.; Edwards, J.C.; Glazier, S.S.; Ray, S.K.; et al. Hippocampal tissue of patients with refractory temporal lobe epilepsy is associated with astrocyte activation, inflammation, and altered expression of channels and receptors. Neuroscience 2012, 220, 237–246. [Google Scholar] [CrossRef]
- Kirschstein, T.; Bauer, M.; Müller, L.; Rüschenschmidt, C.; Reitze, M.; Becker, A.J.; Schoch, S.; Beck, H. Loss of metabotropic glutamate receptor-dependent long-term depression via downregulation of mGluR5 after status epilepticus. J. Neurosci. 2007, 27, 7696–7704. [Google Scholar] [CrossRef]
- Dammann, F.; Kirschstein, T.; Guli, X.; Müller, S.; Porath, K.; Rohde, M.; Tokay, T.; Köhling, R. Bidirectional shift of group III metabotropic glutamate receptor-mediated synaptic depression in the epileptic hippocampus. Epilepsy Res. 2018, 139, 157–163. [Google Scholar] [CrossRef]
- Pitsch, J.; Schoch, S.; Gueler, N.; Flor, P.J.; van der Putten, H.; Becker, A.J. Functional role of mGluR1 and mGluR4 in pilocarpine-induced temporal lobe epilepsy. Neurobiol. Dis. 2007, 26, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Jovanovic, M.; Rooney, M.S.; Mertins, P.; Przybylski, D.; Chevrier, N.; Satija, R.; Rodriguez, E.H.; Fields, A.P.; Schwartz, S.; Raychowdhury, R.; et al. Dynamic profiling of the protein life cycle in response to pathogens. Science 2015, 347. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Beyer, A.; Aebersold, R. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell 2016, 165, 535–550. [Google Scholar] [CrossRef] [PubMed]
- Zubareva, O.E.; Kovalenko, A.A.; Kalemenev, S.V.; Schwarz, A.P.; Karyakin, V.B.; Zaitsev, A.V. Alterations in mRNA expression of glutamate receptor subunits and excitatory amino acid transporters following pilocarpine-induced seizures in rats. Neurosci. Lett. 2018, 686, 94–100. [Google Scholar] [CrossRef]
- Müller, L.; Tokay, T.; Porath, K.; Köhling, R.; Kirschstein, T. Enhanced NMDA receptor-dependent LTP in the epileptic CA1 area via upregulation of NR2B. Neurobiol. Dis. 2013, 54, 183–193. [Google Scholar] [CrossRef]
- Malkin, S.L.; Amakhin, D.V.; Veniaminova, E.A.; Kim, K.K.; Zubareva, O.E.; Magazanik, L.G.; Zaitsev, A. V Changes of AMPA receptor properties in the neocortex and hippocampus following pilocarpine-induced status epilepticus in rats. Neuroscience 2016, 327, 146–155. [Google Scholar] [CrossRef]
- Di Maio, R.; Mastroberardino, P.G.; Hu, X.; Montero, L.; Greenamyre, J.T. Pilocapine alters NMDA receptor expression and function in hippocampal neurons: NADPH oxidase and ERK1/2 mechanisms. Neurobiol. Dis. 2011, 42, 482–495. [Google Scholar] [CrossRef]
- Peng, W.-F.; Ding, J.; Li, X.; Fan, F.; Zhang, Q.-Q.; Wang, X. N-methyl-D-aspartate receptor NR2B subunit involved in depression-like behaviours in lithium chloride-pilocarpine chronic rat epilepsy model. Epilepsy Res. 2016, 119, 77–85. [Google Scholar] [CrossRef]
- Zaitsev, A.V.; Amakhin, D.V.; Dyomina, A.V.; Zakharova, M.V.; Ergina, J.L.; Postnikova, T.Y.; Diespirov, G.P.; Magazanik, L.G. Synaptic Dysfunction in Epilepsy. J. Evol. Biochem. Physiol. 2021, 57, 542–563. [Google Scholar] [CrossRef]
- Ure, J.; Baudry, M.; Perassolo, M. Metabotropic glutamate receptors and epilepsy. J. Neurol. Sci. 2006, 247, 1–9. [Google Scholar] [CrossRef]
- Moldrich, R.X.; Chapman, A.G.; De Sarro, G.; Meldrum, B.S. Glutamate metabotropic receptors as targets for drug therapy in epilepsy. Eur. J. Pharmacol. 2003, 476, 3–16. [Google Scholar] [CrossRef]
- Jesse, C.R.; Savegnago, L.; Rocha, J.B.T.; Nogueira, C.W. Neuroprotective effect caused by MPEP, an antagonist of metabotropic glutamate receptor mGluR5, on seizures induced by pilocarpine in 21-day-old rats. Brain Res. 2008, 1198, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Domin, H.; Zięba, B.; Gołembiowska, K.; Kowalska, M.; Dziubina, A.; Śmiałowska, M. Neuroprotective potential of mGluR5 antagonist MTEP: Effects on kainate-induced excitotoxicity in the rat hippocampus. Pharmacol. Rep. 2010, 62, 1051–1061. [Google Scholar] [CrossRef]
- Dyomina, A.V.; Kovalenko, A.A.; Zakharova, M.V.; Postnikova, T.Y.; Griflyuk, A.V.; Smolensky, I.V.; Antonova, I.V.; Zaitsev, A.V. MTEP, a Selective mGluR5 Antagonist, Had a Neuroprotective Effect but Did Not Prevent the Development of Spontaneous Recurrent Seizures and Behavioral Comorbidities in the Rat Lithium-Pilocarpine Model of Epilepsy. Int. J. Mol. Sci. 2022, 23, 497. [Google Scholar] [CrossRef]
- Hanak, T.J.; Libbey, J.E.; Doty, D.J.; Sim, J.T.; DePaula-Silva, A.B.; Fujinami, R.S. Positive modulation of mGluR5 attenuates seizures and reduces TNF-α+ macrophages and microglia in the brain in a murine model of virus-induced temporal lobe epilepsy. Exp. Neurol. 2019, 311, 194–204. [Google Scholar] [CrossRef]
- Loane, D.J.; Stoica, B.A.; Byrnes, K.R.; Jeong, W.; Faden, A.I. Activation of mGluR5 and Inhibition of NADPH Oxidase Improves Functional Recovery after Traumatic Brain Injury. J. Neurotrauma 2013, 30, 403–412. [Google Scholar] [CrossRef]
- Maj, M.; Bruno, V.; Dragic, Z.; Yamamoto, R.; Battaglia, G.; Inderbitzin, W.; Stoehr, N.; Stein, T.; Gasparini, F.; Vranesic, I.; et al. (−)-PHCCC, a positive allosteric modulator of mGluR4: Characterization, mechanism of action, and neuroprotection. Neuropharmacology 2003, 45, 895–906. [Google Scholar] [CrossRef]
- Szczurowska, E.; Mareš, P. Positive Allosteric Modulator of mGluR4 PHCCC Exhibits Proconvulsant Action in Three Models of Epileptic Seizures in Immature Rats. Physiol. Res. 2012, 61, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, Y.; Yang, J.; Xu, T.; Tan, C.; Zhang, P.; Liu, Q.; Chen, Y. G-alpha interacting protein interacting protein, C terminus 1 regulates epileptogenesis by increasing the expression of metabotropic glutamate receptor 7. CNS Neurosci. Ther. 2022, 28, 126–138. [Google Scholar] [CrossRef]
- Jantas, D.; Lech, T.; Gołda, S.; Pilc, A.; Lasoń, W. New evidences for a role of mGluR7 in astrocyte survival: Possible implications for neuroprotection. Neuropharmacology 2018, 141, 223–237. [Google Scholar] [CrossRef]
- Girard, B.; Tuduri, P.; Moreno, M.P.; Sakkaki, S.; Barboux, C.; Bouschet, T.; Varrault, A.; Vitre, J.; McCort-Tranchepain, I.; Dairou, J.; et al. The mGlu7 receptor provides protective effects against epileptogenesis and epileptic seizures. Neurobiol. Dis. 2019, 129, 13–28. [Google Scholar] [CrossRef] [PubMed]
- Dyomina, A.V.; Zubareva, O.E.; Smolensky, I.V.; Vasilev, D.S.; Zakharova, M.V.; Kovalenko, A.A.; Schwarz, A.P.; Ischenko, A.M.; Zaitsev, A.V. Anakinra Reduces Epileptogenesis, Provides Neuroprotection, and Attenuates Behavioral Impairments in Rats in the Lithium–Pilocarpine Model of Epilepsy. Pharmaceuticals 2020, 13, 340. [Google Scholar] [CrossRef] [PubMed]
- Paxinos, G.; Watson, C. The Rat Brain in Stereotaxic Coordinates; Elsevier: Amsterdam, The Netherlands, 2007. [Google Scholar]
- Schwarz, A.P.; Malygina, D.A.; Kovalenko, A.A.; Trofimov, A.N.; Zaitsev, A.V. Multiplex qPCR assay for assessment of reference gene expression stability in rat tissues/samples. Mol. Cell. Probes 2020, 53, 101611. [Google Scholar] [CrossRef] [PubMed]
- Svec, D.; Tichopad, A.; Novosadova, V.; Pfaffl, M.W.; Kubista, M. How good is a PCR efficiency estimate: Recommendations for precise and robust qPCR efficiency assessments. Biomol. Detect. Quantif. 2015, 3, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Dulman, R.S.; Auta, J.; Teppen, T.; Pandey, S.C. Acute Ethanol Produces Ataxia and Induces Fmr1 Expression via Histone Modifications in the Rat Cerebellum. Alcohol. Clin. Exp. Res. 2019, 43, 1191–1198. [Google Scholar] [CrossRef]
- Xu, J.; Yan, C.-H.; Wu, S.-H.; Yu, X.-D.; Yu, X.-G.; Shen, X.-M. Developmental lead exposure alters gene expression of metabotropic glutamate receptors in rat hippocampal neurons. Neurosci. Lett. 2007, 413, 222–226. [Google Scholar] [CrossRef]
- Bonefeld, B.E.; Elfving, B.; Wegener, G. Reference genes for normalization: A study of rat brain tissue. Synapse 2008, 62, 302–309. [Google Scholar] [CrossRef]
- Lin, W.; Burks, C.A.; Hansen, D.R.; Kinnamon, S.C.; Gilbertson, T.A. Taste receptor cells express pH-sensitive leak K+ channels. J. Neurophysiol. 2004, 92, 2909–2919. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Yamauchi, A.; Nishimura, M.; Ueda, N.; Naito, S. Soybean Oil Fat Emulsion Prevents Cytochrome P450 mRNA Down-Regulation Induced by Fat-Free Overdose Total Parenteral Nutrition in Infant Rats. Biol. Pharm. Bull. 2005, 28, 143–147. [Google Scholar] [CrossRef]
- Swijsen, A.; Nelissen, K.; Janssen, D.; Rigo, J.-M.; Hoogland, G. Validation of reference genes for quantitative real-time PCR studies in the dentate gyrus after experimental febrile seizures. BMC Res. Notes 2012, 5, 685. [Google Scholar] [CrossRef] [PubMed]
- Pohjanvirta, R.; Niittynen, M.; Lindén, J.; Boutros, P.C.; Moffat, I.D.; Okey, A.B. Evaluation of various housekeeping genes for their applicability for normalization of mRNA expression in dioxin-treated rats. Chem. Biol. Interact. 2006, 160, 134–149. [Google Scholar] [CrossRef] [PubMed]
- Cook, N.L.; Vink, R.; Donkin, J.J.; van den Heuvel, C. Validation of reference genes for normalization of real-time quantitative RT-PCR data in traumatic brain injury. J. Neurosci. Res. 2009, 87, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Langnaese, K.; John, R.; Schweizer, H.; Ebmeyer, U.; Keilhoff, G. Selection of reference genes for quantitative real-time PCR in a rat asphyxial cardiac arrest model. BMC Mol. Biol. 2008, 9, 53. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kovalenko, A.A.; Zakharova, M.V.; Schwarz, A.P.; Dyomina, A.V.; Zubareva, O.E.; Zaitsev, A.V. Changes in Metabotropic Glutamate Receptor Gene Expression in Rat Brain in a Lithium–Pilocarpine Model of Temporal Lobe Epilepsy. Int. J. Mol. Sci. 2022, 23, 2752. https://doi.org/10.3390/ijms23052752
Kovalenko AA, Zakharova MV, Schwarz AP, Dyomina AV, Zubareva OE, Zaitsev AV. Changes in Metabotropic Glutamate Receptor Gene Expression in Rat Brain in a Lithium–Pilocarpine Model of Temporal Lobe Epilepsy. International Journal of Molecular Sciences. 2022; 23(5):2752. https://doi.org/10.3390/ijms23052752
Chicago/Turabian StyleKovalenko, Anna A., Maria V. Zakharova, Alexander P. Schwarz, Alexandra V. Dyomina, Olga E. Zubareva, and Aleksey V. Zaitsev. 2022. "Changes in Metabotropic Glutamate Receptor Gene Expression in Rat Brain in a Lithium–Pilocarpine Model of Temporal Lobe Epilepsy" International Journal of Molecular Sciences 23, no. 5: 2752. https://doi.org/10.3390/ijms23052752
APA StyleKovalenko, A. A., Zakharova, M. V., Schwarz, A. P., Dyomina, A. V., Zubareva, O. E., & Zaitsev, A. V. (2022). Changes in Metabotropic Glutamate Receptor Gene Expression in Rat Brain in a Lithium–Pilocarpine Model of Temporal Lobe Epilepsy. International Journal of Molecular Sciences, 23(5), 2752. https://doi.org/10.3390/ijms23052752






