Biological Determinants of Metabolic Syndrome in Visceral and Subcutaneous Adipose Tissue from Severely Obese Women
Abstract
:1. Introduction
2. Results
2.1. Anthropometric and Clinical Data
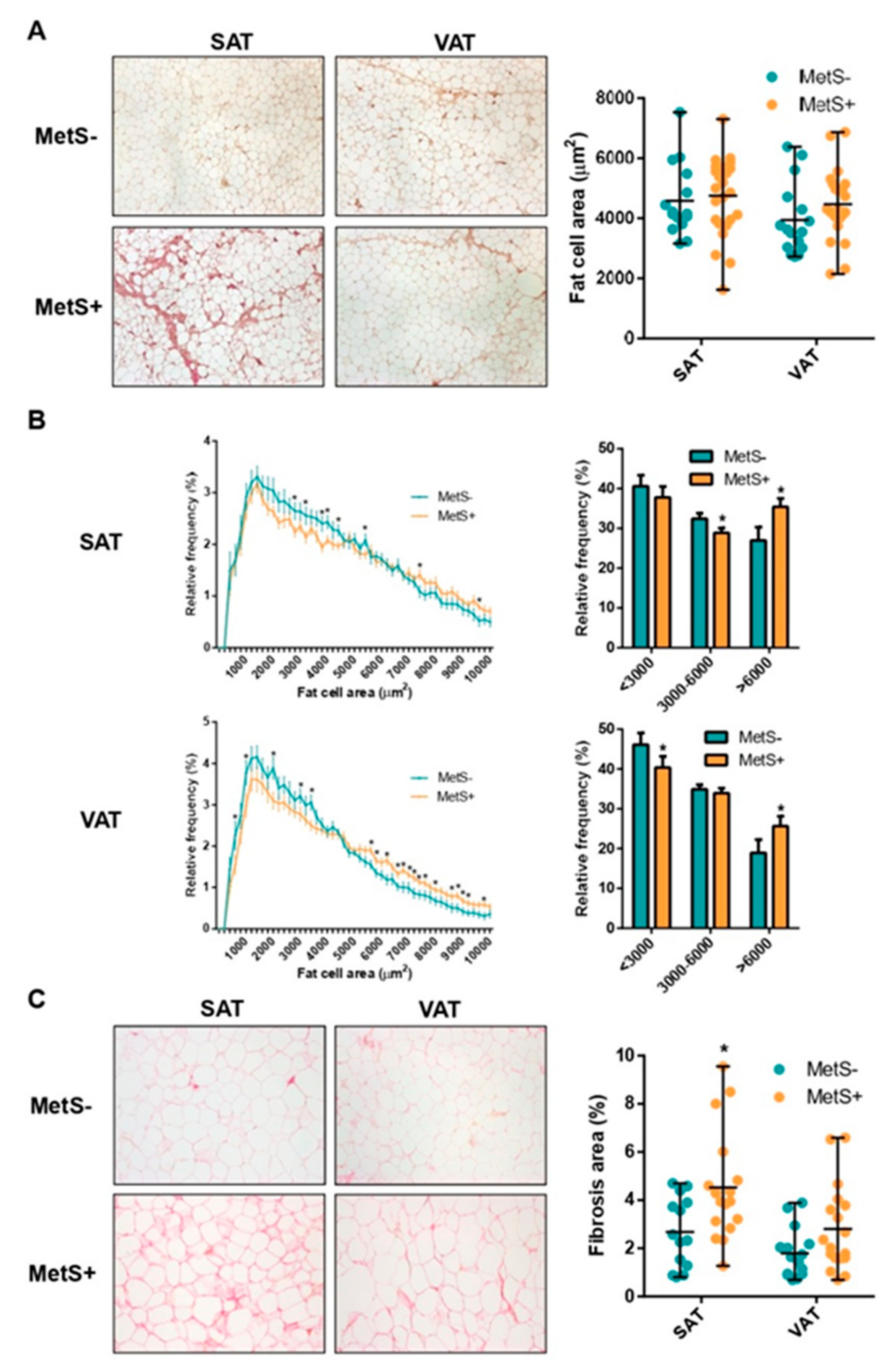
2.2. Fat Cell Size Distribution and Fibrosis
2.3. Differential Gene Expression Analysis
2.4. Expression of Inflammation Markers, Adipokines and Genes Implicated in WAT Expansion
2.5. Expression of Genes Implicated in Glucose and Lipid Metabolism and Fatty Acid Oxidation (FAO)
2.6. Expression of ECM Components and Modifiers
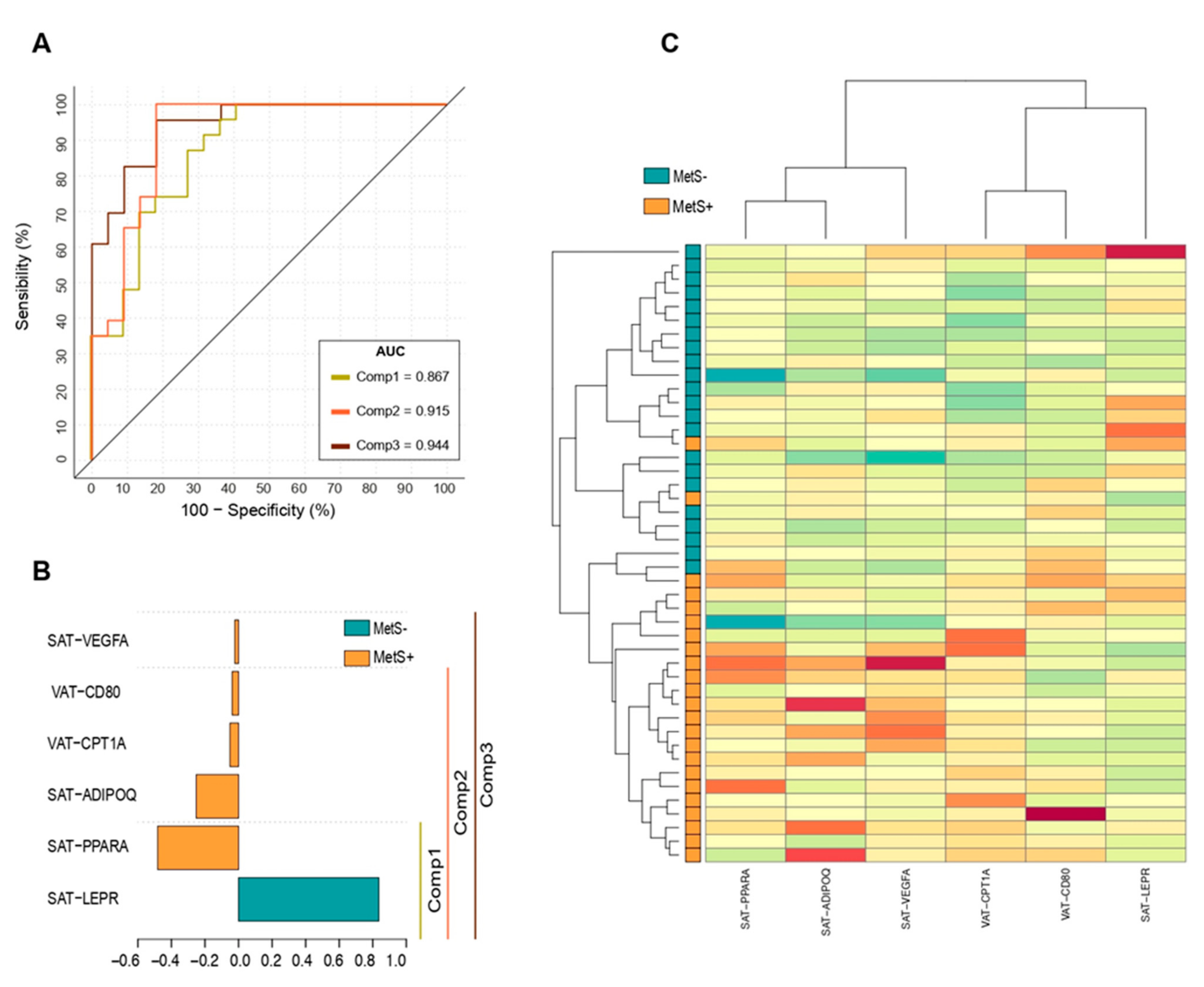
2.7. Principal Component Analysis (PCA) to Identify MetS Subjects
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Clinical and Anthropometric Data
4.3. Determination of Metabolic Health Status
4.4. Body Composition
4.5. White Adipose Tissue Biopsies
4.6. Morphometry and Histopathology
4.7. RNA Extraction and Real Time PCR
4.8. Circulating Levels
4.9. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cornier, M.A.; Dabelea, D.; Hernandez, T.L.; Lindstrom, R.C.; Steig, A.J.; Stob, N.R.; Van Pelt, R.E.; Wang, H.; Eckel, R.H. The metabolic syndrome. Endocr. Rev. 2008, 29, 777–822. [Google Scholar] [CrossRef] [PubMed]
- Alberti, K.G.M.M.; Zimmet, P.; Shaw, J. Metabolic syndrome—A new world-wide definition. A Consensus Statement from the International Diabetes Federation. Diabet. Med. 2006, 23, 469–480. [Google Scholar] [CrossRef] [PubMed]
- Sims, E.A.H. Are there persons who are obese, but metabolically healthy? Metabolism 2001, 50, 1499–1504. [Google Scholar] [CrossRef] [PubMed]
- Oda, E. The metabolic syndrome as a concept of adipose tissue disease. Hypertens. Res. 2008, 31, 1283–1291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klimčáková, E.; Roussel, B.; Márquez-Quiñones, A.; Kováčová, Z.; Kováčiková, M.; Combes, M.; Šiklová-Vítková, M.; Hejnová, J.; Šrámková, P.; Bouloumié, A.; et al. Worsening of obesity and metabolic status yields similar molecular adaptations in human subcutaneous and visceral adipose tissue: Decreased metabolism and increased immune response. J. Clin. Endocrinol. Metab. 2011, 96, E73–E82. [Google Scholar] [CrossRef] [Green Version]
- Mechanick, J.I.; Hurley, D.L.; Garvey, W.T. Adiposity-based chronic disease as a new diagnostic term: The american association of clinical endocrinologists and american college of endocrinology position statement. Endocr. Pract. 2017, 23, 372–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frühbeck, G.; Busetto, L.; Dicker, D.; Yumuk, V.; Goossens, G.H.; Hebebrand, J.; Halford, J.G.C.; Farpour-Lambert, N.J.; Blaak, E.E.; Woodward, E.; et al. The ABCD of Obesity: An EASO Position Statement on a Diagnostic Term with Clinical and Scientific Implications. Obes. Facts 2019, 12, 131–136. [Google Scholar] [CrossRef]
- Crewe, C.; An, Y.A.; Scherer, P.E. The ominous triad of adipose tissue dysfunction: Inflammation, fibrosis, and impaired angiogenesis. J. Clin. Investig. 2017, 127, 74–82. [Google Scholar] [CrossRef] [Green Version]
- O’Rourke, R.W. Inflammation in obesity-related diseases. Surgery 2009, 145, 255–259. [Google Scholar] [CrossRef] [Green Version]
- Zorena, K.; Jachimowicz-Duda, O.; Ślęzak, D.; Robakowska, M.; Mrugacz, M. Adipokines and obesity. Potential link to metabolic disorders and chronic complications. Int. J. Mol. Sci. 2020, 21, 3570. [Google Scholar] [CrossRef]
- Pardo, M.; Roca-Rivada, A.; Seoane, L.M.; Casanueva, F.F. Obesidomics: Contribution of adipose tissue secretome analysis to obesity research. Endocrine 2012, 41, 374–383. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.M. Subcutaneous and visceral adipose tissue: Structural and functional differences. Obes. Rev. 2010, 11, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Stȩpień, M.; Stȩpień, A.; Wlazeł, R.N.; Paradowski, M.; Rizzo, M.; Banach, M.; Rysz, J. Predictors of insulin resistance in patients with obesity: A pilot study. Angiology 2014, 65, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Kahn, C.R.; Wang, G.; Lee, K.Y. Altered adipose tissue and adipocyte function in the pathogenesis of metabolic syndrome. J. Clin. Investig. 2019, 129, 3990–4000. [Google Scholar] [CrossRef]
- Heyn, G.S.; Corrêa, L.H.; Magalhães, K.G. The Impact of Adipose Tissue-Derived miRNAs in Metabolic Syndrome, Obesity, and Cancer. Front. Endocrinol. 2020, 11, 563816. [Google Scholar] [CrossRef]
- Bae, Y.-J.; Kim, S.-H.; Chung, J.-H.; Song, S.-W.; Kim, K.-S.; Kim, M.-K.; Kwon, O.; Choi, M.-S.; Sung, M.-K. Evaluation of adiposity-related biomarkers as metabolic syndrome indicators. Clin. Nutr. Res. 2013, 2, 91. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.Y.; Ahn, S.V.; Yoon, J.H.; Koh, S.B.; Yoon, J.; Yoo, B.S.; Lee, S.H.; Park, J.K.; Choe, K.H.; Guallar, E. Prospective study of serum adiponectin and incident metabolic syndrome: The ARIRANG study. Diabetes Care 2013, 36, 1547–1553. [Google Scholar] [CrossRef] [Green Version]
- Bremer, A.A.; Jialal, I. Adipose tissue dysfunction in nascent metabolic syndrome. J. Obes. 2013, 2013, 393192. [Google Scholar] [CrossRef] [Green Version]
- Ramakrishanan, N.; Denna, T.; Devaraj, S.; Adams-Huet, B.; Jialal, I. Exploratory lipidomics in patients with nascent Metabolic Syndrome. J. Diabetes Complicat. 2018, 32, 791–794. [Google Scholar] [CrossRef]
- Jialal, I.; Devaraj, S. Subcutaneous adipose tissue biology in metabolic syndrome. Horm. Mol. Biol. Clin. Investig. 2018, 33. [Google Scholar] [CrossRef]
- Brochu, M.; Tchernof, A.; Dionne, I.J.; Sites, C.K.; Eltabbakh, G.H.; Sims, E.A.H.; Poehlman, E.T. What are the physical characteristics associated with a normal metabolic profile despite a high level of obesity in postmenopausal women? J. Clin. Endocrinol. Metab. 2001, 86, 1020–1025. [Google Scholar] [PubMed]
- Stefan, N.; Kantartzis, K.; Machann, J.; Schick, F.; Thamer, C.; Rittig, K.; Balletshofer, B.; Machicao, F.; Fritsche, A.; Häring, H.U. Identification and characterization of metabolically benign obesity in humans. Arch. Intern. Med. 2008, 168, 1609–1616. [Google Scholar] [CrossRef] [PubMed]
- Karelis, A.D.; Faraj, M.; Bastard, J.P.; St-Pierre, D.H.; Brochu, M.; Prud’homme, D.; Rabasa-Lhoret, R. The metabolically healthy but obese individual presents a favorable inflammation profile. J. Clin. Endocrinol. Metab. 2005, 90, 4145–4150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobson, R.; Burgess, M.I.; Sprung, V.S.; Irwin, A.; Hamer, M.; Jones, J.; Daousi, C.; Adams, V.; Kemp, G.J.; Shojaee-Moradie, F.; et al. Metabolically healthy and unhealthy obesity: Differential effects on myocardial function according to metabolic syndrome, rather than obesity. Int. J. Obes. 2016, 40, 153–161. [Google Scholar] [CrossRef]
- Bremer, A.A.; Devaraj, S.; Afify, A.; Jialal, I. Adipose tissue dysregulation in patients with metabolic syndrome. J. Clin. Endocrinol. Metab. 2011, 96, E1782–E1788. [Google Scholar] [CrossRef] [Green Version]
- Jialal, I.; Adams-Huet, B.; Major, A.; Devaraj, S. Increased fibrosis and angiogenesis in subcutaneous gluteal adipose tissue in nascent metabolic syndrome. Diabetes Metab. 2017, 43, 364–367. [Google Scholar] [CrossRef]
- Viguerie, N.; Montastier, E.; Maoret, J.J.; Roussel, B.; Combes, M.; Valle, C.; Villa-Vialaneix, N.; Iacovoni, J.S.; Martinez, J.A.; Holst, C.; et al. Determinants of human adipose tissue gene expression: Impact of diet, sex, metabolic status, and cis genetic regulation. PLoS Genet. 2012, 8, e1002959. [Google Scholar] [CrossRef]
- Valencak, T.G.; Osterrieder, A.; Schulz, T.J. Sex matters: The effects of biological sex on adipose tissue biology and energy metabolism. Redox Biol. 2017, 12, 806–813. [Google Scholar] [CrossRef]
- Anderson, W.D.; Soh, J.Y.; Innis, S.E.; Dimanche, A.; Ma, L.; Langefeld, C.D.; Comeau, M.E.; Das, S.K.; Schadt, E.E.; Björkegren, J.L.M.; et al. Sex differences in human adipose tissue gene expression and genetic regulation involve adipogenesis. Genome Res. 2020, 30, 1379–1392. [Google Scholar] [CrossRef]
- Moreira-Pais, A.; Ferreira, R.; Neves, J.S.; Vitorino, R.; Moreira-Gonçalves, D.; Nogueira-Ferreira, R. Sex differences on adipose tissue remodeling: From molecular mechanisms to therapeutic interventions. J. Mol. Med. 2020, 98, 483–493. [Google Scholar] [CrossRef]
- Rizvi, A.A.; Stoian, A.P.; Rizzo, M. Metabolic Syndrome: From Molecular Mechanisms to Novel Therapies. Int. J. Mol. Sci. 2021, 22, 10038. [Google Scholar] [CrossRef] [PubMed]
- Hammarstedt, A.; Gogg, S.; Hedjazifar, S.; Nerstedt, A.; Smith, U. Impaired Adipogenesis and Dysfunctional Adipose Tissue in Human Hypertrophic Obesity. Physiol. Rev. 2018, 98, 1911–1941. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Tordjman, J.; Clément, K.; Scherer, P.E. Fibrosis and adipose tissue dysfunction. Cell Metab. 2013, 18, 470–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klöting, N.; Blüher, M. Adipocyte dysfunction, inflammation and metabolic syndrome. Rev. Endocr. Metab. Disord. 2014, 15, 277–287. [Google Scholar] [CrossRef]
- Vishvanath, L.; Gupta, R.K. Contribution of adipogenesis to healthy adipose tissue expansion in obesity. J. Clin. Investig. 2019, 129, 4022–4031. [Google Scholar] [CrossRef]
- Trayhurn, P.; Wang, B.; Wood, I.S. Hypoxia in adipose tissue: A basis for the dysregulation of tissue function in obesity? Br. J. Nutr. 2008, 100, 227–235. [Google Scholar] [CrossRef] [Green Version]
- Riis-Vestergaard, M.J.; Richelsen, B.; Bruun, J.M.; Li, W.; Hansen, J.B.; Pedersen, S.B. Beta-1 and Not Beta-3 Adrenergic Receptors May Be the Primary Regulator of Human Brown Adipocyte Metabolism. J. Clin. Endocrinol. Metab. 2020, 105, E994–E1005. [Google Scholar] [CrossRef]
- Jiang, Y.; Berry, D.C.; Graff, J.M. Distinct cellular and molecular mechanisms for β3 adrenergic receptor-induced beige adipocyte formation. eLife 2017, 6, e30329. [Google Scholar] [CrossRef]
- Goto, T.; Lee, J.Y.; Teraminami, A.; Kim, Y.I.; Hirai, S.; Uemura, T.; Inoue, H.; Takahashi, N.; Kawada, T. Activation of peroxisome proliferator-activated receptor-alpha stimulates both differentiation and fatty acid oxidation in adipocytes. J. Lipid Res. 2011, 52, 873–884. [Google Scholar] [CrossRef] [Green Version]
- Miranda, C.S.; Silva-Veiga, F.; Martins, F.F.; Rachid, T.L.; Mandarim-De-Lacerda, C.A.; Souza-Mello, V. PPAR-α activation counters brown adipose tissue whitening: A comparative study between high-fat- and high-fructose-fed mice. Nutrition 2020, 78, 110791. [Google Scholar] [CrossRef]
- Kajimura, S. Promoting brown and beige adipocyte biogenesis through the PRDM16 pathway. Int. J. Obes. Suppl. 2015, 5, S11–S14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikeda, K.; Yamada, T. UCP1 Dependent and Independent Thermogenesis in Brown and Beige Adipocytes. Front. Endocrinol. 2020, 11, 498. [Google Scholar] [CrossRef] [PubMed]
- Jash, S.; Banerjee, S.; Lee, M.J.; Farmer, S.R.; Puri, V. CIDEA Transcriptionally Regulates UCP1 for Britening and Thermogenesis in Human Fat Cells. iScience 2019, 20, 73–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz-Ojeda, F.J.; Méndez-Gutiérrez, A.; Aguilera, C.M.; Plaza-Díaz, J. Extracellular Matrix Remodeling of Adipose Tissue in Obesity and Metabolic Diseases. Int. J. Mol. Sci. 2019, 20, 4888. [Google Scholar] [CrossRef] [Green Version]
- Berg, G.; Barchuk, M.; Miksztowicz, V. Behavior of Metalloproteinases in Adipose Tissue, Liver and Arterial Wall: An Update of Extracellular Matrix Remodeling. Cells 2019, 8, 158. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Crewe, C.; Scherer, P.E. Hyaluronan in adipose tissue: Beyond dermal filler and therapeutic carrier. Sci. Transl. Med. 2016, 8, 323ps4. [Google Scholar] [CrossRef] [Green Version]
- Nasias, D.; Dalakoura-Karagkouni, K.; Vassou, D.; Papagiannakis, G.; Papadaki, A.; Kardassis, D. Transcriptome analysis of the adipose tissue in a mouse model of metabolic syndrome identifies gene signatures related to disease pathogenesis. Genomics 2020, 112, 4053–4062. [Google Scholar] [CrossRef]
- Obradovic, M.; Sudar-Milovanovic, E.; Soskic, S.; Essack, M.; Arya, S.; Stewart, A.J.; Gojobori, T.; Isenovic, E.R. Leptin and Obesity: Role and Clinical Implication. Front. Endocrinol. 2021, 12, 585887. [Google Scholar] [CrossRef]
- Kielar, D.; Clark, J.S.C.; Ciechanowicz, A.; Kurzawski, G.; Sulikowski, T.; Naruszewicz, M. Leptin receptor isoforms expressed in human adipose tissue. Metabolism 1998, 47, 844–847. [Google Scholar] [CrossRef]
- Kutoh, E.; Boss, O.; Levasseur, F.; Giacobino, J.P. Quantification of the full length leptin receptor (OB-Rb) in human brown and white adipose tissue. Life Sci. 1998, 62, 445–451. [Google Scholar] [CrossRef]
- Bornstein, S.R.; Abu-Asab, M.; Glasow, A.; Päth, G.; Hauner, H.; Tsokos, M.; Chrousos, G.P.; Scherbaum, W.A. Immunohistochemical and ultrastructural localization of leptin and leptin receptor in human white adipose tissue and differentiating human adipose cells in primary culture. Diabetes 2000, 49, 532–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Séron, K.; Corset, L.; Vasseur, F.; Boutin, P.; Gómez-Ambrosi, J.; Salvador, J.; Frühbeck, G.; Froguel, P. Distinct impaired regulation of SOCS3 and long and short isoforms of the leptin receptor in visceral and subcutaneous fat of lean and obese women. Biochem. Biophys. Res. Commun. 2006, 348, 1232–1238. [Google Scholar] [CrossRef] [PubMed]
- Harris, R.B.S. Direct and indirect effects of leptin on adipocyte metabolism. Biochim. Biophys. Acta 2014, 1842, 414–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palhinha, L.; Liechocki, S.; Hottz, E.D.; Pereira, J.A.d.S.; de Almeida, C.J.; Moraes-Vieira, P.M.M.; Bozza, P.T.; Maya-Monteiro, C.M. Leptin Induces Proadipogenic and Proinflammatory Signaling in Adipocytes. Front. Endocrinol. 2019, 10, 841. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Sánchez, N. There and Back Again: Leptin Actions in White Adipose Tissue. Int. J. Mol. Sci. 2020, 21, 6039. [Google Scholar] [CrossRef]
- Pawlak, M.; Lefebvre, P.; Staels, B. Molecular mechanism of PPARα action and its impact on lipid metabolism, inflammation and fibrosis in non-alcoholic fatty liver disease. J. Hepatol. 2015, 62, 720–733. [Google Scholar] [CrossRef] [Green Version]
- Staels, B.; Maes, M.; Zambon, A. Fibrates and future PPARalpha agonists in the treatment of cardiovascular disease. Nat. Clin. Pract. Cardiovasc. Med. 2008, 5, 542–553. [Google Scholar] [CrossRef]
- Achari, A.E.; Jain, S.K. Adiponectin, a Therapeutic Target for Obesity, Diabetes, and Endothelial Dysfunction. Int. J. Mol. Sci. 2017, 18, 1321. [Google Scholar] [CrossRef] [Green Version]
- Hotta, K.; Funahashi, T.; Arita, Y.; Takahashi, M.; Matsuda, M.; Okamoto, Y.; Iwahashi, H.; Kuriyama, H.; Ouchi, N.; Maeda, K.; et al. Plasma concentrations of a novel, adipose-specific protein, adiponectin, in type 2 diabetic patients. Arterioscler. Thromb. Vasc. Biol. 2000, 20, 1595–1599. [Google Scholar] [CrossRef] [Green Version]
- Arita, Y.; Kihara, S.; Ouchi, N.; Takahashi, M.; Maeda, K.; Miyagawa, J.I.; Hotta, K.; Shimomura, I.; Nakamura, T.; Miyaoka, K.; et al. Paradoxical decrease of an adipose-specific protein, adiponectin, in obesity. Biochem. Biophys. Res. Commun. 1999, 257, 79–83. [Google Scholar] [CrossRef]
- Okamoto, Y.; Kihara, S.; Ouchi, N.; Nishida, M.; Arita, Y.; Kumada, M.; Ohashi, K.; Sakai, N.; Shimomura, I.; Kobayashi, H.; et al. Adiponectin reduces atherosclerosis in apolipoprotein E-deficient mice. Circulation 2002, 106, 2767–2770. [Google Scholar] [CrossRef] [Green Version]
- Ghadge, A.A.; Khaire, A.A.; Kuvalekar, A.A. Adiponectin: A potential therapeutic target for metabolic syndrome. Cytokine Growth Factor Rev. 2018, 39, 151–158. [Google Scholar] [CrossRef]
- Lihn, A.S.; Østergard, T.; Nyholm, B.; Pedersen, S.B.; Richelsen, B.; Schmitz, O. Adiponectin expression in adipose tissue is reduced in first-degree relatives of type 2 diabetic patients. Am. J. Physiol. Endocrinol. Metab. 2003, 284, E443–E448. [Google Scholar] [CrossRef]
- Sadashiv; Tiwari, S.; Paul, B.N.; Kumar, S.; Chandra, A.; Dhananjai, S.; Negi, M.P.S. Adiponectin mRNA in adipose tissue and its association with metabolic risk factors in postmenopausal obese women. Hormones 2013, 12, 119–127. [Google Scholar] [CrossRef]
- Litvinova, L.; Atochin, D.; Vasilenko, M.; Fattakhov, N.; Zatolokin, P.; Vaysbeyn, I.; Kirienkova, E. Role of adiponectin and proinflammatory gene expression in adipose tissue chronic inflammation in women with metabolic syndrome. Diabetol. Metab. Syndr. 2014, 6, 137. [Google Scholar] [CrossRef] [Green Version]
- Motoshima, H.; Wu, X.; Sinha, M.K.; Hardy, V.E.; Rosato, E.L.; Barbot, D.J.; Rosato, F.E.; Goldstein, B.J. Differential regulation of adiponectin secretion from cultured human omental and subcutaneous adipocytes: Effects of insulin and rosiglitazone. J. Clin. Endocrinol. Metab. 2002, 87, 5662–5667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomas, E.; Tsao, T.S.; Saha, A.K.; Murrey, H.E.; Cheng Zhang, C.; Itani, S.I.; Lodish, H.F.; Ruderman, N.B. Enhanced muscle fat oxidation and glucose transport by ACRP30 globular domain: Acetyl-CoA carboxylase inhibition and AMP-activated protein kinase activation. Proc. Natl. Acad. Sci. USA 2002, 99, 16309–16313. [Google Scholar] [CrossRef] [Green Version]
- Myeong, J.Y.; Gha, Y.L.; Chung, J.J.; Young, H.A.; Seung, H.H.; Jae, B.K. Adiponectin increases fatty acid oxidation in skeletal muscle cells by sequential activation of AMP-activated protein kinase, p38 mitogen-activated protein kinase, and peroxisome proliferator-activated receptor alpha. Diabetes 2006, 55, 2562–2570. [Google Scholar]
- Ohashi, K.; Parker, J.L.; Ouchi, N.; Higuchi, A.; Vita, J.A.; Gokce, N.; Pedersen, A.A.; Kalthoff, C.; Tullin, S.; Sams, A.; et al. Adiponectin promotes macrophage polarization toward an anti-inflammatory phenotype. J. Biol. Chem. 2010, 285, 6153–6160. [Google Scholar] [CrossRef] [Green Version]
- Lovren, F.; Pan, Y.; Quan, A.; Szmitko, P.E.; Singh, K.K.; Shukla, P.C.; Gupta, M.; Chan, L.; Al-Omran, M.; Teoh, H.; et al. Adiponectin primes human monocytes into alternative anti-inflammatory M2 macrophages. Am. J. Physiol. Heart Circ. Physiol. 2010, 299, H656–H663. [Google Scholar] [CrossRef] [Green Version]
- Malandrino, M.I.; Fucho, R.; Weber, M.; Calderon-Dominguez, M.; Mir, J.F.; Valcarcel, L.; Escoté, X.; Gómez-Serrano, M.; Peral, B.; Salvadó, L.; et al. Enhanced fatty acid oxidation in adipocytes and macrophages reduces lipid-induced triglyceride accumulation and inflammation. Am. J. Physiol. Endocrinol. Metab. 2015, 308, E756–E769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, M.; Sha, J.; Hidalgo, B.; Aslibekyan, S.; Do, A.N.; Zhi, D.; Sun, D.; Zhang, T.; Li, S.; Chen, W.; et al. Association of DNA Methylation at CPT1A Locus with Metabolic Syndrome in the Genetics of Lipid Lowering Drugs and Diet Network (GOLDN) Study. PLoS ONE 2016, 11, e0145789. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Asterholm, I.W.; Kusminski, C.M.; Bueno, A.C.; Wang, Z.V.; Pollard, J.W.; Brekken, R.A.; Scherer, P.E. Dichotomous effects of VEGF-A on adipose tissue dysfunction. Proc. Natl. Acad. Sci. USA 2012, 109, 5874–5879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elias, I.; Franckhauser, S.; Ferré, T.; Vilà, L.; Tafuro, S.; Muñoz, S.; Roca, C.; Ramos, D.; Pujol, A.; Riu, E.; et al. Adipose tissue overexpression of vascular endothelial growth factor protects against diet-induced obesity and insulin resistance. Diabetes 2012, 61, 1801–1813. [Google Scholar] [CrossRef] [Green Version]
- Park, J.; Kim, M.; Sun, K.; An, Y.A.; Gu, X.; Scherer, P.E. VEGF-A-Expressing Adipose Tissue Shows Rapid Beiging and Enhanced Survival After Transplantation and Confers IL-4-Independent Metabolic Improvements. Diabetes 2017, 66, 1479–1490. [Google Scholar] [CrossRef] [Green Version]
- Silha, J.V.; Krsek, M.; Sucharda, P.; Murphy, L.J. Angiogenic factors are elevated in overweight and obese individuals. Int. J. Obes. 2005, 29, 1308–1314. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.X.; Magovern, C.J.; Mack, C.A.; Budenbender, K.T.; Wilson, K.; Rosengart, T.K. Vascular endothelial growth factor is the major angiogenic factor in omentum: Mechanism of the omentum-mediated angiogenesis. J. Surg. Res. 1997, 67, 147–154. [Google Scholar] [CrossRef] [Green Version]
- Zafar, M.I.; Mills, K.; Ye, X.; Blakely, B.; Min, J.; Kong, W.; Zhang, N.; Gou, L.; Regmi, A.; Hu, S.Q.; et al. Association between the expression of vascular endothelial growth factors and metabolic syndrome or its components: A systematic review and meta-analysis. Diabetol. Metab. Syndr. 2018, 10, 62. [Google Scholar] [CrossRef]
- Menendez, J.A.; Vazquez-Martin, A.; Ortega, F.J.; Fernandez-Real, J.M. Fatty acid synthase: Association with insulin resistance, type 2 diabetes, and cancer. Clin. Chem. 2009, 55, 425–438. [Google Scholar] [CrossRef] [Green Version]
- Goedecke, J.H.; Utzschneider, K.; Faulenbach, M.V.; Rizzo, M.; Berneis, K.; Spinas, G.A.; Dave, J.A.; Levitt, N.S.; Lambert, E.V.; Olsson, T.; et al. Ethnic differences in serum lipoproteins and their determinants in South African women. Metabolism 2010, 59, 1341–1350. [Google Scholar] [CrossRef]
- Andreu, A.; Moizé, V.; Rodríguez, L.; Flores, L.; Vidal, J. Protein intake, body composition, and protein status following bariatric surgery. Obes. Surg. 2010, 20, 1509–1515. [Google Scholar] [CrossRef] [PubMed]
- Osorio-Conles, Ó.; Vega-Beyhart, A.; Ibarzabal, A.; Balibrea, J.M.; Graupera, I.; Rimola, J.; Vidal, J.; de Hollanda, A. A Distinctive NAFLD Signature in Adipose Tissue from Women with Severe Obesity. Int. J. Mol. Sci. 2021, 22, 10541. [Google Scholar] [CrossRef] [PubMed]
- Divoux, A.; Tordjman, J.; Lacasa, D.; Veyrie, N.; Hugol, D.; Aissat, A.; Basdevant, A.; Guerre-Millo, M.; Poitou, C.; Zucker, J.D.; et al. Fibrosis in human adipose tissue: Composition, distribution, and link with lipid metabolism and fat mass loss. Diabetes 2010, 59, 2817–2825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rohart, F.; Gautier, B.; Singh, A.; Lê Cao, K.A. mixOmics: An R package for ’omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| MetS− (n = 33) | MetS+ (n = 33) | p-Value | |
|---|---|---|---|
| Age (years) a | 47.97 ± 8.81 | 49.24 ± 10.65 | 0.599 |
| BMI (kg/m2) ab | 43.60 ± 5.12 | 44.12 ± 4.06 | 0.651 |
| Waist (cm) b | 123.29 ± 13.45 | 122.2 ± 10.51 | 0.785 |
| Hip (cm) | 137.64 ± 9.67 | 134.48 ± 8.73 | 0.312 |
| Waist-to-hip ratio | 0.90 ± 0.07 | 0.91 ± 0.06 | 0.516 |
| HTN b | 2 (6.06%) | 18 (54.54%) | <0.0001 c |
| TG (mg/dL) b | 109.34 ± 38.26 | 147.15 ± 45.7 | 0.001 |
| Total cholesterol (mg/dL) | 193.82 ± 24.44 | 202.55 ± 36.2 | 0.337 |
| HDL (mg/dL) b | 53.85 ± 9.01 | 47.15 ± 8.9 | 0.003 |
| LDL (mg/dL) | 118 ± 22.18 | 123.13 ± 26.96 | 0.408 |
| FPG (mg/dL) b | 96.36 ± 22.68 | 110.12 ± 22.49 | <0.0001 |
| T2D | 1 (3.03%) | 12 (36.36%) | 0.001 |
| HbA1c (%) | 5.69 ± 0.78 | 6.04 ± 0.87 | 0.037 |
| AST (IU/L) | 20.84 ± 7.47 | 23.3 ± 6.83 | 0.103 |
| ALT (IU/L) | 25.48 ± 14.62 | 29.42 ± 18.42 | 0.185 |
| GGT (IU/L) | 24.81 ± 17.24 | 37.15 ± 25.5 | 0.009 |
| AST:ALT ratio | 0.94 ± 0.35 | 0.95 ± 0.42 | 0.914 |
| Platelets (×109/L) | 273.88 ± 54.16 | 305.61 ± 75.74 | 0.268 |
| hs-CRP (mg/dL) | 0.82 ± 0.69 | 1.36 ± 1.67 | 0.134 |
| GM-CSF (pg/mL) | 11.92 (9.52–19.59) | 19.44 (7.28–29.38) | 0.403 |
| IL-1ß (pg/mL) | 0.85 ± 0.55 | 0.90 ± 0.48 | 0.849 |
| IL-6 (pg/mL) | 0.89 ± 0.53 | 1.14 ± 0.95 | 0.48 |
| TNFα (pg/mL) | 2.90 ± 0.91 | 3.64 ± 1.85 | 0.526 |
| NAFLD | 19 (57.58%) | 28 (87.5%) | 0.028 c |
| FIB-4 Score | 0.81 ± 0.38 | 0.79 ± 0.33 | 0.769 |
| APRI Score | 0.20 ± 0.11 | 0.20 ± 0.07 | 0.312 |
| HSI Index | 54.73 ± 7.02 | 56.6 ± 4.42 | 0.09 |
| TyG Index | 4.59 ± 0.21 | 4.81 ± 0.19 | <0.0001 |
| FLI Index | 94.57 ± 5.24 | 97.51 ± 1.87 | 0.018 |
| MetS− | MetS+ | p-Value | |
|---|---|---|---|
| Total mass (kg) | 105.5 (97.65–119.4) | 108.8 (105.6–122.9) | 0.394 |
| Lean mass (g) | 48,399 ± 6280 | 49,637 ± 5737 | 0.591 |
| Fat-free mass (g) | 50,829 ± 6444 | 52,037 ± 5968 | 0.611 |
| Fat mass (g) | 56,637 (50,181–63,045) | 58,213 (51,129–66,602) | 0.546 |
| Android fat mass (%) | 60.5 (57–63.55) | 61.3 (58.2–65.6) | 0.446 |
| Gynoid fat mass (%) | 56 (52.6–59.4) | 55.6 (51.8–58) | 0.675 |
| Total fat mass (%) | 54.9 (50.2–57.05) | 54.7 (51.5–57.7) | 0.742 |
| eVAT (g) | 1904 ± 699 | 2276 ± 696.1 | 0.179 |
| eVAT (cm3) | 2018 ± 741 | 2412 ± 738.1 | 0.179 |
| SAT fat cell area (µm2) | 4585 ± 1195 | 4747 ± 1290 | 0.689 |
| VAT fat cell area (µm2) | 3941 ± 1182 | 4470 ± 1130 | 0.168 |
| SAT:VAT fat cell area ratio | 1.25 ± 0.29 | 1.06 ± 0.19 | 0.041 |
| SAT Total fibrosis (% area) | 0.94 ± 0.39 | 1.10 ± 0.39 | 0.277 |
| VAT Total fibrosis (% area) | 0.74 ± 0.35 | 0.92 ± 0.44 | 0.259 |
| SAT Pericellular fibrosis (% area) | 2.68 ± 1.45 | 4.52 ± 2.21 | 0.029 |
| VAT Pericellular fibrosis (% area) | 1.80 ± 1.06 | 2.80 ± 1.91 | 0.097 |
| SAT:VAT fibrosis ratio | 1.44 ± 1.31 | 1.70 ± 0.92 | 0.222 |
| MetS+ vs. MetS− | ||||
|---|---|---|---|---|
| SAT | VAT | |||
| Inflammation | FC | p-value | FC | p-value |
| CD68 | 1.47 | 0.019 | 1.27 | 0.33 |
| CD80 * | 3.00 | 0.004 | 2.00 | 0.125 |
| MRC1 | 1.35 | 0.03 | 1.11 | 0.338 |
| MSR1 | 1.49 | 0.131 | 1.28 | 0.045 |
| HIF1A | 1.52 | 0.024 | 0.94 | 0.675 |
| Adipokines | ||||
| ADIPOQ * | 1.39 | 0.042 | 0.94 | 0.315 |
| LEPR * | 0.62 | 0.009 | 1.23 | 0.436 |
| FABP4 | 1.22 | 0.295 | 0.53 | 0.003 |
| Angiogenesis | ||||
| VEGFA * | 1.21 | 0.115 | 0.96 | 0.826 |
| Glucose metabolism | ||||
| GLUT1 | 1.00 | 0.085 | 1.33 | 0.044 |
| Lipogenesis | ||||
| FASN | 0.53 | 0.012 | 0.64 | 0.009 |
| MOGAT1 | 1.50 | 0.028 | 2.00 | 0.313 |
| FAO/Beiging | ||||
| PPARA * | 1.25 | 0.008 | 1.03 | 0.825 |
| UCP1 | 1.00 | 0.603 | 0.40 | 0.017 |
| ADRB1 | 1.73 | 0.028 | 1.12 | 0.477 |
| ADRB3 | 1.00 | 0.842 | 0.33 | 0.004 |
| PRDM16 | 2.00 | 0.031 | 1.00 | 0.434 |
| CIDEA | 1.13 | 0.406 | 0.65 | 0.002 |
| CPT1A * | 1.37 | 0.006 | 1.41 | 0.003 |
| ECM remodelling | ||||
| TGFB1 | 1.34 | 0.033 | 1.32 | 0.01 |
| F13A1 | 1.71 | 0.026 | 1.61 | 0.028 |
| COL5A1 | 1.36 | 0.168 | 0.68 | 0.036 |
| MMP2 | 1.09 | 0.74 | 0.51 | 0.016 |
| MMP13 | 1.00 | 0.882 | 0.10 | 0.0005 |
| TIMP1 | 1.70 | 0.331 | 0.45 | 0.002 |
| HYAL1 | 1.00 | 0.185 | 0.30 | <0.0001 |
| HYAL2 | 1.36 | 0.603 | 0.29 | 0.001 |
| LOXL2 | 1.48 | 0.025 | 1.02 | 0.91 |
| LOXL4 | 1.44 | 0.044 | 2.75 | 0.194 |
| Genes | B | S.E. (B) | Exp B (OR) (95% CI) | Sig. | R2 (%) | Correct Prediction (%) | M. Sig. |
|---|---|---|---|---|---|---|---|
| Component 1 | |||||||
| SAT-LEPR | −15.798 | 6.450 | 0.12 [0.04—0.43] | 0.014 | 64.4 | 85.2 | 0.000 |
| SAT-PPARA | 17.9 | 7.365 | 6.01 [1.27—12.95] | 0.015 | |||
| Constant | −3.509 | 2.649 | 0.30 | 0.185 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Osorio-Conles, Ó.; Vega-Beyhart, A.; Ibarzabal, A.; Balibrea, J.M.; Vidal, J.; de Hollanda, A. Biological Determinants of Metabolic Syndrome in Visceral and Subcutaneous Adipose Tissue from Severely Obese Women. Int. J. Mol. Sci. 2022, 23, 2394. https://doi.org/10.3390/ijms23042394
Osorio-Conles Ó, Vega-Beyhart A, Ibarzabal A, Balibrea JM, Vidal J, de Hollanda A. Biological Determinants of Metabolic Syndrome in Visceral and Subcutaneous Adipose Tissue from Severely Obese Women. International Journal of Molecular Sciences. 2022; 23(4):2394. https://doi.org/10.3390/ijms23042394
Chicago/Turabian StyleOsorio-Conles, Óscar, Arturo Vega-Beyhart, Ainitze Ibarzabal, José María Balibrea, Josep Vidal, and Ana de Hollanda. 2022. "Biological Determinants of Metabolic Syndrome in Visceral and Subcutaneous Adipose Tissue from Severely Obese Women" International Journal of Molecular Sciences 23, no. 4: 2394. https://doi.org/10.3390/ijms23042394
APA StyleOsorio-Conles, Ó., Vega-Beyhart, A., Ibarzabal, A., Balibrea, J. M., Vidal, J., & de Hollanda, A. (2022). Biological Determinants of Metabolic Syndrome in Visceral and Subcutaneous Adipose Tissue from Severely Obese Women. International Journal of Molecular Sciences, 23(4), 2394. https://doi.org/10.3390/ijms23042394







