MicroRNAs and ‘Sponging’ Competitive Endogenous RNAs Dysregulated in Colorectal Cancer: Potential as Noninvasive Biomarkers and Therapeutic Targets
Abstract
1. Introduction
2. miRNAs and Colorectal Cancer
2.1. miR-21
2.2. miR-17/92 Cluster
2.3. miR-143 and miR-145
2.4. miR-200 Family
2.5. miR-203
2.6. miR-135 Family
2.7. miR-96 and miR-183
2.8. miR-150
2.9. miR-195
3. Discussion
miRNAs, ceRNAs, and Chemosensitivity in CRC: A War of Attrition
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nigro, J.M.; Cho, K.R.; Fearon, E.R.; Kern, S.E.; Ruppert, J.M.; Oliner, J.D.; Kinzler, K.W.; Vogelstein, B. Scrambled exons. Cell 1991, 64, 607–613. [Google Scholar] [CrossRef]
- Calin, G.A.; Liu, C.; Ferracin, M.; Hyslop, T.; Spizzo, R.; Sevignani, C.; Fabbri, M.; Cimmino, A.; Lee, E.J.; Wojcik, S.E.; et al. Ultraconserved Regions Encoding ncRNAs Are Altered in Human Leukemias and Carcinomas. Cancer Cell 2007, 12, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Hansen, T.B.; Venø, M.T.; Kjems, J. Circular RNAs in cancer: Opportunities and challenges in the field. Oncogene 2018, 37, 555–565. [Google Scholar] [CrossRef]
- Takaoka, Y.; Shimizu, Y.; Hasegawa, H.; Ouchi, Y.; Qiao, S.; Nagahara, M.; Ichihara, M.; Lee, J.D.; Adachi, K.; Hamaguchi, M.; et al. Forced expression of miR-143 represses ERK5/c-Myc and p68/p72 signaling in concert with miR-145 in gut tumors of Apcmin mice. PLoS ONE 2012, 7, e42137. [Google Scholar] [CrossRef]
- Sachdeva, M.; Zhu, S.; Wu, F.; Wu, H.; Walia, V.; Kumar, S.; Elble, R.; Watabe, K.; Mo, Y.-Y. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc. Natl. Acad. Sci. USA 2009, 106, 3207–3212. [Google Scholar] [CrossRef]
- Pan, H.; Li, T.; Jiang, Y.; Pan, C.; Ding, Y.; Huang, Z.; Yu, H.; Kong, D. Overexpression of Circular RNA ciRS-7 Abrogates the Tumor Suppressive Effect of miR-7 on Gastric Cancer via PTEN/PI3K/AKT Signaling Pathway. J. Cell. Biochem. 2018, 119, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Xue, J.-L.; Shen, Q.; Chen, J.; Tian, L. MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatology 2012, 55, 1852–1862. [Google Scholar] [CrossRef]
- Kino, T.; Hurt, D.E.; Ichijo, T.; Nader, N.; Chrousos, G.P. Noncoding RNA Gas5 Is a Growth Arrest– and Starvation-Associated Repressor of the Glucocorticoid Receptor. Sci. Signal. 2010, 3, 1–16. [Google Scholar] [CrossRef]
- Pickard, M.; Williams, G. Molecular and Cellular Mechanisms of Action of Tumour Suppressor GAS5 LncRNA. Genes 2015, 6, 484–499. [Google Scholar] [CrossRef]
- Ren, T.; Hou, J.; Liu, C.; Shan, F.; Xiong, X.; Qin, A.; Chen, J.; Ren, W. The long non-coding RNA HOTAIRM1 suppresses cell progression via sponging endogenous miR-17-5p/ B-cell translocation gene 3 (BTG3) axis in 5-fluorouracil resistant colorectal cancer cells. Biomed. Pharmacother. 2019, 117, 109171. [Google Scholar] [CrossRef]
- Huang, G.; Zhu, H.; Shi, Y.; Wu, W.; Cai, H.; Chen, X. cir-ITCH Plays an Inhibitory Role in Colorectal Cancer by Regulating the Wnt/β-Catenin Pathway. PLoS ONE 2015, 10, e0131225. [Google Scholar] [CrossRef] [PubMed]
- Huels, D.J.; Sansom, O.J. Stem vs. non-stem cell origin of colorectal cancer. Br. J. Cancer 2015, 113, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Blanpain, C. Tracing the cellular origin of cancer. Nat. Cell Biol. 2013, 15, 126–134. [Google Scholar] [CrossRef] [PubMed]
- Mármol, I.; Sánchez-de-Diego, C.; Dieste, A.P.; Cerrada, E.; Yoldi, M.J.R. Colorectal carcinoma: A general overview and future perspectives in colorectal cancer. Int. J. Mol. Sci. 2017, 18, 197. [Google Scholar] [CrossRef] [PubMed]
- Zauber, A.G.; Winawer, S.J.; O’Brien, M.J.; Lansdorp-Vogelaar, I.; van Ballegooijen, M.; Hankey, B.F.; Shi, W.; Bond, J.H.; Schapiro, M.; Panish, J.F.; et al. Colonoscopic Polypectomy and Long-Term Prevention of Colorectal-Cancer Deaths. N. Engl. J. Med. 2012, 366, 687–696. [Google Scholar] [CrossRef]
- Van Dam, J. Prevention of Colorectal Cancer by Endoscopic Polypectomy. Ann. Intern. Med. 1995, 123, 949. [Google Scholar] [CrossRef] [PubMed]
- Baxter, N.N.; Goldwasser, M.A.; Paszat, L.F.; Saskin, R.; Urbach, D.R.; Rabeneck, L. Association of colonoscopy and death from colorectal cancer. Ann. Intern. Med. 2009, 150, 1–8. [Google Scholar] [CrossRef]
- Baran, B.; Mert Ozupek, N.; Yerli Tetik, N.; Acar, E.; Bekcioglu, O.; Baskin, Y. Difference Between Left-Sided and Right-Sided Colorectal Cancer: A Focused Review of Literature. Gastroenterol. Res. 2018, 11, 264–273. [Google Scholar] [CrossRef]
- Luo, X.; Burwinkel, B.; Tao, S.; Brenner, H. MicroRNA signatures: Novel biomarker for colorectal cancer? Cancer Epidemiol. Biomarkers Prev. 2011, 20, 1272–1286. [Google Scholar] [CrossRef]
- Wang, S.; Xiang, J.; Li, Z.; Lu, S.; Hu, J.; Gao, X.; Yu, L.; Wang, L.; Wang, J.; Wu, Y.; et al. A plasma microRNA panel for early detection of colorectal cancer. Int. J. Cancer 2015, 136, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, F.E.; Jeffries, C.D.; Vos, P.W.; Flake, G.; Nuovo, G.J.; Sinar, D.R.; Naziri, W.; Marcuard, S.P. Diagnostic microRNA markers for screening sporadic human colon cancer and active ulcerative colitis in stool and tissue. Cancer Genom. Proteom. 2009, 6, 281–295. [Google Scholar]
- Fan, Y.-H.; Ye, M.-H.; Wu, L.; Wu, M.-J.; Lu, S.-G.; Zhu, X.-G. BRAF-activated lncRNA predicts gastrointestinal cancer patient prognosis: A meta-analysis. Oncotarget 2017, 8, 6295–6303. [Google Scholar] [CrossRef]
- Xu, W.; Zhou, G.; Wang, H.; Liu, Y.; Chen, B.; Chen, W.; Lin, C.; Wu, S.; Gong, A.; Xu, M. Circulating lncRNA SNHG11 as a novel biomarker for early diagnosis and prognosis of colorectal cancer. Int. J. Cancer 2020, 146, 2901–2912. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Z.; Xu, S.; Guo, J. Novel potential tumor biomarkers: Circular RNAs and exosomal circular RNAs in gastrointestinal malignancies. J. Clin. Lab. Anal. 2020, 34, e23359. [Google Scholar] [CrossRef]
- Naeli, P.; Pourhanifeh, M.H.; Karimzadeh, M.R.; Shabaninejad, Z.; Movahedpour, A.; Tarrahimofrad, H.; Mirzaei, H.R.; Bafrani, H.H.; Savardashtaki, A.; Mirzaei, H.; et al. Circular RNAs and gastrointestinal cancers: Epigenetic regulators with a prognostic and therapeutic role. Crit. Rev. Oncol. Hematol. 2020, 145, 102854. [Google Scholar] [CrossRef]
- Liu, J.; Carmell, M.A.; Rivas, F.V.; Marsden, C.G.; Thomson, J.M.; Song, J.-J.; Hammond, S.M.; Joshua-Tor, L.; Hannon, G.J. Argonaute2 Is the Catalytic Engine of Mammalian RNAi. Science 2004, 305, 1437–1441. [Google Scholar] [CrossRef]
- Lewis, B.P.; Shih, I.; Jones-Rhoades, M.W.; Bartel, D.P.; Burge, C.B. Prediction of Mammalian MicroRNA Targets. Cell 2003, 115, 787–798. [Google Scholar] [CrossRef]
- Easow, G.; Teleman, A.A.; Cohen, S.M. Isolation of microRNA targets by miRNP immunopurification. RNA 2007, 13, 1198–1204. [Google Scholar] [CrossRef]
- Baek, D.; Villén, J.; Shin, C.; Camargo, F.D.; Gygi, S.P.; Bartel, D.P. The impact of microRNAs on protein output. Nature 2008, 455, 64–71. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, S.; Steitz, J.A. AU-Rich-Element-Mediated Upregulation of Translation by FXR1 and Argonaute 2. Cell 2007, 128, 1105–1118. [Google Scholar] [CrossRef]
- Cordes, K.R.; Sheehy, N.T.; White, M.P.; Berry, E.C.; Morton, S.U.; Muth, A.N.; Lee, T.H.; Miano, J.M.; Ivey, K.N.; Srivastava, D. MiR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature 2009, 460, 705–710. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-C.; Liu, L.-Z.; Addison, J.B.; Wonderlin, W.F.; Ivanov, A.V.; Ruppert, J.M. A KLF4-miRNA-206 Autoregulatory Feedback Loop Can Promote or Inhibit Protein Translation Depending upon Cell Context. Mol. Cell. Biol. 2011, 31, 2513–2527. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, M.; Paone, A.; Calore, F.; Galli, R.; Gaudio, E.; Santhanam, R.; Lovat, F.; Fadda, P.; Mao, C.; Nuovo, G.J.; et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. USA 2012, 109. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhu, Z.; Watabe, K.; Zhang, X.; Bai, C.; Xu, M.; Wu, F.; Mo, Y.-Y. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ. 2013, 20, 1558–1568. [Google Scholar] [CrossRef]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Volinia, S.; Calin, G.A.; Liu, C.G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef]
- Bandrés, E.; Cubedo, E.; Agirre, X.; Malumbres, R.; Zárate, R.; Ramirez, N.; Abajo, A.; Navarro, A.; Moreno, I.; Monzó, M.; et al. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol. Cancer 2006, 5, 1–10. [Google Scholar] [CrossRef]
- Ren, A.; Dong, Y.; Tsoi, H.; Yu, J. Detection of miRNA as Non-Invasive Biomarkers of Colorectal Cancer. Int. J. Mol. Sci. 2015, 16, 2810–2823. [Google Scholar] [CrossRef]
- Niu, L.; Yang, W.; Duan, L.; Wang, X.; Li, Y.; Xu, C.; Liu, C.; Zhang, Y.; Zhou, W.; Liu, J.; et al. Biological Implications and Clinical Potential of Metastasis-Related miRNA in Colorectal Cancer. Mol. Ther. Nucleic Acids 2021, 23, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Vautrot, V.; Chanteloup, G.; Elmallah, M.; Cordonnier, M.; Aubin, F.; Garrido, C.; Gobbo, J. Exosomal miRNA: Small Molecules, Big Impact in Colorectal Cancer. J. Oncol. 2019, 2019, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Marcuello, M.; Duran-Sanchon, S.; Moreno, L.; Lozano, J.J.; Bujanda, L.; Castells, A.; Gironella, M. Analysis of A 6-Mirna Signature in Serum from Colorectal Cancer Screening Participants as Non-Invasive Biomarkers for Advanced Adenoma and Colorectal Cancer Detection. Cancers 2019, 11, 1542. [Google Scholar] [CrossRef] [PubMed]
- Slattery, M.L.; Mullany, L.E.; Sakoda, L.; Samowitz, W.S.; Wolff, R.K.; Stevens, J.R.; Herrick, J.S. The NF-κB signalling pathway in colorectal cancer: Associations between dysregulated gene and miRNA expression. J. Cancer Res. Clin. Oncol. 2018, 144, 269–283. [Google Scholar] [CrossRef]
- Tang, X.J.; Wang, W.; Hann, S.S. Interactions among lncRNAs, miRNAs and mRNA in colorectal cancer. Biochimie 2019, 163, 58–72. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, M.; Shan, X.; Zhou, X.; Wang, T.; Zhang, J.; Tao, J.; Cheng, W.; Chen, G.; Li, J.; et al. A panel of seven-miRNA signature in plasma as potential biomarker for colorectal cancer diagnosis. Gene 2019, 687, 246–254. [Google Scholar] [CrossRef]
- Rapado-González, Ó.; Majem, B.; Álvarez-Castro, A.; Díaz-Peña, R.; Abalo, A.; Suárez-Cabrera, L.; Gil-Moreno, A.; Santamaría, A.; López-López, R.; Muinelo-Romay, L.; et al. A Novel Saliva-Based miRNA Signature for Colorectal Cancer Diagnosis. J. Clin. Med. 2019, 8, 2029. [Google Scholar] [CrossRef]
- Yang, L.; Belaguli, N.; Berger, D.H. MicroRNA and Colorectal Cancer. World J. Surg. 2009, 33, 638–646. [Google Scholar] [CrossRef]
- Huang, X.; Zhu, X.; Yu, Y.; Zhu, W.; Jin, L.; Zhang, X.; Li, S.; Zou, P.; Xie, C.; Cui, R. Dissecting miRNA signature in colorectal cancer progression and metastasis. Cancer Lett. 2021, 501, 66–82. [Google Scholar] [CrossRef]
- Falzone, L.; Scola, L.; Zanghì, A.; Biondi, A.; Di Cataldo, A.; Libra, M.; Candido, S. Integrated analysis of colorectal cancer microRNA datasets: Identification of microRNAs associated with tumor development. Aging 2018, 10, 1000–1014. [Google Scholar] [CrossRef]
- Carter, J.V.; Roberts, H.L.; Pan, J.; Rice, J.D.; Burton, J.F.; Galbraith, N.J.; Eichenberger, M.R.; Jorden, J.; Deveaux, P.; Farmer, R.; et al. A Highly Predictive Model for Diagnosis of Colorectal Neoplasms Using Plasma MicroRNA. Ann. Surg. 2016, 264, 575–584. [Google Scholar] [CrossRef] [PubMed]
- Lai, X.; Friedman, A. Exosomal microRNA concentrations in colorectal cancer: A mathematical model. J. Theor. Biol. 2017, 415, 70–83. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Zhang, Y.; Yang, J. A Five-microRNA Signature as Prognostic Biomarker in Colorectal Cancer by Bioinformatics Analysis. Front. Oncol. 2019, 9, 1207. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.W.; Ng, S.S.M.; Dong, Y.J.; Ng, S.C.; Leung, W.W.; Lee, C.W.; Wong, Y.N.; Chan, F.K.L.; Yu, J.; Sung, J.J.Y. Detection of miR-92a and miR-21 in stool samples as potential screening biomarkers for colorectal cancer and polyps. Gut 2012, 61, 739–745. [Google Scholar] [CrossRef] [PubMed]
- Yau, T.O.; Tang, C.-M.; Harriss, E.K.; Dickins, B.; Polytarchou, C. Faecal microRNAs as a non-invasive tool in the diagnosis of colonic adenomas and colorectal cancer: A meta-analysis. Sci. Rep. 2019, 9, 9491. [Google Scholar] [CrossRef]
- Link, A.; Balaguer, F.; Shen, Y.; Nagasaka, T.; Lozano, J.J.; Boland, C.R.; Goel, A. Fecal MicroRNAs as Novel Biomarkers for Colon Cancer Screening. Cancer Epidemiol. Biomarkers Prev. 2010, 19, 1766–1774. [Google Scholar] [CrossRef]
- Toiyama, Y.; Takahashi, M.; Hur, K.; Nagasaka, T.; Tanaka, K.; Inoue, Y.; Kusunoki, M.; Boland, C.R.; Goel, A. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J. Natl. Cancer Inst. 2013, 105, 849–859. [Google Scholar] [CrossRef]
- Yamada, A.; Horimatsu, T.; Okugawa, Y.; Nishida, N.; Honjo, H.; Ida, H.; Kou, T.; Kusaka, T.; Sasaki, Y.; Yagi, M.; et al. Serum MIR-21, MIR-29a, and MIR-125b are promising biomarkers for the early detection of colorectal neoplasia. Clin. Cancer Res. 2015, 21, 4234–4242. [Google Scholar] [CrossRef]
- Zhang, G.; Zhou, H.; Xiao, H.; Liu, Z.; Tian, H.; Zhou, T. MicroRNA-92a Functions as an Oncogene in Colorectal Cancer by Targeting PTEN. Dig. Dis. Sci. 2014, 59, 98–107. [Google Scholar] [CrossRef]
- Jin, X.-H.; Lu, S.; Wang, A.-F. Expression and clinical significance of miR-4516 and miR-21-5p in serum of patients with colorectal cancer. BMC Cancer 2020, 20, 241. [Google Scholar] [CrossRef]
- Zhu, M.; Huang, Z.; Zhu, D.; Zhou, X.; Shan, X.; Qi, L.W.; Wu, L.; Cheng, W.; Zhu, J.; Zhang, L.; et al. A panel of microRNA signature in serum for colorectal cancer diagnosis. Oncotarget 2017, 8, 17081–17091. [Google Scholar] [CrossRef] [PubMed]
- Gmerek, L.; Martyniak, K.; Horbacka, K.; Krokowicz, P.; Scierski, W.; Golusinski, P.; Golusinski, W.; Schneider, A.; Masternak, M.M. MicroRNA regulation in colorectal cancer tissue and serum. PLoS ONE 2019, 14, e0222013. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.-H.; Zhou, Z.-G.; Chen, R.; Wang, M.-J.; Zhou, B.; Li, Y.; Sun, X.-F. Serum miR-21 and miR-92a as biomarkers in the diagnosis and prognosis of colorectal cancer. Tumour Biol. 2013, 34, 2175–2181. [Google Scholar] [CrossRef] [PubMed]
- Sarlinova, M.; Halasa, M.; Mistuna, D.; Musak, L.; Iliev, R.; Slaby, O.; Mazuchova, J.; Valentova, V.; Plank, L.; Halasova, E. Mir-21, mir-221 and mir-150 are deregulated in peripheral blood of patients with colorectal cancer. Anticancer Res. 2016, 36, 5449–5454. [Google Scholar] [CrossRef]
- Zekri, A.R.N.; Youssef, A.S.E.D.; Lotfy, M.M.; Gabr, R.; Ahmed, O.S.; Nassar, A.; Hussein, N.; Omran, D.; Medhat, E.; Eid, S.; et al. Circulating serum miRNAs as diagnostic markers for colorectal cancer. PLoS ONE 2016, 11, e0154130. [Google Scholar] [CrossRef]
- Li, J.; Liu, Y.; Wang, C.; Deng, T.; Liang, H.; Wang, Y.; Huang, D.; Fan, Q.; Wang, X.; Ning, T.; et al. Serum miRNA expression profile as a prognostic biomarker of stage II/III colorectal adenocarcinoma. Sci. Rep. 2015, 5, 1–13. [Google Scholar] [CrossRef]
- Zhu, J.; Dong, H.; Zhang, Q.; Zhang, S. Combined assays for serum carcinoembryonic antigen and microRNA-17-3p offer improved diagnostic potential for stage I/II colon cancer. Mol. Clin. Oncol. 2015, 3, 1315–1318. [Google Scholar] [CrossRef]
- Zhang, G.-J.; Zhou, T.; Liu, Z.-L.; Tian, H.-P.; Xia, S.-S. Plasma miR-200c and miR-18a as potential biomarkers for the detection of colorectal carcinoma. Mol. Clin. Oncol. 2013, 1, 379–384. [Google Scholar] [CrossRef]
- Vega, A.B.; Pericay, C.; Moya, I.; Ferrer, A.; Dotor, E.; Pisa, A.; Casalots, À.; Serra-Aracil, X.; Oliva, J.-C.; Ruiz, A.; et al. microRNA expression profile in stage III colorectal cancer: Circulating miR-18a and miR-29a as promising biomarkers. Oncol. Rep. 2013, 30, 320–326. [Google Scholar] [CrossRef]
- Farace, C.; Pisano, A.; Griñan-Lison, C.; Solinas, G.; Jiménez, G.; Serra, M.; Carrillo, E.; Scognamillo, F.; Attene, F.; Montella, A.; et al. Deregulation of cancer-stem-cell-associated miRNAs in tissues and sera of colorectal cancer patients. Oncotarget 2020, 11, 116–130. [Google Scholar] [CrossRef]
- Matsumura, T.; Sugimachi, K.; Iinuma, H.; Takahashi, Y.; Kurashige, J.; Sawada, G.; Ueda, M.; Uchi, R.; Ueo, H.; Takano, Y.; et al. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br. J. Cancer 2015, 113, 275–281. [Google Scholar] [CrossRef]
- Chen, Q.; Xia, H.W.; Ge, X.J.; Zhang, Y.C.; Tang, Q.L.; Bi, F. Serum miR-19a predicts resistance to FOLFOX chemotherapy in advanced colorectal cancer cases. Asian Pacific J. Cancer Prev. 2013, 14, 7421–7426. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.-B.; Sheng, J.; Yu, J.-M.; Liu, J.-H.; Qin, X.-X.; Mou, B. MiR-19a-3p regulates the Forkhead box F2-mediated Wnt/β-catenin signaling pathway and affects the biological functions of colorectal cancer cells. World J. Gastroenterol. 2020, 26, 627–644. [Google Scholar] [CrossRef]
- Maminezhad, H.; Ghanadian, S.; Pakravan, K.; Razmara, E.; Rouhollah, F.; Mossahebi-Mohammadi, M.; Babashah, S. A panel of six-circulating miRNA signature in serum and its potential diagnostic value in colorectal cancer. Life Sci. 2020, 258, 118226. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Du, L.; Yang, X.; Zhang, X.; Wang, L.; Yang, Y.; Li, J.; Wang, C. Serum microRNA panel as biomarkers for early diagnosis of colorectal adenocarcinoma. Br. J. Cancer 2014, 111, 1985–1992. [Google Scholar] [CrossRef] [PubMed]
- Dokhanchi, M.; Pakravan, K.; Zareian, S.; Hussen, B.M.; Farid, M.; Razmara, E.; Mossahebi-Mohammadi, M.; Cho, W.C.; Babashah, S. Colorectal cancer cell-derived extracellular vesicles transfer miR-221-3p to promote endothelial cell angiogenesis via targeting suppressor of cytokine signaling 3. Life Sci. 2021, 285, 119937. [Google Scholar] [CrossRef]
- Yang, Q.; Wang, S.; Huang, J.; Xia, C.; Jin, H.; Fan, Y. Serum miR-20a and miR-486 are potential biomarkers for discriminating colorectal neoplasia: A pilot study. J. Cancer Res. Ther. 2018, 14, 1572. [Google Scholar] [CrossRef]
- Huang, G.; Chen, X.; Cai, Y.; Wang, X.; Xing, C. miR-20a-directed regulation of BID is associated with the TRAIL sensitivity in colorectal cancer. Oncol. Rep. 2017, 37, 571–578. [Google Scholar] [CrossRef]
- Huang, Z.; Huang, D.; Ni, S.; Peng, Z.; Sheng, W.; Du, X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int. J. Cancer 2010, 127, 118–126. [Google Scholar] [CrossRef]
- Shi, Y.; Liu, Z. Serum miR-92a-1 is a novel diagnostic biomarker for colorectal cancer. J. Cell. Mol. Med. 2020, 24, 8363–8367. [Google Scholar] [CrossRef]
- Elshafei, A.; Shaker, O.; Abd El-motaal, O.; Salman, T. The expression profiling of serum miR-92a, miR-375, and miR-760 in colorectal cancer: An Egyptian study. Tumor Biol. 2017, 39, 101042831770576. [Google Scholar] [CrossRef] [PubMed]
- Shiosaki, J.; Tiirikainen, M.; Peplowska, K.; Shaeffer, D.; Machida, M.; Sakamoto, K.; Takahashi, M.; Kojima, K.; Machi, J.; Bryant-Greenwood, P.; et al. Serum micro-RNA Identifies Early Stage Colorectal Cancer in a Multi-Ethnic Population. Asian Pacific J. Cancer Prev. 2020, 21, 3019–3026. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Wang, H.; Zhang, X.; Yang, Y.; Wang, L.; Du, L.; Li, W.; Li, J.; Qu, A.; Liu, Y.; et al. Identification and validation of reference genes for qPCR detection of serum microRNAs in colorectal adenocarcinoma patients. PLoS ONE 2013, 8, e83025. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Huang, S.K.; Zhao, M.; Yang, M.; Zhong, J.L.; Gu, Y.Y.; Peng, H.; Che, Y.Q.; Huang, C.Z. Identification of a circulating microRNA signature for colorectal cancer detection. PLoS ONE 2014, 9, e87451. [Google Scholar] [CrossRef]
- Luo, X.; Stock, C.; Burwinkel, B.; Brenner, H. Identification and Evaluation of Plasma MicroRNAs for Early Detection of Colorectal Cancer. PLoS ONE 2013, 8, e62880. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Wei, B.; Chen, Z.; Wang, J.; Zhao, L.; Peng, X.; Liu, K.; Lai, Y.; Ni, L. Identification of a four-microRNA panel in serum as promising biomarker for colorectal carcinoma detection. Biomark. Med. 2020, 14, 749–760. [Google Scholar] [CrossRef]
- Hiyoshi, Y.; Akiyoshi, T.; Inoue, R.; Murofushi, K.; Yamamoto, N.; Fukunaga, Y.; Ueno, M.; Baba, H.; Mori, S.; Yamaguchi, T. Serum miR-143 levels predict the pathological response to neoadjuvant chemoradiotherapy in patients with locally advanced rectal cancer. Oncotarget 2017, 8, 79201–79211. [Google Scholar] [CrossRef]
- Ramzy, I.; Hasaballah, M.; Marzaban, R.; Shaker, O.; Soliman, Z.A. Evaluation of microRNAs-29a, 92a and 145 in colorectal carcinoma as candidate diagnostic markers: An Egyptian pilot study. Clin. Res. Hepatol. Gastroenterol. 2015, 39, 508–515. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, Y.; Cogdell, D.; Calin, G.A.; Sun, B.; Kopetz, S.; Hamilton, S.R.; Zhang, W. Examining plasma microRNA markers for colorectal cancer at different stages. Oncotarget 2016, 7, 11434–11449. [Google Scholar] [CrossRef]
- Kingham, T.P.; Nguyen, H.C.B.; Zheng, J.; Konstantinidis, I.T.; Sadot, E.; Shia, J.; Kuk, D.; Zhang, S.; Saltz, L.; D’Angelica, M.I.; et al. MicroRNA-203 predicts human survival after resection of colorectal liver metastasis. Oncotarget 2017, 8, 18821–18831. [Google Scholar] [CrossRef]
- Takano, Y.; Masuda, T.; Iinuma, H.; Yamaguchi, R.; Sato, K.; Tobo, T.; Hirata, H.; Kuroda, Y.; Nambara, S.; Hayashi, N.; et al. Circulating exosomal microRNA-203 is associated with metastasis possibly via inducing tumor-associated macrophages in colorectal cancer. Oncotarget 2017, 8, 78598–78613. [Google Scholar] [CrossRef] [PubMed]
- Deng, B.; Wang, B.; Fang, J.; Zhu, X.; Cao, Z.; Lin, Q.; Zhou, L.; Sun, X. MiRNA-203 suppresses cell proliferation, migration and invasion in colorectal cancer via targeting of EIF5A2. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hur, K.; Toiyama, Y.; Okugawa, Y.; Ide, S.; Imaoka, H.; Boland, C.R.; Goel, A. Circulating microRNA-203 predicts prognosis and metastasis in human colorectal cancer. Gut 2017, 66, 654–664. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Shen, Y.; Zhao, P.; Cai, S.; Feng, Z.; Cheng, M.; Wu, Y.; Zhu, Y. Biomarker exploration of microRNA-203 as a promising substrate for predicting poor survival outcome in colorectal cancer. BMC Cancer 2020, 20, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Baker, K.; Redman, M.W.; Wang, L.; Adams, S.V.; Yu, M.; Dickinson, B.; Makar, K.; Ulrich, N.; Böhm, J.; et al. Dynamic plasma microRNAs are biomarkers for prognosis and early detection of recurrence in colorectal cancer. Br. J. Cancer 2017, 117, 1202–1210. [Google Scholar] [CrossRef] [PubMed]
- Nassar, F.J.; Msheik, Z.S.; Itani, M.M.; El Helou, R.; Hadla, R.; Kreidieh, F.; Bejjany, R.; Mukherji, D.; Shamseddine, A.; Nasr, R.R.; et al. Circulating mirna as biomarkers for colorectal cancer diagnosis and liver metastasis. Diagnostics 2021, 11, 341. [Google Scholar] [CrossRef] [PubMed]
- Toiyama, Y.; Hur, K.; Tanaka, K.; Inoue, Y.; Kusunoki, M.; Boland, C.R.; Goel, A. Serum miR-200c Is a Novel Prognostic and Metastasis-Predictive Biomarker in Patients with Colorectal Cancer. Ann. Surg. 2014, 259, 735–743. [Google Scholar] [CrossRef]
- Ardila, H.J.; Sanabria-Salas, M.C.; Meneses, X.; Rios, R.; Huertas-Salgado, A.; Serrano, M.L. Circulating miR-141-3p, miR-143-3p and miR-200c-3p are differentially expressed in colorectal cancer and advanced adenomas. Mol. Clin. Oncol. 2019, 11, 201–207. [Google Scholar] [CrossRef]
- Ding, M.; Zhang, T.; Li, S.; Zhang, Y.; Qiu, Y.; Zhang, B. Correlation analysis between liver metastasis and serum levels of MIR-200 and MIR-141 in patients with colorectal cancer. Mol. Med. Rep. 2017, 16, 7791–7795. [Google Scholar] [CrossRef]
- Tayel, S.I.; Fouda, E.A.M.; Gohar, S.F.; Elshayeb, E.I.; El-sayed, E.H.; El-kousy, S.M. Potential role of MicroRNA 200c gene expression in assessment of colorectal cancer. Arch. Biochem. Biophys. 2018, 647, 41–46. [Google Scholar] [CrossRef]
- Santasusagna, S.; Moreno, I.; Navarro, A.; Martinez Rodenas, F.; Hernández, R.; Castellano, J.J.; Muñoz, C.; Monzo, M. Prognostic impact of miR-200 family members in plasma and exosomes from tumor-draining versus peripheral veins of colon cancer patients. Oncology 2018, 95, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.J.; Cai, X.J.; Li, S.J. The clinical significance of MiR-429 as a predictive biomarker in colorectal cancer patients receiving 5-fluorouracil treatment. Med. Sci. Monit. 2016, 22, 3352–3361. [Google Scholar] [CrossRef]
- Koga, Y.; Yasunaga, M.; Takahashi, A.; Kuroda, J.; Moriya, Y.; Akasu, T.; Fujita, S.; Yamamoto, S.; Baba, H.; Matsumura, Y. MicroRNA expression profiling of exfoliated colonocytes isolated from feces for colorectal cancer screening. Cancer Prev. Res. 2010, 3, 1435–1442. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, A.; Cai, M.; Tong, M.; Chen, F.; Huang, L. Identification of stool miR-135b-5p as a non-invasive diaognostic biomarker in later tumor stage of colorectal cancer. Life Sci. 2020, 260, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhang, H.; Shen, X.; Ju, S. Serum microRNA-135a-5p as an auxiliary diagnostic biomarker for colorectal cancer. Ann. Clin. Biochem. 2017, 54, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Yang, D.; Ding, X.; Xu, P. Clinical value of microRNA-135a and MMP-13 in colon cancer. Oncol. Lett. 2021, 22, 1–9. [Google Scholar] [CrossRef]
- Wu, C.W.; Ng, S.C.; Dong, Y.; Tian, L.; Ng, S.S.M.; Leung, W.W.; Law, W.T.; Yau, T.O.; Chan, F.K.L.; Sung, J.J.Y.; et al. Identification of microrna-135b in stool as a potential noninvasive biomarker for colorectal cancer and adenoma. Clin. Cancer Res. 2014, 20, 2994–3002. [Google Scholar] [CrossRef]
- Phua, L.C.; Chue, X.P.; Koh, P.K.; Cheah, P.Y.; Chan, E.C.Y.; Ho, H.K. Global fecal microRNA profiling in the identification of biomarkers for colorectal cancer screening among Asians. Oncol. Rep. 2014, 32, 97–104. [Google Scholar] [CrossRef]
- Qin, Y.; Li, L.; Wang, F.; Zhou, X.; Liu, Y.; Yin, Y.; Qi, X. Knockdown of miR-135b sensitizes colorectal cancer cells to oxaliplatin-induced apoptosis through increase of FOXO1. Cell. Physiol. Biochem. 2018, 48, 1627–1637. [Google Scholar] [CrossRef]
- Jin, G.; Liu, Y.; Zhang, J.; Bian, Z.; Yao, S.; Fei, B.; Zhou, L.; Yin, Y.; Huang, Z. A panel of serum exosomal microRNAs as predictive markers for chemoresistance in advanced colorectal cancer. Cancer Chemother. Pharmacol. 2019, 84, 315–325. [Google Scholar] [CrossRef]
- Faltejskova, P.; Bocanek, O.; Sachlova, M.; Svoboda, M.; Kiss, I.; Vyzula, R.; Slaby, O. Circulating miR-17-3p, miR-29a, miR-92a and miR-135b in serum: Evidence against their usage as biomarkers in colorectal cancer. Cancer Biomarkers 2012, 12, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Yuan, D.; Li, K.; Zhu, K.; Yan, R.; Dang, C. Plasma miR-183 predicts recurrence and prognosis in patients with colorectal cancer. Cancer Biol. Ther. 2015, 16, 268–275. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.J.; Song, X.; Niu, L.; Tang, Y.; Song, X.; Xie, L. Circulating exosomal mir-150-5p and mir-99b-5p as diagnostic biomarkers for colorectal cancer. Front. Oncol. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Xiao, Y.; Zhong, J.; Zhong, B.; Huang, J.; Jiang, L.; Jiang, Y.; Yuan, J.; Sun, J.; Dai, L.; Yang, C.; et al. Exosomes as potential sources of biomarkers in colorectal cancer. Cancer Lett. 2020, 476, 13–22. [Google Scholar] [CrossRef]
- Aherne, S.T.; Madden, S.F.; Hughes, D.J.; Pardini, B.; Naccarati, A.; Levy, M.; Vodicka, P.; Neary, P.; Dowling, P.; Clynes, M. Circulating miRNAs miR-34a and miR-150 associated with colorectal cancer progression. BMC Cancer 2015, 15, 1–13. [Google Scholar] [CrossRef]
- Ogata-Kawata, H.; Izumiya, M.; Kurioka, D.; Honma, Y.; Yamada, Y.; Furuta, K.; Gunji, T.; Ohta, H.; Okamoto, H.; Sonoda, H.; et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS ONE 2014, 9, e92921. [Google Scholar] [CrossRef]
- Al-Sheikh, Y.A.; Ghneim, H.K.; Softa, K.I.; Al-Jobran, A.A.; Al-Obeed, O.; Mohamed, M.A.V.; Abdulla, M.; Aboul-Soud, M.A.M. Expression profiling of selected microRNA signatures in plasma and tissues of Saudi colorectal cancer patients by qPCR. Oncol. Lett. 2016, 11, 1406–1412. [Google Scholar] [CrossRef] [PubMed]
- Wada, Y.; Shimada, M.; Murano, T.; Takamaru, H.; Morine, Y.; Ikemoto, T.; Saito, Y.; Balaguer, F.; Bujanda, L.; Pellise, M.; et al. A Liquid Biopsy Assay for Noninvasive Identification of Lymph Node Metastases in T1 Colorectal Cancer. Gastroenterology 2021, 161, 151–162.e1. [Google Scholar] [CrossRef]
- Iorio, M.V.; Ferracin, M.; Liu, C.G.; Veronese, A.; Spizzo, R.; Sabbioni, S.; Magri, E.; Pedriali, M.; Fabbri, M.; Campiglio, M.; et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005, 65, 7065–7070. [Google Scholar] [CrossRef]
- Meng, F.; Henson, R.; Wehbe–Janek, H.; Ghoshal, K.; Jacob, S.T.; Patel, T. MicroRNA-21 Regulates Expression of the PTEN Tumor Suppressor Gene in Human Hepatocellular Cancer. Gastroenterology 2007, 133, 647–658. [Google Scholar] [CrossRef]
- Si, M.L.; Zhu, S.; Wu, H.; Lu, Z.; Wu, F.; Mo, Y.Y. miR-21-mediated tumor growth. Oncogene 2007, 26, 2799–2803. [Google Scholar] [CrossRef] [PubMed]
- Kasashima, K.; Nakamura, Y.; Kozu, T. Altered expression profiles of microRNAs during TPA-induced differentiation of HL-60 cells. Biochem. Biophys. Res. Commun. 2004, 322, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Cekaite, L.; Clancy, T.; Sioud, M. Increased miR-21 expression during human monocyte differentiation into DCs. Front. Biosci. 2010, E2, 143. [Google Scholar] [CrossRef]
- Jenike, A.E.; Halushka, M.K. miR-21: A non-specific biomarker of all maladies. Biomark. Res. 2021, 9, 18. [Google Scholar] [CrossRef]
- Ma, X.; Kumar, M.; Choudhury, S.N.; Becker Buscaglia, L.E.; Barker, J.R.; Kanakamedala, K.; Liu, M.-F.; Li, Y. Loss of the miR-21 allele elevates the expression of its target genes and reduces tumorigenesis. Proc. Natl. Acad. Sci. USA 2011, 108, 10144–10149. [Google Scholar] [CrossRef]
- Guo, X.-Z.; Ye, X.-L.; Xiao, W.-Z.; Wei, X.-N.; You, Q.-H.; Che, X.-H.; Cai, Y.-J.; Chen, F.; Yuan, H.; Liu, X.-J.; et al. Downregulation of VMP1 confers aggressive properties to colorectal cancer. Oncol. Rep. 2015, 34, 2557–2566. [Google Scholar] [CrossRef][Green Version]
- Sauermann, M.; Sahin, Ö.; Sültmann, H.; Hahne, F.; Blaszkiewicz, S.; Majety, M.; Zatloukal, K.; Füzesi, L.; Poustka, A.; Wiemann, S.; et al. Reduced expression of vacuole membrane protein 1 affects the invasion capacity of tumor cells. Oncogene 2008, 27, 1320–1326. [Google Scholar] [CrossRef]
- Wang, C.; Peng, R.; Zeng, M.; Zhang, Z.; Liu, S.; Jiang, D.; Lu, Y.; Zou, F. An autoregulatory feedback loop of miR-21/VMP1 is responsible for the abnormal expression of miR-21 in colorectal cancer cells. Cell Death Dis. 2020, 11, 1067. [Google Scholar] [CrossRef]
- Meng, F.; Henson, R.; Lang, M.; Wehbe, H.; Maheshwari, S.; Mendell, J.T.; Jiang, J.; Schmittgen, T.D.; Patel, T. Involvement of Human Micro-RNA in Growth and Response to Chemotherapy in Human Cholangiocarcinoma Cell Lines. Gastroenterology 2006, 130, 2113–2129. [Google Scholar] [CrossRef]
- Asangani, I.A.; Rasheed, S.A.K.; Nikolova, D.A.; Leupold, J.H.; Colburn, N.H.; Post, S.; Allgayer, H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 2008, 27, 2128–2136. [Google Scholar] [CrossRef]
- Sheedy, F.J.; Palsson-McDermott, E.; Hennessy, E.J.; Martin, C.; O’Leary, J.J.; Ruan, Q.; Johnson, D.S.; Chen, Y.; O’Neill, L.A.J. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat. Immunol. 2010, 11, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Song, Y.; Xiong, Y.; Wang, X.; Xu, K.; Han, B.; Bai, Y.; Li, L.; Zhang, Y.; Zhou, L. MicroRNA-21 (Mir-21) Promotes Cell Growth and Invasion by Repressing Tumor Suppressor PTEN in Colorectal Cancer. Cell. Physiol. Biochem. 2017, 43, 945–958. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Yin, Y.; Guo, H.; Li, S.; Sun, Y.; Zeng, C.; Zhu, W. The mechanisms and clinical significance of PDCD4 in colorectal cancer. Gene 2019, 680, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Xiong, B.; Cheng, Y.; Ma, L.; Zhang, C. MiR-21 regulates biological behavior through the PTEN/PI-3 K/Akt signaling pathway in human colorectal cancer cells. Int. J. Oncol. 2013, 42, 219–228. [Google Scholar] [CrossRef]
- Shi, L.; Chen, J.; Yang, J.; Pan, T.; Zhang, S.; Wang, Z. MiR-21 protected human glioblastoma U87MG cells from chemotherapeutic drug temozolomide induced apoptosis by decreasing Bax/Bcl-2 ratio and caspase-3 activity. Brain Res. 2010, 1352, 255–264. [Google Scholar] [CrossRef]
- Buscaglia, L.E.B.; Li, Y. Apoptosis and the target genes of microRNA-21. Chin. J. Cancer 2011, 30, 371–380. [Google Scholar] [CrossRef]
- Ferraro, A.; Kontos, C.K.; Boni, T.; Bantounas, I.; Siakouli, D.; Kosmidou, V.; Vlassi, M.; Spyridakis, Y.; Tsipras, I.; Zografos, G.; et al. Epigenetic regulation of miR-21 in colorectal cancer. Epigenetics 2014, 9, 129–141. [Google Scholar] [CrossRef]
- Chang, K.H.; Miller, N.; Kheirelseid, E.A.H.; Ingoldsby, H.; Hennessy, E.; Curran, C.E.; Curran, S.; Smith, M.J.; Regan, M.; McAnena, O.J.; et al. MicroRNA-21 and PDCD4 expression in colorectal cancer. Eur. J. Surg. Oncol. 2011, 37, 597–603. [Google Scholar] [CrossRef]
- Hayashita, Y.; Osada, H.; Tatematsu, Y.; Yamada, H.; Yanagisawa, K.; Tomida, S.; Yatabe, Y.; Kawahara, K.; Sekido, Y.; Takahashi, T. A Polycistronic MicroRNA Cluster, miR-17-92, Is Overexpressed in Human Lung Cancers and Enhances Cell Proliferation. Cancer Res. 2005, 65, 9628–9632. [Google Scholar] [CrossRef]
- Mendell, J.T. miRiad Roles for the miR-17-92 Cluster in Development and Disease. Cell 2008, 133, 217–222. [Google Scholar] [CrossRef]
- Diosdado, B.; Van De Wiel, M.A.; Terhaar Sive Droste, J.S.; Mongera, S.; Postma, C.; Meijerink, W.J.H.J.; Carvalho, B.; Meijer, G.A. MiR-17-92 cluster is associated with 13q gain and c-myc expression during colorectal adenoma to adenocarcinoma progression. Br. J. Cancer 2009, 101, 707–714. [Google Scholar] [CrossRef]
- Sun, R.; Liang, Y.; Yuan, F.; Nie, X.; Sun, H.; Wang, Y.; Yu, T.; Gao, L.; Zhang, L. Functional polymorphisms in the promoter region of miR-17-92 cluster are associated with a decreased risk of colorectal cancer. Oncotarget 2017, 8, 82531–82540. [Google Scholar] [CrossRef]
- Humphreys, K.J.; Cobiac, L.; Le Leu, R.K.; Van der Hoek, M.B.; Michael, M.Z. Histone deacetylase inhibition in colorectal cancer cells reveals competing roles for members of the oncogenic miR-17-92 cluster. Mol. Carcinog. 2013, 52, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Ventura, A.; Young, A.G.; Winslow, M.M.; Lintault, L.; Meissner, A.; Erkeland, S.J.; Newman, J.; Bronson, R.T.; Crowley, D.; Stone, J.R.; et al. Targeted Deletion Reveals Essential and Overlapping Functions of the miR-17~92 Family of miRNA Clusters. Cell 2008, 132, 875–886. [Google Scholar] [CrossRef] [PubMed]
- Lujambio, A.; Lowe, S.W. The microcosmos of cancer. Nature 2012, 482, 347–355. [Google Scholar] [CrossRef] [PubMed]
- de Pontual, L.; Yao, E.; Callier, P.; Faivre, L.; Drouin, V.; Cariou, S.; Van Haeringen, A.; Geneviève, D.; Goldenberg, A.; Oufadem, M.; et al. Germline deletion of the miR-17∼92 cluster causes skeletal and growth defects in humans. Nat. Genet. 2011, 43, 1026–1030. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Zhang, P.; Wang, F.; Zhang, H.; Yang, Y.; Shi, C.; Xia, Y.; Peng, J.; Liu, W.; Yang, Z.; et al. Elevated oncofoetal miR-17-5p expression regulates colorectal cancer progression by repressing its target gene P130. Nat. Commun. 2012, 3, 1291. [Google Scholar] [CrossRef]
- Ast, V.; Kordaß, T.; Oswald, M.; Kolte, A.; Eisel, D.; Osen, W.; Eichmüller, S.B.; Berndt, A.; König, R. MiR-192, miR-200c and miR-17 are fibroblast-mediated inhibitors of colorectal cancer invasion. Oncotarget 2018, 9, 35559–35580. [Google Scholar] [CrossRef]
- Humphreys, K.J.; McKinnon, R.A.; Michael, M.Z. miR-18a Inhibits CDC42 and Plays a Tumour Suppressor Role in Colorectal Cancer Cells. PLoS ONE 2014, 9, e112288. [Google Scholar] [CrossRef]
- Huang, G.; Wu, X.; Li, S.; Xu, X.; Zhu, H.; Chen, X. The long noncoding RNA CASC2 functions as a competing endogenous RNA by sponging miR-18a in colorectal cancer. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef]
- Olive, V.; Bennett, M.J.; Walker, J.C.; Ma, C.; Jiang, I.; Cordon-Cardo, C.; Li, Q.-J.; Lowe, S.W.; Hannon, G.J.; He, L. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009, 23, 2839–2849. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, X.; Zhang, J.; Xu, Y.; Shao, J.; Hu, Y.; Shu, P.; Cheng, H. Inhibition of miR-19a partially reversed the resistance of colorectal cancer to oxaliplatin via PTEN/PI3K/AKT pathway. Aging 2020, 12, 5640–5650. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, R.; Yang, F.; Cheng, R.; Chen, X.; Cui, S.; Gu, Y.; Sun, W.; You, C.; Liu, Z.; et al. miR-19a promotes colorectal cancer proliferation and migration by targeting TIA1. Mol. Cancer 2017, 16, 53. [Google Scholar] [CrossRef]
- Chen, M.; Lin, M.; Wang, X. Overexpression of miR-19a inhibits colorectal cancer angiogenesis by suppressing KRAS expression. Oncol. Rep. 2018, 39, 619–626. [Google Scholar] [CrossRef]
- Jia, Z.; Wang, K.; Zhang, A.; Wang, G.; Kang, C.; Han, L.; Pu, P. miR-19a and miR-19b Overexpression in Gliomas. Pathol. Oncol. Res. 2013, 19, 847–853. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Tang, Y.; Bei, Y.; Ding, S.; Che, L.; Yao, J.; Wang, H.; Lv, D.; Xiao, J. miR-19b attenuates H2O2-induced apoptosis in rat H9C2 cardiomyocytes via targeting PTEN. Oncotarget 2016, 7, 10870–10878. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Yin, S.; Hao, Y.; Yang, J.; Zhang, H.; Sun, C.; Ma, M.; Chang, Q.; Xi, J.J. MiR-19b promotes tumor growth and metastasis via targeting TP53. RNA 2014, 20, 765–772. [Google Scholar] [CrossRef]
- Zaporozhchenko, I.A.; Morozkin, E.S.; Skvortsova, T.E.; Ponomaryova, A.A.; Rykova, E.Y.; Cherdyntseva, N.V.; Polovnikov, E.S.; Pashkovskaya, O.A.; Pokushalov, E.A.; Vlassov, V.V.; et al. Plasma miR-19b and miR-183 as Potential Biomarkers of Lung Cancer. PLoS ONE 2016, 11, e0165261. [Google Scholar] [CrossRef]
- Wu, C.; Cao, Y.; He, Z.; He, J.; Hu, C.; Duan, H.; Jiang, J. Serum Levels of miR-19b and miR-146a as Prognostic Biomarkers for Non-Small Cell Lung Cancer. Tohoku J. Exp. Med. 2014, 232, 85–95. [Google Scholar] [CrossRef]
- Copier, C.U.; León, L.; Fernández, M.; Contador, D.; Calligaris, S.D. Circulating miR-19b and miR-181b are potential biomarkers for diabetic cardiomyopathy. Sci. Rep. 2017, 7, 13514. [Google Scholar] [CrossRef]
- Ke, T.-W.; Wei, P.-L.; Yeh, K.-T.; Chen, W.T.-L.; Cheng, Y.-W. MiR-92a Promotes Cell Metastasis of Colorectal Cancer Through PTEN-Mediated PI3K/AKT Pathway. Ann. Surg. Oncol. 2015, 22, 2649–2655. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.J.; Li, L.F.; Yang, G.D.; Xia, S.S.; Wang, R.; Leng, Z.W.; Liu, Z.L.; Tian, H.P.; He, Y.; Meng, C.Y.; et al. MiR-92a promotes stem cell-like properties by activating Wnt/β-catenin signaling in colorectal cancer. Oncotarget 2017, 8, 101760–101770. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.-D.; Zheng, W.-B.; Sun, K.; Xue, Q.; Yang, C.-Z.; Li, G.-X. MiR-92a promotes the invasion and migration of colorectal cancer by targeting RECK. Int. J. Clin. Exp. Pathol. 2019, 12, 1565–1577. [Google Scholar] [PubMed]
- Lv, H.; Zhang, Z.; Wang, Y.; Li, C.; Gong, W.; Wang, X. MicroRNA-92a promotes colorectal cancer cell growth and migration by inhibiting KLF4. Oncol. Res. 2016, 23, 283–290. [Google Scholar] [CrossRef]
- Ma, H.; Pan, J.-S.; Jin, L.-X.; Wu, J.; Ren, Y.-D.; Chen, P.; Xiao, C.; Han, J. MicroRNA-17~92 inhibits colorectal cancer progression by targeting angiogenesis. Cancer Lett. 2016, 376, 293–302. [Google Scholar] [CrossRef]
- Cheng, D.; Zhao, S.; Tang, H.; Zhang, D.; Sun, H.; Yu, F.; Jiang, W.; Yue, B.; Wang, J.; Zhang, M.; et al. MicroRNA-20a-5p promotes colorectal cancer invasion and metastasis by downregulating Smad4. Oncotarget 2016, 7, 45199–45213. [Google Scholar] [CrossRef]
- Sarshar, M.; Scribano, D.; Ambrosi, C.; Palamara, A.T.; Masotti, A. Fecal microRNAs as Innovative Biomarkers of Intestinal Diseases and Effective Players in Host-Microbiome Interactions. Cancers 2020, 12, 2174. [Google Scholar] [CrossRef]
- Zhou, T.; Zhang, G.; Liu, Z.; Xia, S.; Tian, H. Overexpression of miR-92a correlates with tumor metastasis and poor prognosis in patients with colorectal cancer. Int. J. Colorectal Dis. 2013, 28, 19–24. [Google Scholar] [CrossRef]
- Salvi, S.; Molinari, C.; Foca, F.; Teodorani, N.; Saragoni, L.; Puccetti, M.; Passardi, A.; Tamberi, S.; Avanzolini, A.; Lucci, E.; et al. miR-17-92a-1 cluster host gene (MIR17HG) evaluation and response to neoadjuvant chemoradiotherapy in rectal cancer. Onco. Targets. Ther. 2016, 9, 2735. [Google Scholar] [CrossRef][Green Version]
- Yu, X.; Li, Z.; Yu, J.; Chan, M.T.V.; Wu, W.K.K. MicroRNAs predict and modulate responses to chemotherapy in colorectal cancer. Cell Prolif. 2015, 48, 503–510. [Google Scholar] [CrossRef]
- Xu, J.; Meng, Q.; Li, X.; Yang, H.; Xu, J.; Gao, N.; Sun, H.; Wu, S.; Familiari, G.; Relucenti, M.; et al. Long Noncoding RNA MIR17HG Promotes Colorectal Cancer Progression via miR-17-5p. Cancer Res. 2019, 79, 4882–4895. [Google Scholar] [CrossRef] [PubMed]
- Kooshapur, H.; Choudhury, N.R.; Simon, B.; Mühlbauer, M.; Jussupow, A.; Fernandez, N.; Jones, A.N.; Dallmann, A.; Gabel, F.; Camilloni, C.; et al. Structural basis for terminal loop recognition and stimulation of pri-miRNA-18a processing by hnRNP A1. Nat. Commun. 2018, 9, 2479. [Google Scholar] [CrossRef] [PubMed]
- Chaulk, S.G.; Xu, Z.; Glover, M.J.N.; Fahlman, R.P. MicroRNA miR-92a-1 biogenesis and mRNA targeting is modulated by a tertiary contact within the miR-17~92 microRNA cluster. Nucleic Acids Res. 2014, 42, 5234–5244. [Google Scholar] [CrossRef]
- Chakraborty, S.; Mehtab, S.; Patwardhan, A.; Krishnan, Y. Pri-miR-17-92a transcript folds into a tertiary structure and autoregulates its processing. RNA 2012, 18, 1014–1028. [Google Scholar] [CrossRef] [PubMed]
- Ounzain, S.; Micheletti, R.; Arnan, C.; Plaisance, I.; Cecchi, D.; Schroen, B.; Reverter, F.; Alexanian, M.; Gonzales, C.; Ng, S.Y.; et al. CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis. J. Mol. Cell. Cardiol. 2015, 89, 98–112. [Google Scholar] [CrossRef] [PubMed]
- Elia, L.; Quintavalle, M.; Zhang, J.; Contu, R.; Cossu, L.; Latronico, M.V.G.; Peterson, K.L.; Indolfi, C.; Catalucci, D.; Chen, J.; et al. The knockout of miR-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice: Correlates with human disease. Cell Death Differ. 2009, 16, 1590–1598. [Google Scholar] [CrossRef]
- Chen, X.; Guo, X.; Zhang, H.; Xiang, Y.; Chen, J.; Yin, Y.; Cai, X.; Wang, K.; Wang, G.; Ba, Y.; et al. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene 2009, 28, 1385–1392. [Google Scholar] [CrossRef]
- Su, J.; Liang, H.; Yao, W.; Wang, N.; Zhang, S.; Yan, X.; Feng, H.; Pang, W.; Wang, Y.; Wang, X.; et al. MiR-143 and MiR-145 regulate IGF1R to suppress cell proliferation in colorectal cancer. PLoS ONE 2014, 9, e0114420. [Google Scholar] [CrossRef]
- Michael, M.Z.; O’ Connor, S.M.; van Holst Pellekaan, N.G.; Young, G.P.; James, R.J. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol. Cancer Res. 2003, 1, 882–891. [Google Scholar]
- Pagliuca, A.; Valvo, C.; Fabrizi, E.; di Martino, S.; Biffoni, M.; Runci, D.; Forte, S.; De Maria, R.; Ricci-Vitiani, L. Analysis of the combined action of miR-143 and miR-145 on oncogenic pathways in colorectal cancer cells reveals a coordinate program of gene repression. Oncogene 2013, 32, 4806–4813. [Google Scholar] [CrossRef]
- Xing, A.Y.; Wang, Y.W.; Su, Z.X.; Shi, D.B.; Wang, B.; Gao, P. Catenin-δ 1, negatively regulated by miR-145, promotes tumour aggressiveness in gastric cancer. J. Pathol. 2015, 236, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Du, J.; Mao, K.; Wang, X.; Ding, Y.; Wang, F. MicroRNA-143-3p suppresses tumorigenesis by targeting catenin-δ1 in colorectal cancer. Onco. Targets. Ther. 2019, 12, 3255–3265. [Google Scholar] [CrossRef] [PubMed]
- Chivukula, R.R.; Shi, G.; Acharya, A.; Mills, E.W.; Zeitels, L.R.; Anandam, J.L.; Abdelnaby, A.A.; Balch, G.C.; Mansour, J.C.; Yopp, A.C.; et al. An Essential Mesenchymal Function for miR-143/145 in Intestinal Epithelial Regeneration. Cell 2014, 157, 1104–1116. [Google Scholar] [CrossRef] [PubMed]
- Kent, O.A.; McCall, M.N.; Cornish, T.C.; Halushka, M.K. Lessons from miR-143/145: The importance of cell-type localization of miRNAs. Nucleic Acids Res. 2014, 42, 7528–7538. [Google Scholar] [CrossRef]
- Akao, Y.; Nakagawa, Y.; Naoe, T. MicroRNAs 143 and 145 are possible common onco-microRNAs in human cancers. Oncol. Rep. 2006, 16, 845–850. [Google Scholar] [CrossRef]
- Yin, Y.; Yan, Z.-P.; Lu, N.-N.; Xu, Q.; He, J.; Qian, X.; Yu, J.; Guan, X.; Jiang, B.-H.; Liu, L.-Z. Downregulation of miR-145 associated with cancer progression and VEGF transcriptional activation by targeting N-RAS and IRS1. Biochim. Biophys. Acta 2013, 1829, 239–247. [Google Scholar] [CrossRef]
- Feng, Y.; Zhu, J.; Ou, C.; Deng, Z.; Chen, M.; Huang, W.; Li, L. MicroRNA-145 inhibits tumour growth and metastasis in colorectal cancer by targeting fascin-1. Br. J. Cancer 2014, 110, 2300–2309. [Google Scholar] [CrossRef]
- Huang, F.T.; Chen, W.Y.; Gu, Z.Q.; Zhuang, Y.Y.; Li, C.Q.; Wang, L.Y.; Peng, J.F.; Zhu, Z.; Luo, X.; Li, Y.H.; et al. The novel long intergenic noncoding RNA UCC promotes colorectal cancer progression by sponging MIR-143. Cell Death Dis. 2017, 8, e2778. [Google Scholar] [CrossRef]
- Luan, Y.; Li, X.; Luan, Y.; Zhao, R.; Li, Y.; Liu, L.; Hao, Y.; Oleg Vladimir, B.; Jia, L. Circulating lncRNA UCA1 Promotes Malignancy of Colorectal Cancer via the miR-143/MYO6 Axis. Mol. Ther. Nucleic Acids 2020, 19, 790–803. [Google Scholar] [CrossRef]
- Hu, Y.; Ma, Z.; He, Y.; Liu, W.; Su, Y.; Tang, Z. PART-1 functions as a competitive endogenous RNA for promoting tumor progression by sponging miR-143 in colorectal cancer. Biochem. Biophys. Res. Commun. 2017, 490, 317–323. [Google Scholar] [CrossRef]
- Xie, H.; Ren, X.; Xin, S.; Lan, X.; Lu, G.; Lin, Y.; Yang, S.; Zeng, Z.; Liao, W.; Ding, Y.-Q.; et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget 2016, 7, 26680–26691. [Google Scholar] [CrossRef] [PubMed]
- Slaby, O.; Svoboda, M.; Fabian, P.; Smerdova, T.; Knoflickova, D.; Bednarikova, M.; Nenutil, R.; Vyzula, R. Altered Expression of miR-21, miR-31, miR-143 and miR-145 Is Related to Clinicopathologic Features of Colorectal Cancer. Oncology 2007, 72, 397–402. [Google Scholar] [CrossRef] [PubMed]
- Akao, Y.; Nakagawa, Y.; Hirata, I.; Iio, A.; Itoh, T.; Kojima, K.; Nakashima, R.; Kitade, Y.; Naoe, T. Role of anti-oncomirs miR-143 and -145 in human colorectal tumors. Cancer Gene Ther. 2010, 17, 398–408. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, F.E.; Ahmed, N.C.; Vos, P.W.; Bonnerup, C.; Atkins, J.N.; Casey, M.; Nuovo, G.J.; Naziri, W.; Wiley, J.E.; Mota, H.; et al. Diagnostic microRNA markers to screen for sporadic human colon cancer in stool: I. Proof of principle. Cancer Genomics Proteomics 2013, 10, 93–113. [Google Scholar] [PubMed]
- Li, J.M.; Zhao, R.H.; Li, S.T.; Xie, C.X.; Jiang, H.H.; Ding, W.J.; Du, P.; Chen, W.; Yang, M.; Cui, L. Down-regulation of fecal miR-143 and miR-145 as potential markers for colorectal cancer. Saudi Med. J. 2012, 33, 24–29. [Google Scholar] [PubMed]
- Arndt, G.M.; Dossey, L.; Cullen, L.M.; Lai, A.; Druker, R.; Eisbacher, M.; Zhang, C.; Tran, N.; Fan, H.; Retzlaff, K.; et al. Characterization of global microRNA expression reveals oncogenic potential of miR-145 in metastatic colorectal cancer. BMC Cancer 2009, 9, 374. [Google Scholar] [CrossRef] [PubMed]
- Humphries, B.; Yang, C. The microRNA-200 family: Small molecules with novel roles in cancer development, progression and therapy. Oncotarget 2015, 6, 6472–6498. [Google Scholar] [CrossRef] [PubMed]
- Korpal, M.; Lee, E.S.; Hu, G.; Kang, Y. The miR-200 Family Inhibits Epithelial-Mesenchymal Transition and Cancer Cell Migration by Direct Targeting of E-cadherin Transcriptional Repressors ZEB1 and ZEB2. J. Biol. Chem. 2008, 283, 14910–14914. [Google Scholar] [CrossRef]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008, 10, 593–601. [Google Scholar] [CrossRef]
- Browne, G.; Sayan, A.E.; Tulchinsky, E. ZEB proteins link cell motility with cell cycle control and cell survival in cancer. Cell Cycle 2010, 9, 886–891. [Google Scholar] [CrossRef]
- Drápela, S.; Bouchal, J.; Jolly, M.K.; Culig, Z.; Souček, K. ZEB1: A Critical Regulator of Cell Plasticity, DNA Damage Response, and Therapy Resistance. Front. Mol. Biosci. 2020, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Bracken, C.P.; Gregory, P.A.; Kolesnikoff, N.; Bert, A.G.; Wang, J.; Shannon, M.F.; Goodall, G.J. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008, 68, 7846–7854. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, S.J.; Carter, J.V.; Burton, J.F.; Oxford, B.G.; Schmidt, M.N.; Hallion, J.C.; Galandiuk, S. The role of the miR-200 family in epithelial-mesenchymal transition in colorectal cancer: A systematic review. Int. J. Cancer 2018, 142, 2501–2511. [Google Scholar] [CrossRef] [PubMed]
- Hur, K.; Toiyama, Y.; Takahashi, M.; Balaguer, F.; Nagasaka, T.; Koike, J.; Hemmi, H.; Koi, M.; Boland, C.R.; Goel, A. MicroRNA-200c modulates epithelial-tomesenchymal transition (EMT) in human colorectal cancer metastasis. Gut 2013, 62, 1315–1326. [Google Scholar] [CrossRef]
- Carter, J.V.; O’Brien, S.J.; Burton, J.F.; Oxford, B.G.; Stephen, V.; Hallion, J.; Bishop, C.; Galbraith, N.J.; Eichenberger, M.R.; Sarojini, H.; et al. The microRNA-200 family acts as an oncogene in colorectal cancer by inhibiting the tumor suppressor RASSF2. Oncol. Lett. 2019, 18, 3994–4007. [Google Scholar] [CrossRef]
- Pecot, C.V.; Rupaimoole, R.; Yang, D.; Akbani, R.; Ivan, C.; Lu, C.; Wu, S.; Han, H.D.; Shah, M.Y.; Rodriguez-Aguayo, C.; et al. Tumour angiogenesis regulation by the miR-200 family. Nat. Commun. 2013, 4, 1–14. [Google Scholar] [CrossRef]
- Schliekelman, M.J.; Gibbons, D.L.; Faca, V.M.; Creighton, C.J.; Rizvi, Z.H.; Zhang, Q.; Wong, C.H.; Wang, H.; Ungewiss, C.; Ahn, Y.H.; et al. Targets of the tumor suppressor miR-200 in regulation of the epithelial-mesenchymal transition in cancer. Cancer Res. 2011, 71, 7670–7682. [Google Scholar] [CrossRef]
- Yu, C.; Wan, H.; Shan, R.; Wen, W.; Li, J.; Luo, D.; Wan, R. The prognostic value of the MiR-200 family in colorectal cancer: A meta-analysis with 1882 patients. J. Cancer 2019, 10, 4009–4016. [Google Scholar] [CrossRef]
- Sun, Y.; Shen, S.; Liu, X.; Tang, H.; Wang, Z.; Yu, Z.; Li, X.; Wu, M. MiR-429 inhibits cells growth and invasion and regulates EMT-related marker genes by targeting Onecut2 in colorectal carcinoma. Mol. Cell. Biochem. 2014, 390, 19–30. [Google Scholar] [CrossRef]
- Ding, L.; Yu, L.L.; Han, N.; Zhang, B.T. miR-141 promotes colon cancer cell proliferation by inhibiting MAP2K4. Oncol. Lett. 2017, 13, 1665–1671. [Google Scholar] [CrossRef]
- Hu, M.; Xia, M.; Chen, X.; Lin, Z.; Xu, Y.; Ma, Y.; Su, L. MicroRNA-141 regulates smad interacting protein 1 (SIP1) and inhibits migration and invasion of colorectal cancer cells. Dig. Dis. Sci. 2010, 55, 2365–2372. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.P.; Zhu, H.Y.; Sun, X.F.; Chen, L.X.; Zhou, Q.; Chen, J. MicroRNA-141 regulates the tumour suppressor DLC1 in colorectal cancer. Neoplasma 2013, 62, 705–712. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Cao, C.; Li, X.; Zhang, M.; Gu, Q.; Gao, H.; Balic, J.J.; Xu, D.; Zhang, L.; Ying, L.; et al. Complete loss of miR-200 family induces EMT associated cellular senescence in gastric cancer. Oncogene 2021, 41, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Maierthaler, M.; Benner, A.; Hoffmeister, M.; Surowy, H.; Jansen, L.; Knebel, P.; Chang-Claude, J.; Brenner, H.; Burwinkel, B. Plasma miR-122 and miR-200 family are prognostic markers in colorectal cancer. Int. J. Cancer 2017, 140, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Pichler, M.; Ress, A.L.; Winter, E.; Stiegelbauer, V.; Karbiener, M.; Schwarzenbacher, D.; Scheideler, M.; Ivan, C.; Jahn, S.W.; Kiesslich, T.; et al. MiR-200a regulates epithelial to mesenchymal transition-related gene expression and determines prognosis in colorectal cancer patients. Br. J. Cancer 2014, 110, 1614–1621. [Google Scholar] [CrossRef]
- Cheng, H.; Zhang, L.; Cogdell, D.E.; Zheng, H.; Schetter, A.J.; Nykter, M.; Harris, C.C.; Chen, K.; Hamilton, S.R.; Zhang, W. Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS ONE 2011, 6, e17745. [Google Scholar] [CrossRef]
- Liang, W.C.; Fu, W.M.; Wong, C.W.; Wang, Y.; Wang, W.M.; Hu, G.X.; Zhang, L.; Xiao, L.J.; Wan, D.C.C.; Zhang, J.F.; et al. The LncRNA H19 promotes epithelial to mesenchymal transition by functioning as MiRNA sponges in colorectal cancer. Oncotarget 2015, 6, 22513–22525. [Google Scholar] [CrossRef]
- O’Brien, S.J.; Fiechter, C.; Burton, J.; Hallion, J.; Paas, M.; Patel, A.; Patel, A.; Rochet, A.; Scheurlen, K.; Gardner, S.; et al. Long non-coding RNA ZFAS1 is a major regulator of epithelial-mesenchymal transition through miR-200/ZEB1/E-cadherin, vimentin signaling in colon adenocarcinoma. Cell Death Discov. 2021, 7, 1–14. [Google Scholar] [CrossRef]
- Wu, G.; Xue, M.; Zhao, Y.; Han, Y.; Li, C.; Zhang, S.; Zhang, J.; Xu, J. Long noncoding rna zeb1-as1 acts as a sponge of mir-141-3p to inhibit cell proliferation in colorectal cancer. Int. J. Med. Sci. 2020, 17, 1589–1597. [Google Scholar] [CrossRef]
- Cheng, Z.; Li, Z.; Ma, K.; Li, X.; Tian, N.; Duan, J.; Xiao, X.; Wang, Y. Long non-coding RNA XIST promotes glioma tumorigenicity and angiogenesis by acting as a molecular sponge of miR-429. J. Cancer 2017, 8, 4106–4116. [Google Scholar] [CrossRef]
- Schetter, A.J.; Leung, S.Y.; Sohn, J.J.; Zanetti, K.A.; Bowman, E.D.; Yanaihara, N.; Yuen, S.T.; Chan, T.L.; Kwong, D.L.W.; Au, G.K.H.; et al. MicroRNA Expression Profiles Associated With Prognosis and Therapeutic Outcome in Colon Adenocarcinoma. J. Am. Med. Assoc. 2008, 299, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Zhang, J.; Xu, X.; Qian, F.; Feng, K.; Ma, J. miR-203 is a predictive biomarker for colorectal cancer and its expression is associated with BIRC5. Tumor Biol. 2016, 37, 15989–15995. [Google Scholar] [CrossRef] [PubMed]
- Chiang, Y.; Song, Y.; Wang, Z.; Chen, Y.; Yue, Z.; Xu, H.; Xing, C.; Liu, Z. Aberrant Expression of miR-203 and Its Clinical Significance in Gastric and Colorectal Cancers. J. Gastrointest. Surg. 2011, 15, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Yi, R.; Poy, M.N.; Stoffel, M.; Fuchs, E. A skin microRNA promotes differentiation by repressing “stemness”. Nature 2008, 452, 225–229. [Google Scholar] [CrossRef]
- DeCastro, A.J.; Dunphy, K.A.; Hutchinson, J.; Balboni, A.L.; Cherukuri, P.; Jerry, D.J.; DiRenzo, J. MiR203 mediates subversion of stem cell properties during mammary epithelial differentiation via repression of δnP63α and promotes mesenchymal-to-epithelial transition. Cell Death Dis. 2013, 4, 1–10. [Google Scholar] [CrossRef]
- Patturajan, M.; Nomoto, S.; Sommer, M.; Fomenkov, A.; Hibi, K.; Zangen, R.; Poliak, N.; Califano, J.; Trink, B.; Ratovitski, E.; et al. ΔNp63 induces β-catenin nuclear accumulation and signaling. Cancer Cell 2002, 1, 369–379. [Google Scholar] [CrossRef]
- Riemondy, K.; Wang, X.; Torchia, E.C.; Roop, D.R.; Yi, R. MicroRNA-203 represses selection and expansion of oncogenic Hras transformed tumor initiating cells. Elife 2015, 4, e07004. [Google Scholar] [CrossRef]
- Lai, H.T.; Tseng, W.K.; Huang, S.W.; Chao, T.C.; Su, Y. MicroRNA-203 diminishes the stemness of human colon cancer cells by suppressing GATA6 expression. J. Cell. Physiol. 2020, 235, 2866–2880. [Google Scholar] [CrossRef]
- Ju, S.Y.; Chiou, S.H.; Su, Y. Maintenance of the stemness in CD44+ HCT-15 and HCT-116 human colon cancer cells requires miR-203 suppression. Stem Cell Res. 2014, 12, 86–100. [Google Scholar] [CrossRef]
- Li, P.; Zhou, H.; Zhu, X.; Ma, G.; Liu, C.; Lin, B.; Mao, W. High expression of NEDD9 predicts adverse outcomes of colorectal cancer patients. Int. J. Clin. Exp. Pathol. 2014, 7, 2565–2570. [Google Scholar]
- Han, R.; Sun, Q.; Wu, J.; Zheng, P.; Zhao, G. Sodium Butyrate Upregulates miR-203 Expression to Exert Anti-Proliferation Effect on Colorectal Cancer Cells. Cell. Physiol. Biochem. 2016, 39, 1919–1929. [Google Scholar] [CrossRef]
- Li, J.; Chen, Y.; Zhao, J.; Kong, F.; Zhang, Y. MiR-203 reverses chemoresistance in p53-mutated colon cancer cells through downregulation of Akt2 expression. Cancer Lett. 2011, 304, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Gao, S.; Chen, X.; Liu, M.; Mao, C.; Fang, X. Overexpression of miR-203 sensitizes paclitaxel (Taxol)-resistant colorectal cancer cells through targeting the salt-inducible kinase 2 (SIK2). Tumor Biol. 2016, 37, 12231–12239. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Xiao, Y.; Chen, C.; Wei, X.; Hu, C.; Ling, X.; Liu, X. MicroRNA-203-mediated posttranscriptional deregulation of CPEB4 contributes to colorectal cancer progression. Biochem. Biophys. Res. Commun. 2015, 466, 206–213. [Google Scholar] [CrossRef]
- Li, T.; Gao, F.; Zhang, X.P. MiR-203 enhances chemosensitivity to 5-fluorouracil by targeting thymidylate synthase in colorectal cancer. Oncol. Rep. 2015, 33, 607–614. [Google Scholar] [CrossRef]
- Shen, B.; Yuan, Y.; Zhang, Y.; Yu, S.; Peng, W.; Huang, X.; Feng, J. Long non-coding RNA FBXL19-AS1 plays oncogenic role in colorectal cancer by sponging miR-203. Biochem. Biophys. Res. Commun. 2017, 488, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Yang, D.; Liu, Y.; Wang, Y.; Lin, T.; Li, Y.; Yang, S.; Zhang, W.; Zhang, R. LncRNA BANCR promotes tumorigenesis and enhances adriamycin resistance in colorectal cancer. Aging 2018, 10, 2062–2078. [Google Scholar] [CrossRef]
- Zhao, L.; Liu, C.; Yan, S.; Hu, G.; Xiang, K.; Xiang, H.; Yu, H. LINC00657 promotes colorectal cancer stem-like cell invasion by functioning as a miR-203a sponge. Biochem. Biophys. Res. Commun. 2020, 529, 500–506. [Google Scholar] [CrossRef]
- Ye, H.; Hao, H.; Wang, J.; Chen, R.; Huang, Z. miR-203 as a novel biomarker for the diagnosis and prognosis of colorectal cancer: A systematic review and meta-analysis. Onco. Targets. Ther. 2017, 10, 3685–3696. [Google Scholar] [CrossRef]
- Valeri, N.; Braconi, C.; Gasparini, P.; Murgia, C.; Lampis, A.; Paulus-Hock, V.; Hart, J.R.; Ueno, L.; Grivennikov, S.I.; Lovat, F.; et al. MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell 2014, 25, 469–483. [Google Scholar] [CrossRef]
- Nagel, R.; Le Sage, C.; Diosdado, B.; Van Der Waal, M.; Oude Vrielink, J.A.F.; Bolijn, A.; Meijer, G.A.; Agami, R. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res. 2008, 68, 5795–5802. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, X.; Nie, F.; Liu, T.; Yu, X.; Wang, H.; Li, Q.; Peng, R.; Mao, Z.; Zhou, Q.; et al. MiR-135 family members mediate podocyte injury through the activation of Wnt/β-catenin signaling. Int. J. Mol. Med. 2015, 36, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Ishak Gabra, M.B.; Hanse, E.A.; Lowman, X.H.; Tran, T.Q.; Li, H.; Milman, N.; Liu, J.; Reid, M.A.; Locasale, J.W.; et al. MiR-135 suppresses glycolysis and promotes pancreatic cancer cell adaptation to metabolic stress by targeting phosphofructokinase-1. Nat. Commun. 2019, 10, 1–15. [Google Scholar] [CrossRef]
- Zhou, W.; Li, X.; Liu, F.; Xiao, Z.; He, M.; Shen, S.; Liu, S. MiR-135a promotes growth and invasion of colorectal cancer via metastasis suppressor 1 in vitro. Acta Biochim. Biophys. Sin. 2012, 44, 838–846. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liang, H.; Bai, M.; Ning, T.; Wang, C.; Fan, Q.; Wang, Y.; Fu, Z.; Wang, N.; Liu, R.; et al. miR-135b promotes cancer progression by targeting transforming growth factor beta receptor II (TGFBR2) in colorectal cancer. PLoS ONE 2015, 10, e130194. [Google Scholar] [CrossRef]
- Parsons, R.; Myeroff, L.L.; Liu, B.; Willson, J.K.; Markowitz, S.D.; Kinzler, K.W.; Vogelstein, B. Microsatellite instability and mutations of the transforming growth factor beta type II receptor gene in colorectal cancer. Cancer Res. 1995, 55, 5548–5550. [Google Scholar] [PubMed]
- Zhang, Y.; Zhang, Z.; Yi, Y.; Wang, Y.; Fu, J. CircNOL10 acts as a sponge of miR-135a/b-5p in suppressing colorectal cancer progression via regulating KLF9. Onco. Targets. Ther. 2020, 13, 5165–5176. [Google Scholar] [CrossRef]
- Xu, X.M.; Qian, J.C.; Deng, Z.L.; Cai, Z.; Tang, T.; Wang, P.; Zhang, K.H.; Cai, J.P. Expression of miR-21, miR-31, miR-96 and miR-135b is correlated with the clinical parameters of colorectal cancer. Oncol. Lett. 2012, 4, 339–345. [Google Scholar] [CrossRef]
- Vickers, M.M.; Bar, J.; Gorn-Hondermann, I.; Yarom, N.; Daneshmand, M.; Hanson, J.E.L.; Addison, C.L.; Asmis, T.R.; Jonker, D.J.; Maroun, J.; et al. Stage-dependent differential expression of microRNAs in colorectal cancer: Potential role as markers of metastatic disease. Clin. Exp. Metastasis 2012, 29, 123–132. [Google Scholar] [CrossRef]
- Jia, L.; Luo, S.; Ren, X.; Li, Y.; Hu, J.; Liu, B.; Zhao, L.; Shan, Y.; Zhou, H. miR-182 and miR-135b Mediate the Tumorigenesis and Invasiveness of Colorectal Cancer Cells via Targeting ST6GALNAC2 and PI3K/AKT Pathway. Dig. Dis. Sci. 2017, 62, 3447–3459. [Google Scholar] [CrossRef]
- Mencía, A.; Modamio-Høybjør, S.; Redshaw, N.; Morín, M.; Mayo-Merino, F.; Olavarrieta, L.; Aguirre, L.A.; Del Castillo, I.; Steel, K.P.; Dalmay, T.; et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat. Genet. 2009, 41, 609–613. [Google Scholar] [CrossRef]
- Pierce, M.L.; Weston, M.D.; Fritzsch, B.; Gabel, H.W.; Ruvkun, G.; Soukup, G.A. MicroRNA-183 family conservation and ciliated neurosensory organ expression. Evol. Dev. 2008, 10, 106–113. [Google Scholar] [CrossRef]
- Guttilla, I.K.; White, B.A. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J. Biol. Chem. 2009, 284, 23204–23216. [Google Scholar] [CrossRef]
- Gao, F.; Wang, W. MicroRNA-96 promotes the proliferation of colorectal cancer cells and targets tumor protein p53 inducible nuclear protein 1, forkhead box protein O1 (FOXO1) and FOXO3a. Mol. Med. Rep. 2015, 11, 1200–1206. [Google Scholar] [CrossRef]
- Suzuki, R.; Amatya, V.J.; Kushitani, K.; Kai, Y.; Kambara, T.; Takeshima, Y. miR-182 and miR-183 promote cell proliferation and invasion by targeting FOXO1 in mesothelioma. Front. Oncol. 2018, 8, 1–9. [Google Scholar] [CrossRef]
- Huangfu, L.; Liang, H.; Wang, G.; Su, X.; Li, L.; Du, Z.; Hu, M.; Dong, Y.; Bai, X.; Liu, T.; et al. miR-183 regulates autophagy and apoptosis in colorectal cancer through targeting of UVRAG. Oncotarget 2016, 7, 4735–4745. [Google Scholar] [CrossRef]
- Xiang, L.; Chen, X.J.; Wu, K.C.; Zhang, C.J.; Zhou, G.H.; Lv, J.N.; Sun, L.F.; Cheng, F.F.; Cai, X.B.; Jin, Z.B. MiR-183/96 plays a pivotal regulatory role in mouse photoreceptor maturation and maintenance. Proc. Natl. Acad. Sci. USA 2017, 114, 6376–6381. [Google Scholar] [CrossRef]
- He, P.Y.; Yip, W.K.; Jabar, M.F.; Mohtarrudin, N.; Dusa, N.M.; Seow, H.F. Effect of the miR-96-5p inhibitor and mimic on the migration and invasion of the SW480-7 colorectal cancer cell line. Oncol. Lett. 2019, 18, 1949–1960. [Google Scholar] [CrossRef]
- Li, P.; Sheng, C.; Huang, L.; Zhang, H.; Huang, L.; Cheng, Z.; Zhu, Q. MiR-183/-96/-182 cluster is up-regulated in most breast cancers and increases cell proliferation and migration. Breast Cancer Res. 2014, 16, 1–17. [Google Scholar] [CrossRef]
- Iseki, Y.; Shibutani, M.; Maeda, K.; Nagahara, H.; Fukuoka, T.; Matsutani, S.; Hirakawa, K.; Ohira, M. MicroRNA-96 promotes tumor invasion in colorectal cancer via RECK. Anticancer Res. 2018, 38, 2031–2035. [Google Scholar] [CrossRef]
- Bi, D.P.; Yin, C.H.; Zhang, X.Y.; Yang, N.N.; Xu, J.Y. MIR-183 functions as an oncogene by targeting ABCA1 in colon cancer. Oncol. Rep. 2016, 35, 2873–2879. [Google Scholar] [CrossRef]
- Ge, T.; Xiang, P.; Mao, H.; Tang, S.; Zhou, J.; Zhang, Y. Inhibition of miR-96 enhances the sensitivity of colorectal cancer cells to oxaliplatin by targeting TPM1. Exp. Ther. Med. 2020, 20, 2134–2140. [Google Scholar] [CrossRef]
- Zheng, S.; Zhong, Y.F.; Tan, D.M.; Xu, Y.; Chen, H.X.; Wang, D. miR-183-5p enhances the radioresistance of colorectal cancer by directly targeting ATG5. J. Biosci. 2019, 44, 1–11. [Google Scholar] [CrossRef]
- Rapti, S.M.; Kontos, C.K.; Papadopoulos, I.N.; Scorilas, A. High miR-96 levels in colorectal adenocarcinoma predict poor prognosis, particularly in patients without distant metastasis at the time of initial diagnosis. Tumor Biol. 2016, 37, 11815–11824. [Google Scholar] [CrossRef]
- Yue, C.; Chen, J.; Li, Z.; Li, L.; Chen, J.; Guo, Y. microRNA-96 promotes occurrence and progression of colorectal cancer via regulation of the AMPKα2-FTO-m6A/MYC axis. J. Exp. Clin. Cancer Res. 2020, 39, 1–15. [Google Scholar] [CrossRef]
- Zhang, Q.; Ren, W.; Huang, B.; Yi, L.; Zhu, H. MicroRNA-183/182/96 cooperatively regulates the proliferation of colon cancer cells. Mol. Med. Rep. 2015, 12, 668–674. [Google Scholar] [CrossRef]
- Zhou, T.; Zhang, G.J.; Zhou, H.; Xiao, H.X.; Li, Y. Overexpression of microRNA-183 in human colorectal cancer and its clinical significance. Eur. J. Gastroenterol. Hepatol. 2014, 26, 229–233. [Google Scholar] [CrossRef]
- Ress, A.L.; Stiegelbauer, V.; Winter, E.; Schwarzenbacher, D.; Kiesslich, T.; Lax, S.; Jahn, S.; Deutsch, A.; Bauernhofer, T.; Ling, H.; et al. MiR-96-5p influences cellular growth and is associated with poor survival in colorectal cancer patients. Mol. Carcinog. 2015, 54, 1442–1450. [Google Scholar] [CrossRef]
- Ning, S.; Liu, H.; Gao, B.; Wei, W.; Yang, A.; Li, J.; Zhang, L. MiR-155, miR-96 and miR-99a as potential diagnostic and prognostic tools for the clinical management of hepatocellular carcinoma. Oncol. Lett. 2019, 18, 3381–3387. [Google Scholar] [CrossRef]
- Wu, H.; Zhou, J.; Mei, S.; Wu, D.; Mu, Z.; Chen, B.; Xie, Y.; Ye, Y.; Liu, J. Circulating exosomal microRNA-96 promotes cell proliferation, migration and drug resistance by targeting LMO7. J. Cell. Mol. Med. 2017, 21, 1228–1236. [Google Scholar] [CrossRef]
- Shao, S.; Wang, C.; Wang, S.; Zhang, H.; Zhang, Y. LncRNA STXBP5-AS1 suppressed cervical cancer progression via targeting miR-96-5p/PTEN axis. Biomed. Pharmacother. 2019, 117, 109082. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, H.; Du, Y.; Liu, P.; Zhang, J.; Li, Y.; Shen, H.; Xing, L.; Xue, X.; Chen, J.; et al. Long noncoding RNA TP53TG1 promotes pancreatic ductal adenocarcinoma development by acting as a molecular sponge of microRNA-96. Cancer Sci. 2019, 110, 2760–2772. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.; Cheng, X.; Liu, X.; Xia, C.; Zhang, H.; Pan, D.; Zhang, X.; Li, Y. Circ_0026344 restrains metastasis of human colorectal cancer cells via miR-183. Artif. Cells Nanomed. Biotechnol. 2019, 47, 4038–4045. [Google Scholar] [CrossRef] [PubMed]
- Monticelli, S.; Ansel, K.M.; Xiao, C.; Socci, N.D.; Krichevsky, A.M.; Thai, T.H.; Rajewsky, N.; Marks, D.S.; Sander, C.; Rajewsky, K.; et al. MicroRNA profiling of the murine hematopoietic system. Genome Biol. 2005, 6, 1–15. [Google Scholar] [CrossRef]
- Xiao, C.; Calado, D.P.; Galler, G.; Thai, T.H.; Patterson, H.C.; Wang, J.; Rajewsky, N.; Bender, T.P.; Rajewsky, K. MiR-150 Controls B Cell Differentiation by Targeting the Transcription Factor c-Myb. Cell 2007, 131, 146–159. [Google Scholar] [CrossRef]
- Feng, J.; Yang, Y.; Zhang, P.; Wang, F.; Ma, Y.; Qin, H.; Wang, Y. miR-150 functions as a tumour suppressor in human colorectal cancer by targeting c-Myb. J. Cell. Mol. Med. 2014, 18, 2125–2134. [Google Scholar] [CrossRef]
- Ying, W.; Tseng, A.; Chang, R.C.A.; Wang, H.; Lin, Y.L.; Kanameni, S.; Brehm, T.; Morin, A.; Jones, B.; Splawn, T.; et al. MiR-150 regulates obesity-Associated insulin resistance by controlling B cell functions. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef]
- He, Z.; Dang, J.; Song, A.; Cui, X.; Ma, Z.; Zhang, Y. The involvement of miR-150/β-catenin axis in colorectal cancer progression. Biomed. Pharmacother. 2020, 121, 109495. [Google Scholar] [CrossRef]
- Chen, X.; Xu, X.; Pan, B.; Zeng, K.; Xu, M.; Liu, X.; He, B.; Pan, Y.; Sun, H.; Wang, S. miR-150-5p suppresses tumor progression by targeting VEGFA in colorectal cancer. Aging 2018, 10, 3421–3437. [Google Scholar] [CrossRef]
- Wang, W.H.; Chen, J.; Zhao, F.; Zhang, B.R.; Yu, H.S.; Jin, H.Y.; Dai, J.H. MiR-150-5p suppresses colorectal cancer cell migration and invasion through targeting MUC4. Asian Pacific J. Cancer Prev. 2014, 15, 6269–6273. [Google Scholar] [CrossRef]
- Zhou, L.; Qi, X.; Potashkin, J.A.; Abdul-Karim, F.W.; Gorodeski, G.I. MicroRNAs miR-186 and miR-150 down-regulate expression of the pro-apoptotic purinergic P2X7 receptor by activation of instability sites at the 3′-untranslated region of the gene that decrease steady-state levels of the transcript. J. Biol. Chem. 2008, 283, 28274–28286. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Jin, H.; Yang, Z.; Luo, G.; Lu, Y.; Li, K.; Ren, G.; Su, T.; Pan, Y.; Feng, B.; et al. MiR-150 promotes gastric cancer proliferation by negatively regulating the pro-apoptotic gene EGR2. Biochem. Biophys. Res. Commun. 2010, 392, 340–345. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Di Wang, X. miR-150-5p represses TP53 tumor suppressor gene to promote proliferation of colon adenocarcinoma. Sci. Rep. 2019, 9, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Slattery, M.L.; Herrick, J.S.; Mullany, L.E.; Samowitz, W.S.; Sevens, J.R.; Sakoda, L.; Wolff, R.K. The co-regulatory networks of tumor suppressor genes, oncogenes, and miRNAs in colorectal cancer. Genes Chromosom. Cancer 2017, 56, 769–787. [Google Scholar] [CrossRef] [PubMed]
- Yanlei, M.; Zhang, P.; Wang, F.; Zhang, H.; Yang, J.; Peng, J.; Liu, W.; Qin, H. miR-150 as a potential biomarker associated with prognosis and therapeutic outcome in colorectal cancer. Gut 2012, 61, 1447–1453. [Google Scholar] [CrossRef]
- Chen, X.; Zeng, K.; Xu, M.; Hu, X.; Liu, X.; Xu, T.; He, B.; Pan, Y.; Sun, H.; Wang, S. SP1-induced lncRNA-ZFAS1 contributes to colorectal cancer progression via the miR-150-5p/VEGFA axis. Cell Death Dis. 2018, 9, 1–18. [Google Scholar] [CrossRef]
- Zhou, T.; Wu, L.; Ma, N.; Tang, F.; Zong, Z.; Chen, S. LncRNA PART1 regulates colorectal cancer via targeting miR-150-5p/miR-520h/CTNNB1 and activating Wnt/β-catenin pathway. Int. J. Biochem. Cell Biol. 2020, 118, 105637. [Google Scholar] [CrossRef]
- Van Rooij, E.; Sutherland, L.B.; Liu, N.; Williams, A.H.; McAnally, J.; Gerard, R.D.; Richardson, J.A.; Olson, E.N. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc. Natl. Acad. Sci. USA 2006, 103, 18255–18260. [Google Scholar] [CrossRef]
- Porrello, E.R.; Johnson, B.A.; Aurora, A.B.; Simpson, E.; Nam, Y.J.; Matkovich, S.J.; Dorn, G.W.; Van Rooij, E.; Olson, E.N. MiR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circ. Res. 2011, 109, 670–679. [Google Scholar] [CrossRef]
- Murakami, Y.; Yasuda, T.; Saigo, K.; Urashima, T.; Toyoda, H.; Okanoue, T.; Shimotohno, K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene 2006, 25, 2537–2545. [Google Scholar] [CrossRef]
- Xu, T.; Zhu, Y.; Xiong, Y.; Ge, Y.Y.; Yun, J.P.; Zhuang, S.M. MicroRNA-195 suppresses tumorigenicity and regulates G1/S transition of human hepatocellular carcinoma cells. Hepatology 2009, 50, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Chao, Y.M.; Lin, H.F.; Chen, C.J.; Chen, C.S.; Yang, J.L.; Chan, J.Y.H.; Juo, S.H.H. miR-195 reduces age-related blood–brain barrier leakage caused by thrombospondin-1-mediated selective autophagy. Aging Cell 2020, 19, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Wang, S.; Wang, S. MiR-195 suppresses colon cancer proliferation and metastasis by targeting WNT3A. Mol. Genet. Genom. 2018, 293, 1245–1253. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.W.; Min, D.S. Positive feedback regulation between phospholipase D and Wnt signaling promotes Wnt-Driven anchorage-independent growth of colorectal cancer cells. PLoS ONE 2010, 5, e12109. [Google Scholar] [CrossRef]
- Lin, X.; Wang, S.; Sun, M.; Zhang, C.; Wei, C.; Yang, C.; Dou, R.; Liu, Q.; Xiong, B. MiR-195-5p/NOTCH2-mediated EMT modulates IL-4 secretion in colorectal cancer to affect M2-like TAM polarization. J. Hematol. Oncol. 2019, 12, 1–14. [Google Scholar] [CrossRef]
- Liu, L.; Chen, L.; Xu, Y.; Li, R.; Du, X. MicroRNA-195 promotes apoptosis and suppresses tumorigenicity of human colorectal cancer cells. Biochem. Biophys. Res. Commun. 2010, 400, 236–240. [Google Scholar] [CrossRef]
- Wang, X.; Wang, J.; Ma, H.; Zhang, J.; Zhou, X. Downregulation of miR-195 correlates with lymph node metastasis and poor prognosis in colorectal cancer. Med. Oncol. 2012, 29, 919–927. [Google Scholar] [CrossRef]
- Jin, Y.; Wang, M.; Hu, H.; Huang, Q.; Chen, Y.; Wang, G. Overcoming stemness and chemoresistance in colorectal cancer through miR-195-5p-modulated inhibition of notch signaling. Int. J. Biol. Macromol. 2018, 117, 445–453. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, J.; Jiang, T.; Liu, G.; Wang, D.; Lu, Y. MicroRNA-195 suppresses colorectal cancer cells proliferation via targeting FGF2 and regulating Wnt/β-catenin pathway. Am. J. Cancer Res. 2016, 6, 2631–2640. [Google Scholar]
- Wang, L.; Qian, L.; Li, X.; Yan, J. MicroRNA-195 inhibits colorectal cancer cell proliferation, colony-formation and invasion through targeting CARMA3. Mol. Med. Rep. 2014, 10, 473–478. [Google Scholar] [CrossRef]
- Luo, Q.; Zhang, Z.; Dai, Z.; Basnet, S.; Li, S.; Xu, B.; Ge, H. Tumor-suppressive microRNA-195-5p regulates cell growth and inhibits cell cycle by targeting cyclin dependent kinase 8 in colon cancer. Am. J. Transl. Res. 2016, 8, 2088–2096. [Google Scholar] [PubMed]
- Feng, C.; Zhang, L.; Sun, Y.; Li, X.; Zhan, L.; Lou, Y.; Wang, Y.; Liu, L.; Zhang, Y. GDPD5, a target of miR-195-5p, is associated with metastasis and chemoresistance in colorectal cancer. Biomed. Pharmacother. 2018, 101, 945–952. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Zhao, L.; Zhang, P.; Wang, J.; Xu, N.; Mi, W.; Jiang, X.; Zhang, C.; Qu, J. MicroRNA-195 chemosensitizes colon cancer cells to the chemotherapeutic drug doxorubicin by targeting the first binding site of BCL2L2 mRNA. J. Cell. Physiol. 2015, 230, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Chen, J.; Zhou, Z.; He, Z. miR-195 enhances the radiosensitivity of colorectal cancer cells by suppressing CARM1. Onco. Targets. Ther. 2017, 10, 1027–1038. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Xu, J.; Zhao, J.; Zhang, R. lncRNA SNHG1 cooperated with miR-497/miR-195-5p to modify epithelial–mesenchymal transition underlying colorectal cancer exacerbation. J. Cell. Physiol. 2020, 235, 1453–1468. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Lv, C.; Li, J.; Xie, T.; Liu, X.; Zheng, Z.; Qin, Z.; Hui, X.; Yu, Y. LncRNA LINC00473 promoted colorectal cancer cell proliferation and invasion by targeting miR-195 expression. Am. J. Transl. Res. 2021, 13, 6066–6075. [Google Scholar]
- Gu, H.; Xu, Z.; Zhang, J.; Wei, Y.; Cheng, L.; Wang, J. circ_0038718 promotes colon cancer cell malignant progression via the miR-195-5p/Axin2 signaling axis and also effect Wnt/β-catenin signal pathway. BMC Genom. 2021, 22, 1–17. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, Z.; Hou, X.; Sun, Y. LncRNA AFAP1-AS1 Promotes the Progression of Colorectal Cancer through miR-195-5p and WISP1. J. Oncol. 2021, 2021, 6242798. [Google Scholar] [CrossRef]
- Ebert, M.S.; Neilson, J.R.; Sharp, P.A. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 2007, 4, 721–726. [Google Scholar] [CrossRef]
- Dey, N.; Das, F.; Ghosh-Choudhury, N.; Mandal, C.C.; Parekh, D.J.; Block, K.; Kasinath, B.S.; Abboud, H.E.; Choudhury, G.G. microRNA-21 governs TORC1 activation in renal cancer cell proliferation and invasion. PLoS ONE 2012, 7, e37366. [Google Scholar] [CrossRef]
- Liu, X.; Abraham, J.M.; Cheng, Y.; Wang, Z.; Wang, Z.; Zhang, G.; Ashktorab, H.; Smoot, D.T.; Cole, R.N.; Boronina, T.N.; et al. Synthetic Circular RNA Functions as a miR-21 Sponge to Suppress Gastric Carcinoma Cell Proliferation. Mol. Ther. Nucleic Acids 2018, 13, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Monfared, H.; Jahangard, Y.; Nikkhah, M.; Mirnajafi-Zadeh, J.; Mowla, S.J. Potential therapeutic effects of exosomes packed with a miR-21-sponge construct in a rat model of glioblastoma. Front. Oncol. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Wang, Z.; Ma, K.; Cheng, Y.; Abraham, J.M.; Liu, X.; Ke, X.; Wang, Z.; Meltzer, S.J. Synthetic circular multi-miR sponge simultaneously inhibits miR-21 and miR-93 in esophageal carcinoma. Lab. Investig. 2019, 99, 1442–1453. [Google Scholar] [CrossRef] [PubMed]
- Zeng, K.; Chen, X.; Xu, M.; Liu, X.; Hu, X.; Xu, T.; Sun, H.; Pan, Y.; He, B.; Wang, S. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7 article. Cell Death Dis. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Fu, C.; Xu, Q.; Wei, X. Long non-coding RNA CASC7 inhibits the proliferation and migration of colon cancer cells via inhibiting microRNA-21. Biomed. Pharmacother. 2017, 95, 1644–1653. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Liu, W.; Zhang, Y.; Zhang, Y.; Sun, S. CircRNA circ_0026344 as a prognostic biomarker suppresses colorectal cancer progression via microRNA-21 and microRNA-31. Biochem. Biophys. Res. Commun. 2018, 503, 870–875. [Google Scholar] [CrossRef]
- He, J.H.; Li, Y.G.; Han, Z.P.; Zhou, J.B.; Chen, W.M.; Lv, Y.B.; He, M.L.; Zuo, J.D.; Zheng, L. The CircRNA-ACAP2/Hsa-miR-21-5p/Tiam1 Regulatory Feedback Circuit Affects the Proliferation, Migration, and Invasion of Colon Cancer SW480 Cells. Cell. Physiol. Biochem. 2018, 49, 1539–1550. [Google Scholar] [CrossRef]
- Jiang, Z.; Hou, Z.; Li, L.; Liu, W.; Yu, Z.; Chen, S. Exosomal circEPB41L2 serves as a sponge for miR-21-5p and miR-942-5p to suppress colorectal cancer progression by regulating the PTEN/AKT signalling pathway. Eur. J. Clin. Investig. 2021, 51, e13581. [Google Scholar] [CrossRef]
- Huang, H.; Yang, X.; Chen, J.; Fu, J.; Chen, C.; Wen, J.; Mo, Q. lncRNA DGCR5 inhibits the proliferation of colorectal cancer cells by downregulating miR-21. Oncol. Lett. 2019, 18, 3331–3336. [Google Scholar] [CrossRef]
- Li, G.; Wang, C.; Wang, Y.; Xu, B.; Zhang, W. LINC00312 represses proliferation and metastasis of colorectal cancer cells by regulation of miR-21. J. Cell. Mol. Med. 2018, 22, 5565–5572. [Google Scholar] [CrossRef]
- Cai, Y.; Li, Y.; Shi, C.; Zhang, Z.; Xu, J.; Sun, B. LncRNA OTUD6B-AS1 inhibits many cellular processes in colorectal cancer by sponging miR-21-5p and regulating PNRC2. Hum. Exp. Toxicol. 2021, 40, 1463–1473. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Wang, F.W.; Cao, C.H.; Ling, H.; Chen, J.W.; Chen, R.X.; Feng, Z.H.; Luo, J.; Jin, X.H.; Duan, J.L.; et al. CircLONP2 enhances colorectal carcinoma invasion and metastasis through modulating the maturation and exosomal dissemination of microRNA-17. Mol. Cancer 2020, 19, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Wen, R.; Chen, C.; Zhong, X.; Hu, C. PAX6 upstream antisense RNA (PAUPAR) inhibits colorectal cancer progression through modulation of the microRNA (miR)-17-5p / zinc finger protein 750 (ZNF750) axis. Bioengineered 2021, 12, 3886–3899. [Google Scholar] [CrossRef] [PubMed]
- Yin, S.L.; Xiao, F.; Liu, Y.F.; Chen, H.; Guo, G.C. Long non-coding RNA FENDRR restrains the aggressiveness of CRC via regulating miR-18a-5p/ING4 axis. J. Cell. Biochem. 2020, 121, 3973–3985. [Google Scholar] [CrossRef]
- Horita, K.; Kurosaki, H.; Nakatake, M.; Ito, M.; Kono, H.; Nakamura, T. Long noncoding RNA UCA1 enhances sensitivity to oncolytic vaccinia virus by sponging miR-18a/miR-182 and modulating the Cdc42/filopodia axis in colorectal cancer. Biochem. Biophys. Res. Commun. 2019, 516, 831–838. [Google Scholar] [CrossRef]
- Li, Y.; Lv, S.; Ning, H.; Li, K.; Zhou, X.; Xv, H.; Wen, H. Down-regulation of CASC2 contributes to cisplatin resistance in gastric cancer by sponging miR-19a. Biomed. Pharmacother. 2018, 108, 1775–1782. [Google Scholar] [CrossRef]
- Shen, P.; Qu, L.; Wang, J.; Ding, Q.; Zhou, C.; Xie, R.; Wang, H.; Ji, G. LncRNA LINC00342 contributes to the growth and metastasis of colorectal cancer via targeting miR-19a-3p/NPEPL1 axis. Cancer Cell Int. 2021, 21, 1–11. [Google Scholar] [CrossRef]
- Dai, W.; Zeng, W.; Lee, D. lncRNA MCM3AP-AS1 inhibits the progression of colorectal cancer via the miR-19a-3p/FOXF2 axis. J. Gene Med. 2021, 23, 1–13. [Google Scholar] [CrossRef]
- Jiang, Z.; Li, L.; Hou, Z.; Liu, W.; Wang, H.; Zhou, T.; Li, Y.; Chen, S. LncRNA HAND2-AS1 inhibits 5-fluorouracil resistance by modulating miR-20a/PDCD4 axis in colorectal cancer. Cell. Signal. 2020, 66, 109483. [Google Scholar] [CrossRef]
- Zhang, G.; Liu, Z.; Zhong, J.; Lin, L. Circ-ACAP2 facilitates the progression of colorectal cancer through mediating miR-143-3p/FZD4 axis. Eur. J. Clin. Investig. 2021, 51, e13607. [Google Scholar] [CrossRef]
- Huang, F.; Wen, C.; Zhuansun, Y.; Huang, L.; Chen, W.; Yang, X.; Liu, H. A novel long noncoding RNA OECC promotes colorectal cancer development and is negatively regulated by miR-143-3p. Biochem. Biophys. Res. Commun. 2018, 503, 2949–2955. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Li, Y.; Song, A. Inhibition of lncRNA TMPO-AS1 suppresses proliferation, migration and invasion of colorectal cancer cells by targeting miR-143-3p. Mol. Med. Rep. 2020, 22, 3245–3254. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Liu, L. Long non-coding RNA ZEB2-AS1 promotes proliferation and inhibits apoptosis of colon cancer cells via miR-143/bcl-2 axis. Am. J. Transl. Res. 2019, 11, 5240–5248. [Google Scholar] [PubMed]
- Gao, R.; Fang, C.; Xu, J.; Tan, H.; Li, P.; Ma, L. LncRNA CACS15 contributes to oxaliplatin resistance in colorectal cancer by positively regulating ABCC1 through sponging miR-145. Arch. Biochem. Biophys. 2019, 663, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.L.; Li, X.N.; Ye, C.X.; Chen, H.Y.; Wang, Z.J. Elevated levels of circrunx1 in colorectal cancer promote cell growth and metastasis via MiR-145-5p/IGF1 signalling. Onco. Targets. Ther. 2020, 13, 4035–4048. [Google Scholar] [CrossRef] [PubMed]
- Bahreini, F.; Saidijam, M.; Mousivand, Z.; Najafi, R.; Afshar, S. Assessment of lncRNA DANCR, miR-145-5p and NRAS axis as biomarkers for the diagnosis of colorectal cancer. Mol. Biol. Rep. 2021, 48, 3541–3547. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.; Wang, M.; Li, Z.; Wang, Y.; Zhou, Y. Long Non-coding RNA MIR570MG Causes Regorafenib Resistance in Colon Cancer by Repressing miR-145/SMAD3 Signaling. Front. Oncol. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Wang, Z.; Su, M.; Xiang, B.; Zhao, K.; Qin, B. Circular RNA PVT1 promotes metastasis via miR-145 sponging in CRC. Biochem. Biophys. Res. Commun. 2019, 512, 716–722. [Google Scholar] [CrossRef]
- Tian, T.; Qiu, R.; Qiu, X. SNHG1 promotes cell proliferation by acting as a sponge of miR- 145 in colorectal cancer. Oncotarget 2018, 9, 2128–2139. [Google Scholar] [CrossRef]
- Wei, A.W.; Li, L.F. Long non-coding RNA SOX21-AS1 sponges miR-145 to promote the tumorigenesis of colorectal cancer by targeting MYO6. Biomed. Pharmacother. 2017, 96, 953–959. [Google Scholar] [CrossRef]
- Wang, X.; Bai, X.; Yan, Z.; Guo, X.; Zhang, Y. The lncRNA TUG1 promotes cell growth and migration in colorectal cancer via the TUG1–miR-145-5p–TRPC6 pathway. Biochem. Cell Biol. 2021, 99, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Tang, J.; Qian, Y.; Sun, T.; Chen, H.; Chen, Z.; Sun, D.; Zhong, M.; Chen, H.; Hong, J.; et al. Long noncoding RNA BFAL1 mediates enterotoxigenic Bacteroides fragilis-related carcinogenesis in colorectal cancer via the RHEB/mTOR pathway. Cell Death Dis. 2019, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lu, Y.; Chen, Y. Long non-coding RNA SNHG16 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer via sponging MIR-200a-3p. Biosci. Rep. 2019, 39, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Jiao, M.; Ning, S.; Chen, J.; Chen, L.; Jiao, M.; Cui, Z.; Guo, L.; Mu, W.; Yang, H. Long non-coding RNA ZEB1-AS1 predicts a poor prognosis and promotes cancer progression through the miR-200a/ZEB1 signaling pathway in intrahepatic cholangiocarcinoma. Int. J. Oncol. 2020, 56, 1455–1467. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.L.; Chen, L.Z.; Lu, Y.X.; Zhang, D.S.; Zeng, Z.L.; Pan, Z.Z.; Huang, P.; Wang, F.H.; Li, Y.H.; Ju, H.Q.; et al. Long noncoding rna xist expedites metastasis and modulates epithelial–mesenchymal transition in colorectal cancer. Cell Death Dis. 2017, 8, e3011. [Google Scholar] [CrossRef]
- Gao, Z.; Zhou, H.; Wang, Y.; Chen, J.; Ou, Y. Regulatory effects of lncRNA ATB targeting miR-200c on proliferation and apoptosis of colorectal cancer cells. J. Cell. Biochem. 2020, 121, 332–343. [Google Scholar] [CrossRef]
- Liu, X.; Wang, C. Long non-coding RNA ATB is associated with metastases and promotes cell invasion in colorectal cancer via sponging miR-141-3p. Exp. Ther. Med. 2020, 20, 1. [Google Scholar] [CrossRef]
- Ren, J.; Ding, L.; Zhang, D.; Shi, G.; Xu, Q.; Shen, S.; Wang, Y.; Wang, T.; Hou, Y. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics 2018, 8, 3932–3948. [Google Scholar] [CrossRef]
- Sun, Z.; Shao, B.; Liu, Z.; Dang, Q.; Guo, Y.; Chen, C.; Guo, Y.; Chen, Z.; Liu, J.; Hu, S.; et al. LINC01296/miR-141-3p/ZEB1-ZEB2 axis promotes tumor metastasis via enhancing epithelial-mesenchymal transition process. J. Cancer 2021, 12, 2723–2734. [Google Scholar] [CrossRef]
- Wang, H.; Li, H.; Zhang, L.; Yang, D. Overexpression of MEG3 sensitizes colorectal cancer cells to oxaliplatin through regulation of miR-141/PDCD4 axis. Biomed. Pharmacother. 2018, 106, 1607–1615. [Google Scholar] [CrossRef]
- Deng, Z.; Li, X.; Wang, H.; Geng, Y.; Cai, Y.; Tang, Y.; Wang, Y.; Yu, X.; Li, L.; Li, R. Dysregulation of CircRNA_0001946 Contributes to the Proliferation and Metastasis of Colorectal Cancer Cells by Targeting MicroRNA-135a-5p. Front. Genet. 2020, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Shi, M.; Meng, W.; Wang, Y.; Hui, P.; Ma, J. Long noncoding RNA FOXD3-AS1 promotes colon adenocarcinoma progression and functions as a competing endogenous RNA to regulate SIRT1 by sponging miR-135a-5p. J. Cell. Physiol. 2019, 234, 21889–21902. [Google Scholar] [CrossRef]
- Zhou, D.; He, S.; Zhang, D.; Lv, Z.; Yu, J.; Li, Q.; Li, M.; Guo, W.; Qi, F. LINC00857 promotes colorectal cancer progression by sponging miR-150-5p and upregulating HMGB3 (high mobility group box 3) expression. Bioengineered 2021, 12, 12107–12122. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Jiang, G.; Ren, W.; Wang, B.; Yang, C.; Li, M. LncRNA NEAT1 regulates 5-fu sensitivity, apoptosis and invasion in colorectal cancer through the MiR-150-5p/CPSF4 axis. Onco. Targets. Ther. 2020, 13, 6373–6383. [Google Scholar] [CrossRef] [PubMed]
- Shoshan, E.; Mobley, A.K.; Braeuer, R.R.; Kamiya, T.; Huang, L.; Vasquez, M.E.; Salameh, A.; Lee, H.J.; Kim, S.J.; Ivan, C.; et al. Hypo adenosine-to-inosine miR-455-5p editing promotes melanoma growth and metastasis. Nat. Cell Biol. 2015, 17, 311–321. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, S.; He, C.; Xue, P.; Zhang, L.; He, Z.; Zang, L.; Feng, B.; Sun, J.; Zheng, M. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol. Cancer 2020, 19, 1–16. [Google Scholar] [CrossRef]
- Huang, Q.R.; Pan, X. Bin Prognostic lncRNAs, miRNAs, and mRNAs Form a Competing Endogenous RNA Network in Colon Cancer. Front. Oncol. 2019, 9, 712. [Google Scholar] [CrossRef]
- Shuwen, H.; Qing, Z.; Yan, Z.; Xi, Y. Competitive endogenous RNA in colorectal cancer: A systematic review. Gene 2018, 645, 157–162. [Google Scholar] [CrossRef]
- Wang, L.; Cho, K.B.; Li, Y.; Tao, G.; Xie, Z.; Guo, B. Long noncoding RNA (LncRNA)-mediated competing endogenous RNA networks provide novel potential biomarkers and therapeutic targets for colorectal cancer. Int. J. Mol. Sci. 2019, 20, 5758. [Google Scholar] [CrossRef]
- Jung, G.; Hernández-Illán, E.; Moreira, L.; Balaguer, F.; Goel, A. Epigenetics of colorectal cancer: Biomarker and therapeutic potential. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 111–130. [Google Scholar] [CrossRef]
- Qi, X.; Lin, Y.; Liu, X.; Chen, J.; Shen, B. Biomarker Discovery for the Carcinogenic Heterogeneity Between Colon and Rectal Cancers Based on lncRNA-Associated ceRNA Network Analysis. Front. Oncol. 2020, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, H.; Zheng, B.; Sun, L.; Yuan, Y.; Xing, C. Competitive Endogenous RNA (ceRNA) Regulation Network of lncRNA–miRNA–mRNA in Colorectal Carcinogenesis. Dig. Dis. Sci. 2019, 64, 1868–1877. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Li, X.; Liu, L.; Wei, C.; Sun, D.; Peng, S.; Jiang, L. Comprehensive analysis of lncRNA-associated ceRNA network in colorectal cancer. Biochem. Biophys. Res. Commun. 2019, 508, 374–379. [Google Scholar] [CrossRef] [PubMed]
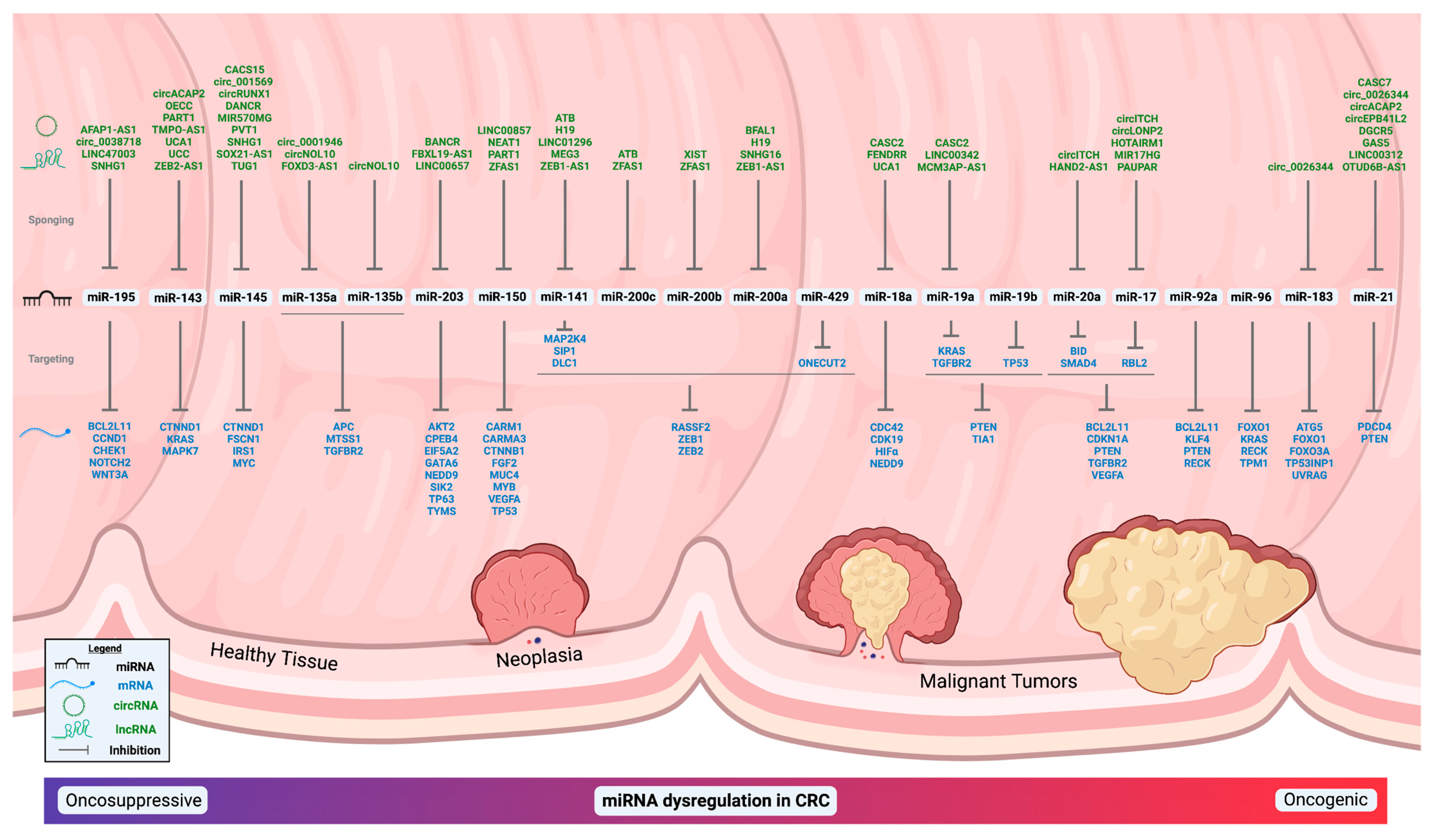
| miRNA | Biofluid Source | Levels in CRC | Reference |
|---|---|---|---|
| miR-21 | Stool | Upregulated | [54] |
| Stool | Upregulated | [55] | |
| Stool | Upregulated | [56] | |
| Serum | Upregulated | [57] | |
| Serum | Upregulated | [58] | |
| Serum | Upregulated | [59] | |
| Serum | Upregulated | [60] | |
| Serum | Upregulated | [61] | |
| Serum | Upregulated | [62] | |
| Serum | Upregulated | [63] | |
| Serum | Upregulated | [64] | |
| miR-17 | Serum | Upregulated | [65] |
| Serum | Upregulated | [66] | |
| Serum | Upregulated | [67] | |
| Serum | Upregulated | [62] | |
| miR-18a | Plasma | Downregulated | [68] |
| Serum | Upregulated | [69] | |
| Serum | Upregulated | [70] | |
| miR-19a | Serum | Upregulated | [65] |
| Serum | Upregulated | [71] | |
| Serum | Upregulated | [43] | |
| Serum | Upregulated | [72] | |
| Serum | Upregulated | [73] | |
| Serum | Upregulated | [74] | |
| Serum | Upregulated | [61] | |
| Serum | Upregulated | [75] | |
| Serum | Upregulated | [76] | |
| miR-20a | Serum | Downregulated | [77] |
| Serum | Upregulated | [78] | |
| Serum | Upregulated | [62] | |
| Serum | Upregulated | [65] | |
| Serum | Upregulated | [74] | |
| miR-19b | Serum | Upregulated | [43] |
| miR-92a | Plasma | Upregulated | [79] |
| Stool | Upregulated | [54] | |
| Serum | Upregulated | [75] | |
| Serum | Upregulated | [80] | |
| Serum | Upregulated | [63] | |
| Serum | Upregulated | [81] | |
| Serum | Upregulated | [82] | |
| Serum | Upregulated | [83] | |
| Serum | Upregulated | [84] | |
| miR-143 | Plasma | Downregulated | [85] |
| Serum | Downregulated | [74] | |
| Serum | Downregulated | [86] | |
| Serum | Upregulated | [87] | |
| Serum | No Significant Change | [84] | |
| miR-145 | Plasma | Downregulated | [85] |
| Serum | Downregulated | [88] | |
| Serum | Downregulated | [74] | |
| Serum | Downregulated | [86] | |
| Serum | No Significant Change | [84] | |
| Serum | Downregulated | [66] | |
| miR-203 | Plasma | Upregulated | [89] |
| Serum | Downregulated | [84] | |
| Serum | Upregulated | [90] | |
| Serum | Upregulated | [91] | |
| Serum | Downregulated | [92] | |
| Serum | Upregulated | [93] | |
| Serum | Upregulated | [94] | |
| Serum | Upregulated | [95] | |
| Serum | Upregulated | [96] | |
| miR-200a | None Reported | ||
| miR-200b | Plasma | Upregulated | [89] |
| miR-200c | Serum | Upregulated | [97] |
| Serum | Downregulated | [86] | |
| Serum | Upregulated | [98] | |
| Serum | Upregulated | [99] | |
| Serum | Upregulated | [100] | |
| Serum | Upregulated | [101] | |
| miR-141 | Plasma | Upregulated | [89] |
| Serum | Upregulated | [98] | |
| Serum | Upregulated | [99] | |
| Serum | Upregulated | [101] | |
| miR-429 | Serum | Upregulated | [102] |
| miR-135a | Stool | Upregulated | [103] |
| Stool | Upregulated | [104] | |
| Serum | Upregulated | [105] | |
| Serum | Downregulated | [106] | |
| miR-135b | Stool | Upregulated | [107] |
| Stool | Upregulated | [108] | |
| Serum | Upregulated | [109] | |
| miR-96 | Plasma | Upregulated | [89] |
| Serum | Upregulated | [110] | |
| Serum | No Significant Change | [69] | |
| miR-183 | Serum | Upregulated | [111] |
| Serum | Upregulated | [112] | |
| miR-150 | Serum | Downregulated | [113] |
| Serum | Upregulated | [74] | |
| Serum | Downregulated | [114] | |
| Serum | Downregulated | [115] | |
| Serum | Upregulated | [116] | |
| Serum | Downregulated | [64] | |
| miR-195 | Plasma | Downregulated | [117] |
| Serum | Downregulated | [118] |
| miRNA | ceRNA in CRC | Reference |
|---|---|---|
| miR-21 | CASC7 | [315] |
| circ_0026344 | [316] | |
| circACAP2 | [317] | |
| circEPB41L2 | [318] | |
| DGCR5 | [319] | |
| GAS5 | [36] | |
| LINC00312 | [320] | |
| OTUD6B-AS1 | [321] | |
| miR-17 | circITCH | [12] |
| circLONP2 | [322] | |
| HOTAIRM1 | [11] | |
| MIR17HG | [171] | |
| PAUPAR | [323] | |
| miR-18a | CASC2 | [150] |
| FENDRR | [324] | |
| UCA1 | [325] | |
| miR-19a | CASC2 | [326] |
| LINC00342 | [327] | |
| MCM3AP-AS1 | [328] | |
| miR-20a | circITCH | [12] |
| HAND2-AS1 | [329] | |
| miR-19b | None Reported | |
| miR-92a | None Reported | |
| miR-143 | circACAP2 | [330] |
| OECC | [331] | |
| PART1 | [190] | |
| TMPO-AS1 | [332] | |
| UCA1 | [189] | |
| UCC | [188] | |
| ZEB2-AS1 | [333] | |
| miR-145 | CACS15 | [334] |
| circ_001569 | [191] | |
| circRUNX1 | [335] | |
| DANCR | [336] | |
| MIR570MG | [337] | |
| PVT1 | [338] | |
| SNHG1 | [339] | |
| SOX2-AS1 | [340] | |
| TUG1 | [341] | |
| miR-203 | BANCR | [237] |
| FBXL19-AS1 | [236] | |
| LINC00657 | [238] | |
| miR-200a | BFAL1 | [342] |
| H19 | [217] | |
| SNHG16 | [343] | |
| ZEB1-AS1 | [344] | |
| miR-200b | XIST | [345] |
| ZFAS1 | [218] | |
| miR-200c | ATB | [346] |
| ZFAS1 | [218] | |
| miR-141 | ATB | [347] |
| H19 | [348] | |
| LINC01296 | [349] | |
| MEG3 | [350] | |
| ZEB1-AS1 | [218] | |
| miR-429 | None Reported | |
| miR-135a | circ_0001946 | [351] |
| circNOL10 | [247] | |
| FOXD3-AS1 | [352] | |
| miR-135b | circNOL10 | [247] |
| miR-96 | None Reported | |
| miR-183 | circ_0026344 | [273] |
| miR-150 | LINC00857 | [353] |
| NEAT1 | [354] | |
| PART1 | [287] | |
| ZFAS1 | [286] | |
| miR-195 | AFAP-AS1 | [308] |
| circ_0038718 | [307] | |
| LINC00473 | [306] | |
| SNHG1 | [305] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jorgensen, B.G.; Ro, S. MicroRNAs and ‘Sponging’ Competitive Endogenous RNAs Dysregulated in Colorectal Cancer: Potential as Noninvasive Biomarkers and Therapeutic Targets. Int. J. Mol. Sci. 2022, 23, 2166. https://doi.org/10.3390/ijms23042166
Jorgensen BG, Ro S. MicroRNAs and ‘Sponging’ Competitive Endogenous RNAs Dysregulated in Colorectal Cancer: Potential as Noninvasive Biomarkers and Therapeutic Targets. International Journal of Molecular Sciences. 2022; 23(4):2166. https://doi.org/10.3390/ijms23042166
Chicago/Turabian StyleJorgensen, Brian G., and Seungil Ro. 2022. "MicroRNAs and ‘Sponging’ Competitive Endogenous RNAs Dysregulated in Colorectal Cancer: Potential as Noninvasive Biomarkers and Therapeutic Targets" International Journal of Molecular Sciences 23, no. 4: 2166. https://doi.org/10.3390/ijms23042166
APA StyleJorgensen, B. G., & Ro, S. (2022). MicroRNAs and ‘Sponging’ Competitive Endogenous RNAs Dysregulated in Colorectal Cancer: Potential as Noninvasive Biomarkers and Therapeutic Targets. International Journal of Molecular Sciences, 23(4), 2166. https://doi.org/10.3390/ijms23042166







