Deficiency in RCAT-1 Function Causes Dopamine Metabolism Related Behavioral Disorders in Caenorhabditis elegans
Abstract
1. Introduction
2. Results
2.1. Behavioral Assays Using R02D3.7(ok1745)
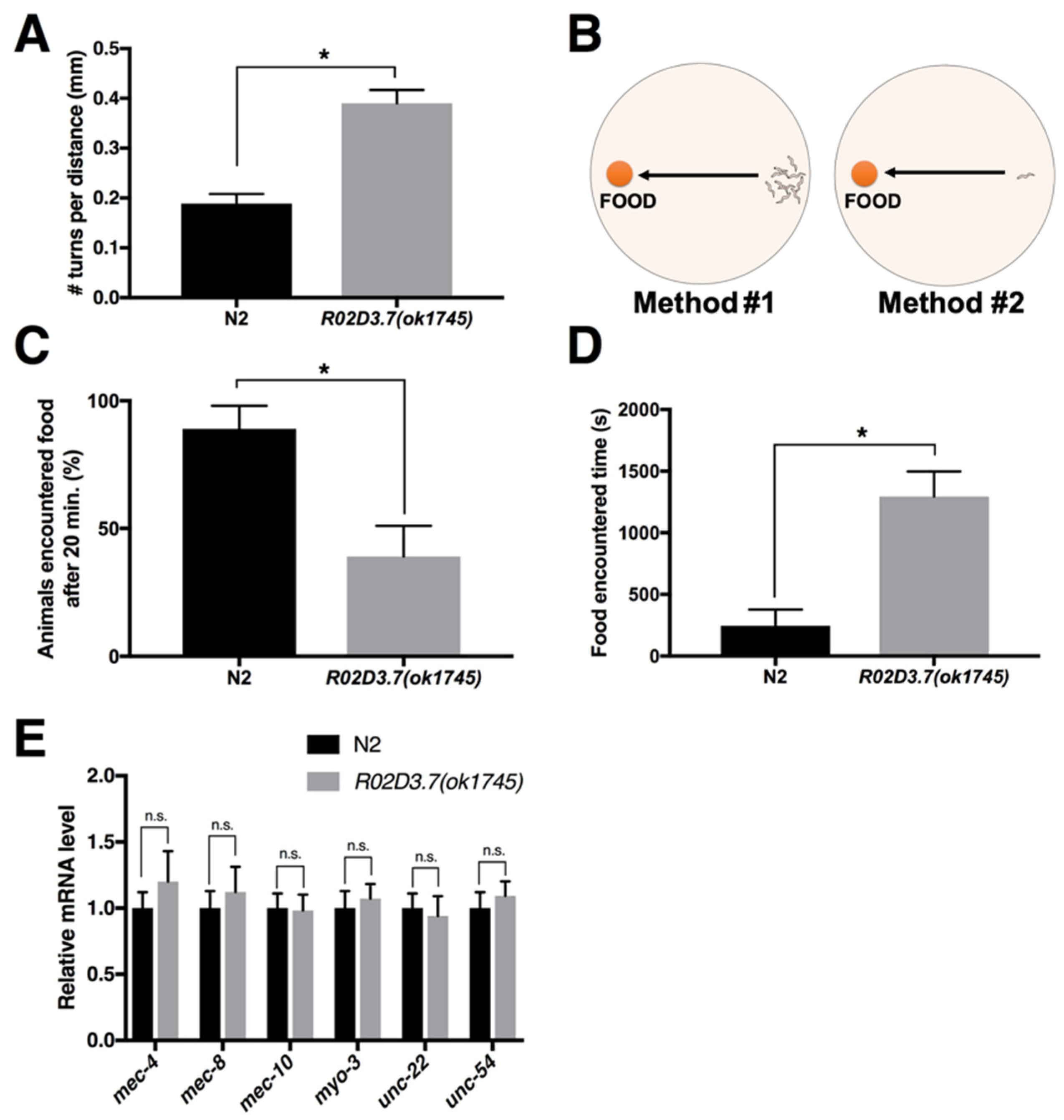
2.1.1. Turning Frequency of R02D3.7(ok1745)
2.1.2. Food Detection Assay of R02D3.7(ok1745)
2.1.3. R02D3.7(ok1745) Shows No Defects in Cuticular Structures
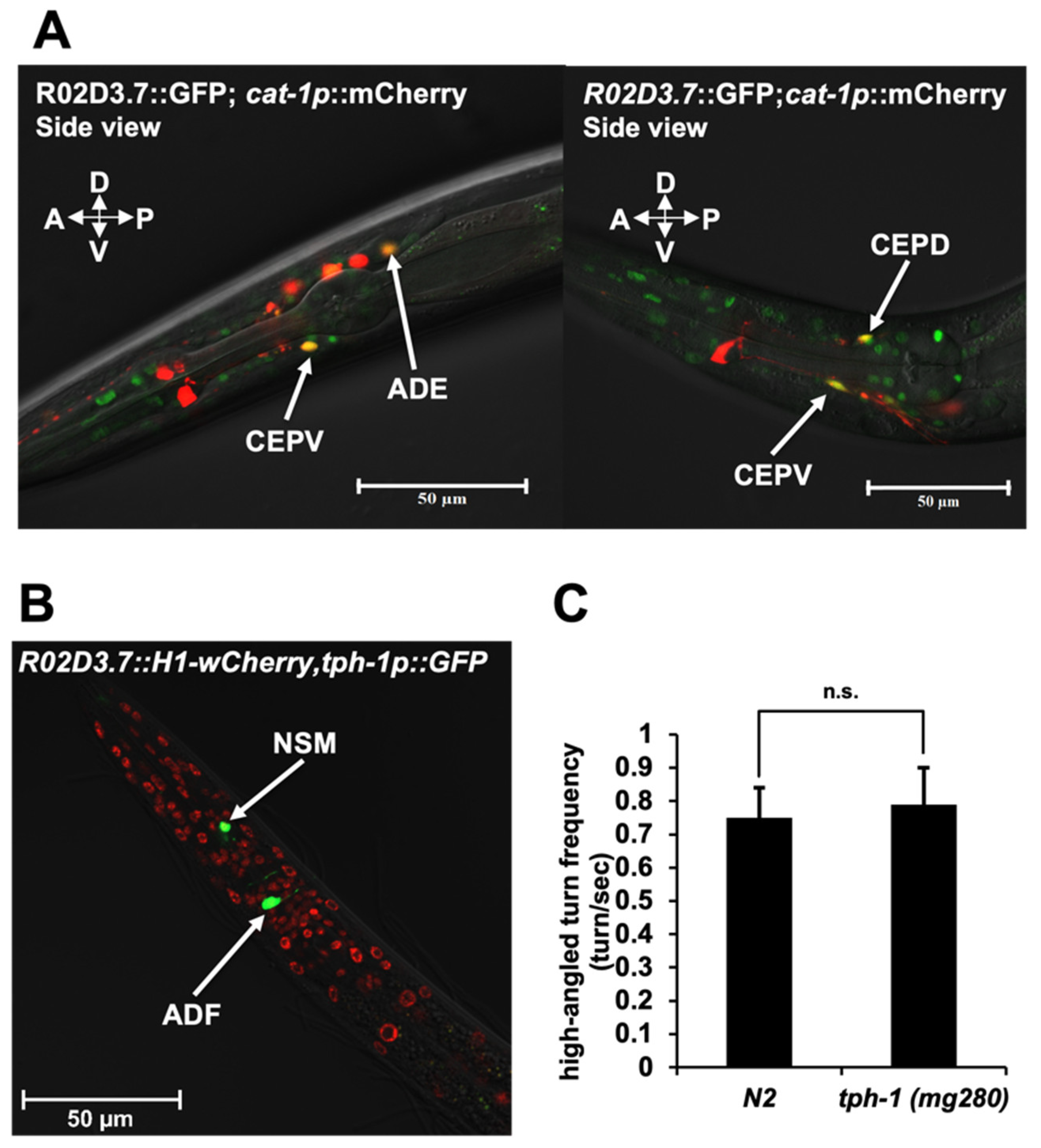
2.2. R02D3.7 Is Expressed in Dopaminergic Neurons
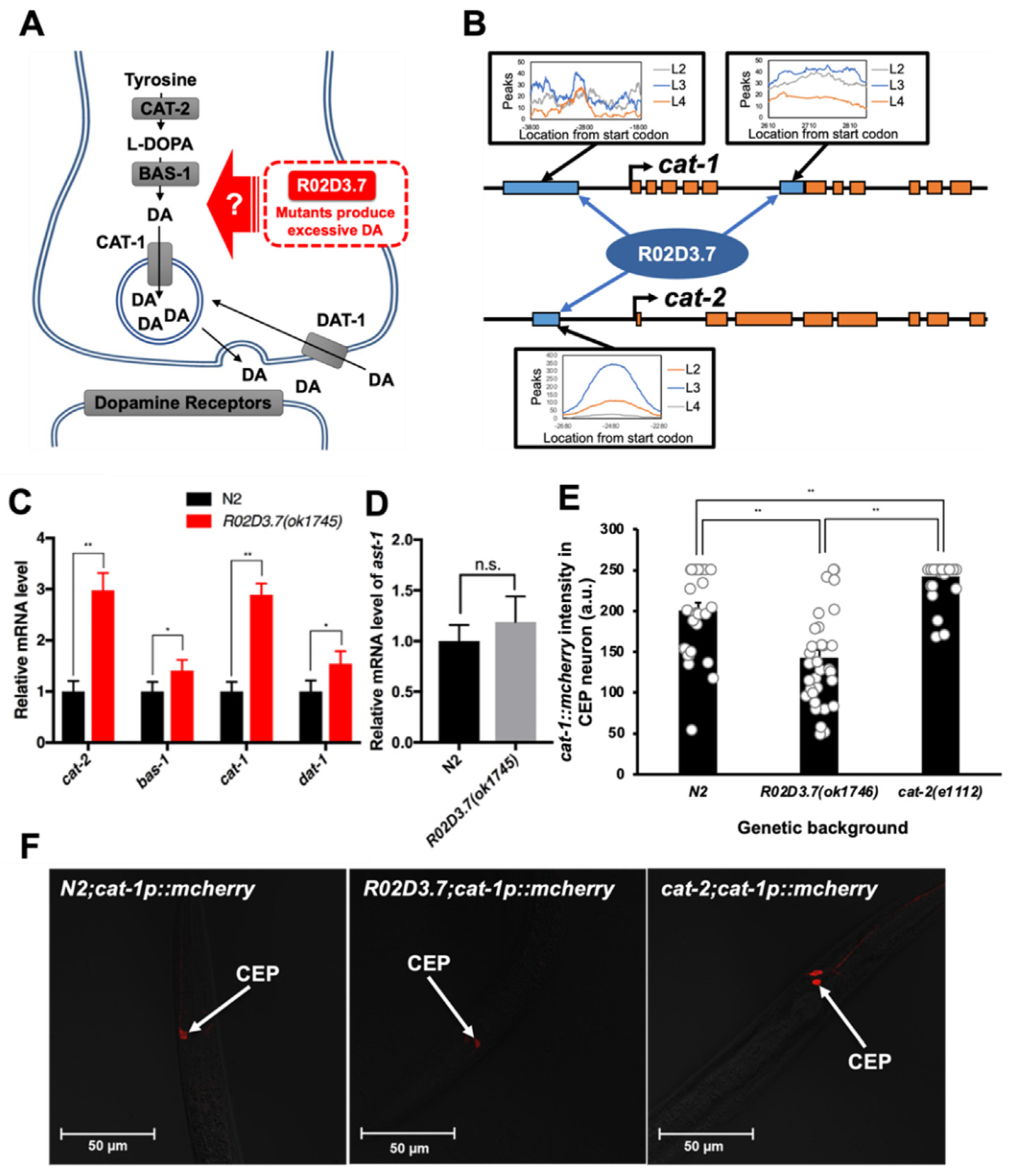
2.3. Potential Mechanisms Underlying Excessive Dopamine Production and Release in R02D3.7(ok1745) Mutants
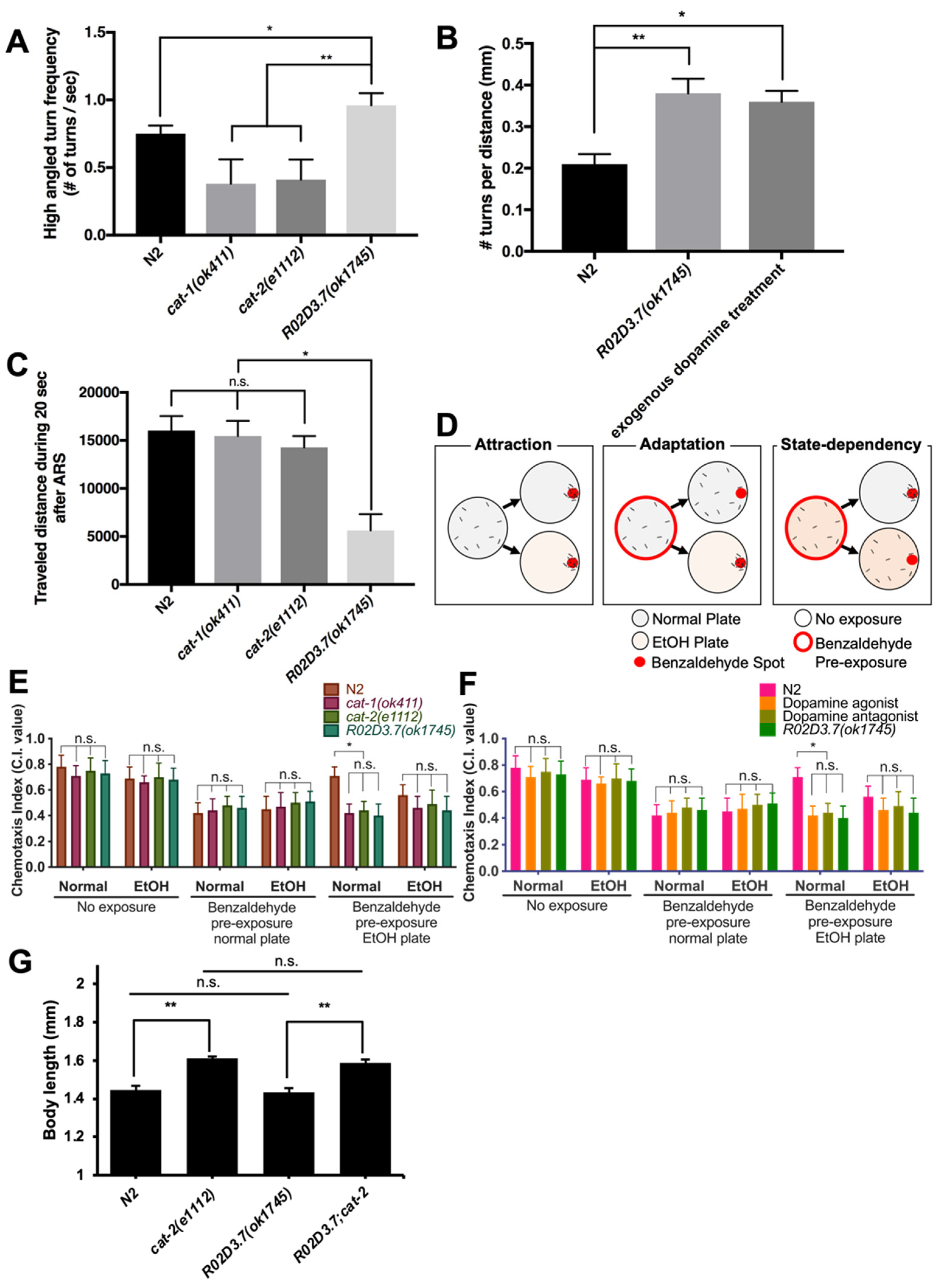
2.3.1. Area Restricted Search (ARS) Behavior of R02D3.7(ok1745) Mutants
2.3.2. State-Dependency Olfactory Adaptation of R02D3.7(ok1745)
2.3.3. Measurement of Body Size
2.4. R02D3.7 Downregulates the Expression of Dopamine Metabolism-Related Genes
3. Discussion
4. Materials and Methods
4.1. Strains
4.2. Behavioral Assays
4.3. Quantitative Reverse Transcription-Polymerase Chain Reaction
4.4. DiI Staining Assay
4.5. Confocal Microscopy
4.6. Body Size Measurement
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sawin, E.R.; Ranganathan, R.; Horvitz, H.R. C. elegans Locomotory Rate Is Modulated by the Environment through a Dopaminergic Pathway and by Experience through a Serotonergic Pathway. Neuron 2000, 26, 619–631. [Google Scholar] [CrossRef]
- Hills, T.; Brockie, P.J.; Maricq, A.V. Dopamine and Glutamate Control Area-Restricted Search Behavior in Caenorhabditis elegans. J. Neurosci. 2004, 24, 1217–1225. [Google Scholar] [CrossRef] [PubMed]
- Chase, D. Biogenic Amine Neurotransmitters in C. elegans. WormBook 2007, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Schafer, W.R.; Kenyon, C.J. A Calcium-Channel Homologue Required for Adaptation to Dopamine and Serotonin in Caenorhabditis elegans. Nature 1995, 375, 73–78. [Google Scholar] [CrossRef]
- Beninger, R.J. The Role of Dopamine in Locomotor Activity and Learning. Brain Res. Rev. 1983, 6, 173–196. [Google Scholar] [CrossRef]
- Sudevan, S.; Muto, K.; Higashitani, N.; Hashizume, T.; Higashibata, A.; Ellwood, R.A.; Deane, C.S.; Rahman, M.; Vanapalli, S.A.; Etheridge, T.; et al. Loss of Physical Contact in Space Alters the Dopamine System in C. elegans. iScience 2022, 25, 103762. [Google Scholar] [CrossRef] [PubMed]
- Baik, J.-H. Dopamine Signaling in Reward-Related Behaviors. Front. Neural Circuits 2013, 7, 152. [Google Scholar] [CrossRef]
- Bettinger, J.C.; McIntire, S.L. State-Dependency in C. elegans. Genes Brain Behav. 2004, 3, 266–272. [Google Scholar] [CrossRef]
- Maulik, M.; Mitra, S.; Bult-Ito, A.; Taylor, B.E.; Vayndorf, E.M. Behavioral Phenotyping and Pathological Indicators of Parkinson’s Disease in C. elegans Models. Front. Genet. 2017, 8, 77. [Google Scholar] [CrossRef]
- Jeong, P.-Y.; Jung, M.; Yim, Y.-H.; Kim, H.; Park, M.; Hong, E.; Lee, W.; Kim, Y.H.; Kim, K.; Paik, Y.-K. Chemical Structure and Biological Activity of the Caenorhabditis elegans Dauer-Inducing Pheromone. Nature 2005, 433, 541–545. [Google Scholar] [CrossRef]
- Park, J.Y.; Joo, H.-J.; Park, S.; Paik, Y.-K. Ascaroside Pheromones: Chemical Biology and Pleiotropic Neuronal Functions. Int. J. Mol. Sci. 2019, 20, 3898. [Google Scholar] [CrossRef] [PubMed]
- Duerr, J.S.; Frisby, D.L.; Gaskin, J.; Duke, A.; Asermely, K.; Huddleston, D.; Eiden, L.E.; Rand, J.B. The Cat-1 Gene of Caenorhabditis elegans Encodes a Vesicular Monoamine Transporter Required for Specific Monoamine-Dependent Behaviors. J. Neurosci. 1999, 19, 72–84. [Google Scholar] [CrossRef] [PubMed]
- Lints, R.; Emmons, S.W. Patterning of Dopaminergic Neurotransmitter Identity among Caenorhabditis elegans Ray Sensory Neurons by a TGFbeta Family Signaling Pathway and a Hox Gene. Development 1999, 126, 5819–5831. [Google Scholar] [CrossRef] [PubMed]
- Lee, J. Identification of a Transcription Factor Which Regulates Daf-7 Expression during Dauer Formation in C. elegans. Master’s Thesis, Graduate School, Yonsei University, Seoul, Korea, 2009. [Google Scholar]
- Kaeberlein, T.L.; Smith, E.D.; Tsuchiya, M.; Welton, K.L.; Thomas, J.H.; Fields, S.; Kennedy, B.K.; Kaeberlein, M. Lifespan Extension in Caenorhabditis elegans by Complete Removal of Food. Aging Cell 2006, 5, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Mair, W.; Dillin, A. Aging and Survival: The Genetics of Life Span Extension by Dietary Restriction. Annu. Rev. Biochem. 2008, 77, 727–754. [Google Scholar] [CrossRef] [PubMed]
- Colbert, H.A.; Bargmann, C.I. Odorant-Specific Adaptation Pathways Generate Olfactory Plasticity in C. elegans. Neuron 1995, 14, 803–812. [Google Scholar] [CrossRef]
- Schultz, R.D.; Gumienny, T.L. Visualization of Caenorhabditis elegans Cuticular Structures Using the Lipophilic Vital Dye DiI. J. Vis. Exp. 2012, 59, e3362. [Google Scholar] [CrossRef]
- Anderson, P. Molecular Genetics of Nematode Muscle. Annu. Rev. Genet. 1989, 23, 507–525. [Google Scholar] [CrossRef]
- Tavernarakis, N.; Driscoll, M. Molecular Modeling of Mechanotransduction in the Nematode Caenorhabditis elegans. Annu. Rev. Physiol. 1997, 59, 659–689. [Google Scholar] [CrossRef]
- Calhoun, A.J.; Tong, A.; Pokala, N.; Fitzpatrick, J.A.J.; Sharpee, T.O.; Chalasani, S.H. Neural Mechanisms for Evaluating Environmental Variability in Caenorhabditis elegans. Neuron 2015, 86, 428–441. [Google Scholar] [CrossRef]
- Ramos, E.J.B.; Meguid, M.M.; Campos, A.C.L.; Coelho, J.C.U. Neuropeptide Y, α-Melanocyte–Stimulating Hormone, and Monoamines in Food Intake Regulation. Nutrition 2005, 21, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Sarov, M.; Murray, J.I.; Schanze, K.; Pozniakovski, A.; Niu, W.; Angermann, K.; Hasse, S.; Rupprecht, M.; Vinis, E.; Tinney, M.; et al. A Genome-Scale Resource for In Vivo Tag-Based Protein Function Exploration in C. elegans. Cell 2012, 150, 855–866. [Google Scholar] [CrossRef] [PubMed]
- Tursun, B.; Patel, T.; Kratsios, P.; Hobert, O. Direct Conversion of C. elegans Germ Cells into Specific Neuron Types. Science 2011, 331, 304–308. [Google Scholar] [CrossRef] [PubMed]
- Sze, J.Y.; Victor, M.; Loer, C.; Shi, Y.; Ruvkun, G. Food and Metabolic Signalling Defects in a Caenorhabditis elegans Serotonin-Synthesis Mutant. Nature 2000, 403, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Packer, J.S.; Zhu, Q.; Huynh, C.; Sivaramakrishnan, P.; Preston, E.; Dueck, H.; Stefanik, D.; Tan, K.; Trapnell, C.; Kim, J.; et al. A Lineage-Resolved Molecular Atlas of C. elegans Embryogenesis at Single-Cell Resolution. Science 2019, 365, eaax1971. [Google Scholar] [CrossRef]
- Lee, J.-E.; Jeong, P.-Y.; Joo, H.-J.; Kim, H.; Lee, T.; Koo, H.-S.; Paik, Y.-K. STR-33, a Novel G Protein-Coupled Receptor That Regulates Locomotion and Egg Laying in Caenorhabditis elegans. J. Biol. Chem. 2011, 286, 39860–39870. [Google Scholar] [CrossRef]
- Crisford, A.; Calahorro, F.; Ludlow, E.; Marvin, J.M.C.; Hibbard, J.K.; Lilley, C.J.; Kearn, J.; Keefe, F.; Johnson, P.; Harmer, R.; et al. Identification and Characterisation of Serotonin Signalling in the Potato Cyst Nematode Globodera Pallida Reveals New Targets for Crop Protection. PLoS Pathog. 2020, 16, e1008884. [Google Scholar] [CrossRef]
- Omura, D.T.; Clark, D.A.; Samuel, A.D.T.; Horvitz, H.R. Dopamine Signaling Is Essential for Precise Rates of Locomotion by C. elegans. PLoS ONE 2012, 7, e38649. [Google Scholar] [CrossRef]
- Hukema, R.K.; Rademakers, S.; Jansen, G. Gustatory Plasticity in C. elegans Involves Integration of Negative Cues and NaCl Taste Mediated by Serotonin, Dopamine, and Glutamate. Learn. Mem. 2008, 15, 829–836. [Google Scholar] [CrossRef]
- Root, C.M.; Ko, K.I.; Jafari, A.; Wang, J.W. Presynaptic Facilitation by Neuropeptide Signaling Mediates Odor-Driven Food Search. Cell 2011, 145, 133–144. [Google Scholar] [CrossRef]
- Bargmann, C.I.; Hartwieg, E.; Horvitz, H.R. Odorant-Selective Genes and Neurons Mediate Olfaction in C. elegans. Cell 1993, 74, 515–527. [Google Scholar] [CrossRef]
- Nagashima, T.; Oami, E.; Kutsuna, N.; Ishiura, S.; Suo, S. Dopamine Regulates Body Size in Caenorhabditis elegans. Dev. Biol. 2016, 412, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Lee, S.H.; Cha, Y.J.; Hong, S.J.; Chung, S.K.; Park, T.H.; Choi, S.S. C. elegans-on-a-Chip for in Situ and in Vivo Ag Nanoparticles’ Uptake and Toxicity Assay. Sci. Rep. 2017, 7, 40225. [Google Scholar] [CrossRef] [PubMed]
- Flames, N.; Hobert, O. Gene Regulatory Logic of Dopamine Neuron Differentiation. Nature 2009, 458, 885–889. [Google Scholar] [CrossRef]
- Loer, C.M.; Kenyon, C.J. Serotonin-Deficient Mutants and Male Mating Behavior in the Nematode Caenorhabditis elegans. J. Neurosci. 1993, 13, 5407–5417. [Google Scholar] [CrossRef]
- Jayanthi, L.D.; Apparsundaram, S.; Malone, M.D.; Ward, E.; Miller, D.M.; Eppler, M.; Blakely, R.D. The Caenorhabditis elegans GeneT23G5.5 Encodes an Antidepressant- and Cocaine-Sensitive Dopamine Transporter. Mol. Pharmacol. 1998, 54, 601–609. [Google Scholar]
- Carvelli, L.; Blakely, R.D.; DeFelice, L.J. Dopamine Transporter/Syntaxin 1A Interactions Regulate Transporter Channel Activity and Dopaminergic Synaptic Transmission. Proc. Natl. Acad. Sci. USA 2008, 105, 14192–14197. [Google Scholar] [CrossRef]
- Celniker, S.E.; Dillon, L.A.L.; Gerstein, M.B.; Gunsalus, K.C.; Henikoff, S.; Karpen, G.H.; Kellis, M.; Lai, E.C.; Lieb, J.D.; MacAlpine, D.M.; et al. Unlocking the Secrets of the Genome. Nature 2009, 459, 927–930. [Google Scholar] [CrossRef]
- Gerstein, M.B.; Lu, Z.J.; Nostrand, E.L.V.; Cheng, C.; Arshinoff, B.I.; Liu, T.; Yip, K.Y.; Robilotto, R.; Rechtsteiner, A.; Ikegami, K.; et al. Integrative Analysis of the Caenorhabditis elegans Genome by the ModENCODE Project. Science 2010, 330, 1775–1787. [Google Scholar] [CrossRef]
- Van Nostrand, E.L.; Kim, S.K. Integrative Analysis of C. elegans ModENCODE ChIP-Seq Data Sets to Infer Gene Regulatory Interactions. Genome Res. 2013, 23, 941–953. [Google Scholar] [CrossRef]
- Lloret-Fernández, C.; Maicas, M.; Mora-Martínez, C.; Artacho, A.; Jimeno-Martín, Á.; Chirivella, L.; Weinberg, P.; Flames, N. A Transcription Factor Collective Defines the HSN Serotonergic Neuron Regulatory Landscape. eLife 2018, 7, e32785. [Google Scholar] [CrossRef] [PubMed]
- Manalo, R.V.M.; Medina, P.M.B. Caffeine Reduces Deficits in Mechanosensation and Locomotion Induced by L-DOPA and Protects Dopaminergic Neurons in a Transgenic Caenorhabditis elegans Model of Parkinson’s Disease. Pharm. Biol. 2020, 58, 721–731. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Gelwix, C.C.; Caldwell, K.A.; Caldwell, G.A. Torsin-Mediated Protection from Cellular Stress in the Dopaminergic Neurons of Caenorhabditis elegans. J. Neurosci. 2005, 25, 3801–3812. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.-Q.; Fan, H.-X.; Li, X.-X.; Li, J.-J.; Sheng, S.; Zhang, F. Resveratrol Alleviates Levodopa-Induced Dyskinesia in Rats. Front. Immunol. 2021, 12, 683577. [Google Scholar] [CrossRef]
- Chase, D.L.; Pepper, J.S.; Koelle, M.R. Mechanism of Extrasynaptic Dopamine Signaling in Caenorhabditis elegans. Nat. Neurosci. 2004, 7, 1096–1103. [Google Scholar] [CrossRef]
- Merims, D.; Giladi, N. Dopamine Dysregulation Syndrome, Addiction and Behavioral Changes in Parkinson’s Disease. Parkinsonism Relat. Disord. 2008, 14, 273–280. [Google Scholar] [CrossRef]
- Suo, S.; Culotti, J.G.; Van Tol, H.H.M. Dopamine Counteracts Octopamine Signalling in a Neural Circuit Mediating Food Response in C. elegans. EMBO J. 2009, 28, 2437–2448. [Google Scholar] [CrossRef]
- Warren, N.; O’Gorman, C.; Lehn, A.; Siskind, D. Dopamine Dysregulation Syndrome in Parkinson’s Disease: A Systematic Review of Published Cases. J. Neurol. Neurosurg. Psychiatry 2017, 88, 1060–1064. [Google Scholar] [CrossRef]
- Cohen, J.D.; Servan-Schreiber, D. A Theory of Dopamine Function and Its Role in Cognitive Deficits in Schizophrenia. Schizophr. Bull. 1993, 19, 85–104. [Google Scholar] [CrossRef]
- Salgado-Pineda, P.; Delaveau, P.; Blin, O.; Nieoullon, A. Dopaminergic Contribution to the Regulation of Emotional Perception. Clin. Neuropharmacol. 2005, 28, 228–237. [Google Scholar] [CrossRef]
- Sourkes, T. Levadopa and Anticholinergic Drugs in Parkinsonism. Br. Med. J. 1971, 3, 703–704. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Poletti, M.; Bonuccelli, U. Acute and Chronic Cognitive Effects of Levodopa and Dopamine Agonists on Patients with Parkinson’s Disease: A Review. Ther. Adv. Psychopharmacol. 2013, 3, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Hayes, M.T. Parkinson’s Disease and Parkinsonism. Am. J. Med. 2019, 132, 802–807. [Google Scholar] [CrossRef] [PubMed]
- Cannas, A.; Solla, P.; Marrosu, M.G.; Marrosu, F. Dopamine Dysregulation Syndrome in Parkinson’s Disease Patients on Duodenal Levodopa Infusion. Mov. Disord. 2013, 28, 840–841. [Google Scholar] [CrossRef]
- Jung Kang, U.; Auinger, P. Activity Enhances Dopaminergic Long-Duration Response in Parkinson Disease. Neurology 2012, 78, 1146. [Google Scholar] [CrossRef]
- Chong, T.T.-J.; Bonnelle, V.; Manohar, S.; Veromann, K.-R.; Muhammed, K.; Tofaris, G.K.; Hu, M.; Husain, M. Dopamine Enhances Willingness to Exert Effort for Reward in Parkinson’s Disease. Cortex 2015, 69, 40–46. [Google Scholar] [CrossRef]
- Beaulieu-Boire, I.; Lang, A.E. Behavioral Effects of Levodopa. Mov. Disord. 2015, 30, 90–102. [Google Scholar] [CrossRef]
- Zurowski, M.; O’Brien, J.D. Developments in Impulse Control Behaviours of Parkinson’s Disease. Curr. Opin. Neurol. 2015, 28, 387–392. [Google Scholar] [CrossRef]
- Lohr, K.M.; Bernstein, A.I.; Stout, K.A.; Dunn, A.R.; Lazo, C.R.; Alter, S.P.; Wang, M.; Li, Y.; Fan, X.; Hess, E.J.; et al. Increased Vesicular Monoamine Transporter Enhances Dopamine Release and Opposes Parkinson Disease-Related Neurodegeneration in Vivo. Proc. Natl. Acad. Sci. USA 2014, 111, 9977–9982. [Google Scholar] [CrossRef]
- Fay, D.S. Classical Genetic Methods. WormBook 2013, 1–58. [Google Scholar] [CrossRef]
- Rieckher, M.; Kourtis, N.; Pasparaki, A.; Tavernarakis, N. Transgenesis in Caenorhabditis elegans. In Transgenesis Techniques: Principles and Protocols; Cartwright, E.J., Ed.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2009; pp. 21–39. ISBN 978-1-60327-019-9. [Google Scholar]
- Brenner, S. The Genetics of Caenorhabditis elegans. Genetics 1974, 77, 71–94. [Google Scholar] [CrossRef] [PubMed]
- Felton, C.M.; Johnson, C.M. Modulation of Dopamine-Dependent Behaviors by the Caenorhabditis elegans Olig Homolog HLH-17. J. Neurosci. Res. 2011, 89, 1627–1636. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, M.; Ishihara, T.; Katsura, I. A Novel WD40 Protein, CHE-2, Acts Cell-Autonomously in the Formation of C. elegans Sensory Cilia. Development 1999, 126, 4839–4848. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Cheong, M.C.; Cho, J.-Y.; Koo, H.-S.; Paik, Y.-K. A Novel Functional Cross-Interaction between Opioid and Pheromone Signaling May Be Involved in Stress Avoidance in Caenorhabditis elegans. Sci. Rep. 2020, 10, 1–15. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeong, H.; Park, J.Y.; Lee, J.-H.; Baik, J.-H.; Kim, C.-Y.; Cho, J.-Y.; Driscoll, M.; Paik, Y.-K. Deficiency in RCAT-1 Function Causes Dopamine Metabolism Related Behavioral Disorders in Caenorhabditis elegans. Int. J. Mol. Sci. 2022, 23, 2393. https://doi.org/10.3390/ijms23042393
Jeong H, Park JY, Lee J-H, Baik J-H, Kim C-Y, Cho J-Y, Driscoll M, Paik Y-K. Deficiency in RCAT-1 Function Causes Dopamine Metabolism Related Behavioral Disorders in Caenorhabditis elegans. International Journal of Molecular Sciences. 2022; 23(4):2393. https://doi.org/10.3390/ijms23042393
Chicago/Turabian StyleJeong, Haelim, Jun Young Park, Ji-Hyun Lee, Ja-Hyun Baik, Chae-Yeon Kim, Jin-Young Cho, Monica Driscoll, and Young-Ki Paik. 2022. "Deficiency in RCAT-1 Function Causes Dopamine Metabolism Related Behavioral Disorders in Caenorhabditis elegans" International Journal of Molecular Sciences 23, no. 4: 2393. https://doi.org/10.3390/ijms23042393
APA StyleJeong, H., Park, J. Y., Lee, J.-H., Baik, J.-H., Kim, C.-Y., Cho, J.-Y., Driscoll, M., & Paik, Y.-K. (2022). Deficiency in RCAT-1 Function Causes Dopamine Metabolism Related Behavioral Disorders in Caenorhabditis elegans. International Journal of Molecular Sciences, 23(4), 2393. https://doi.org/10.3390/ijms23042393






