Identification and Functional Evaluation of a Novel TBX4 Mutation Underlies Small Patella Syndrome
Abstract
:1. Introduction
2. Results
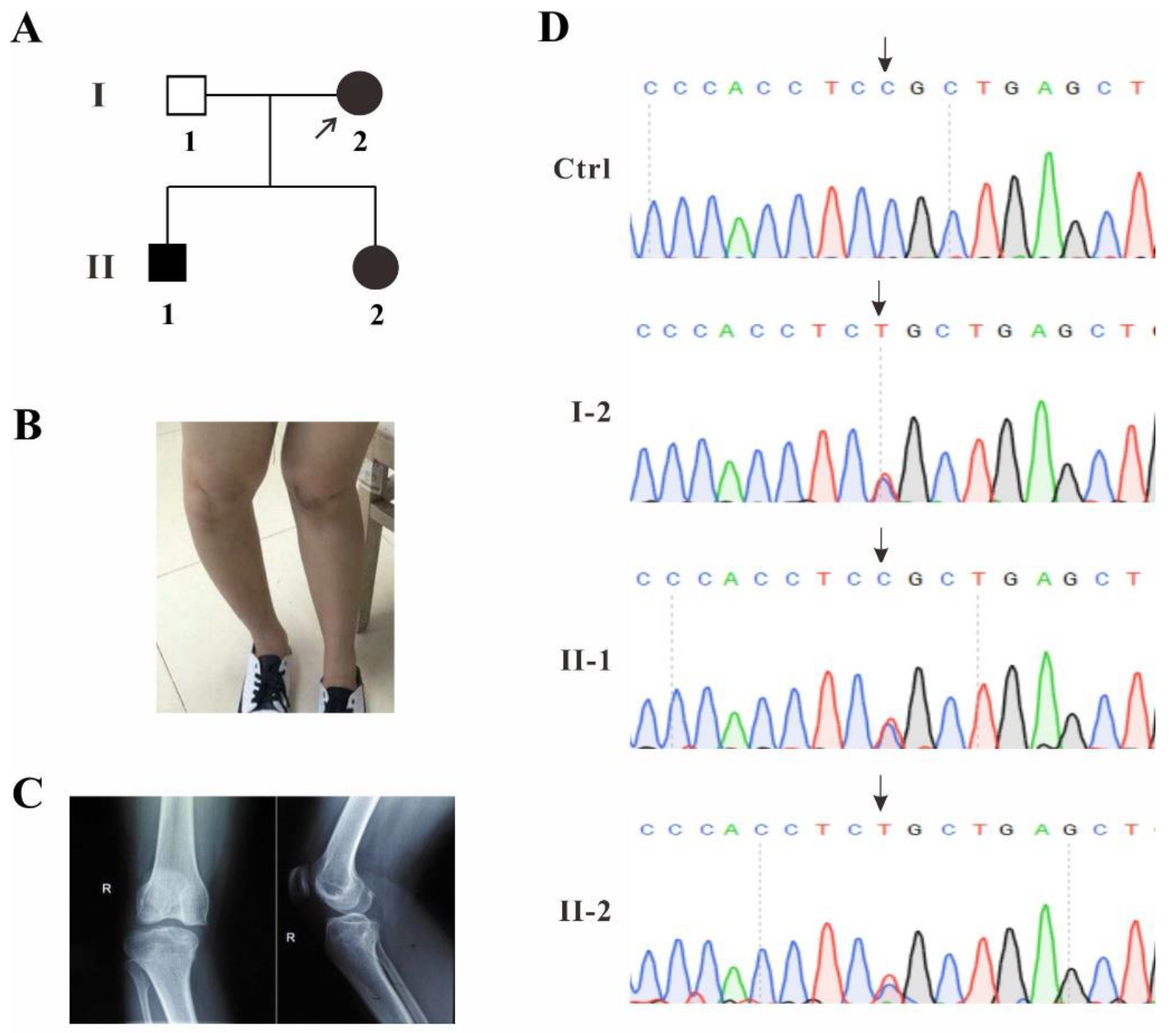
2.1. Identification of a Novel TBX4 Missense Mutation
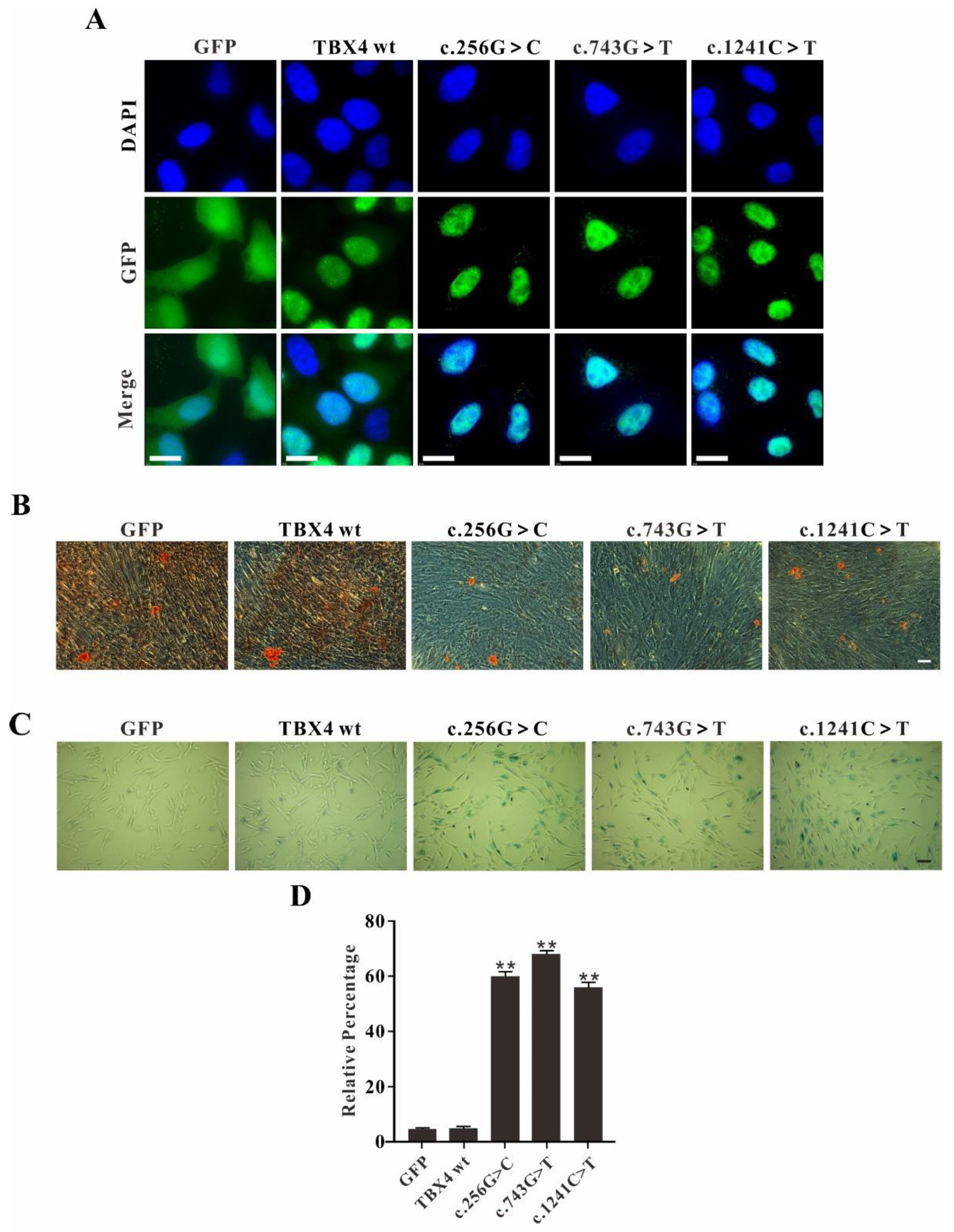
2.2. TBX4 Plasmids Construction and Mesenchymal Stem Cells (MSCs) Cell Lines Screening
2.3. TBX4 Mutations Affect MSCs Osteogenic Differentiation and Promote Cell Senescence
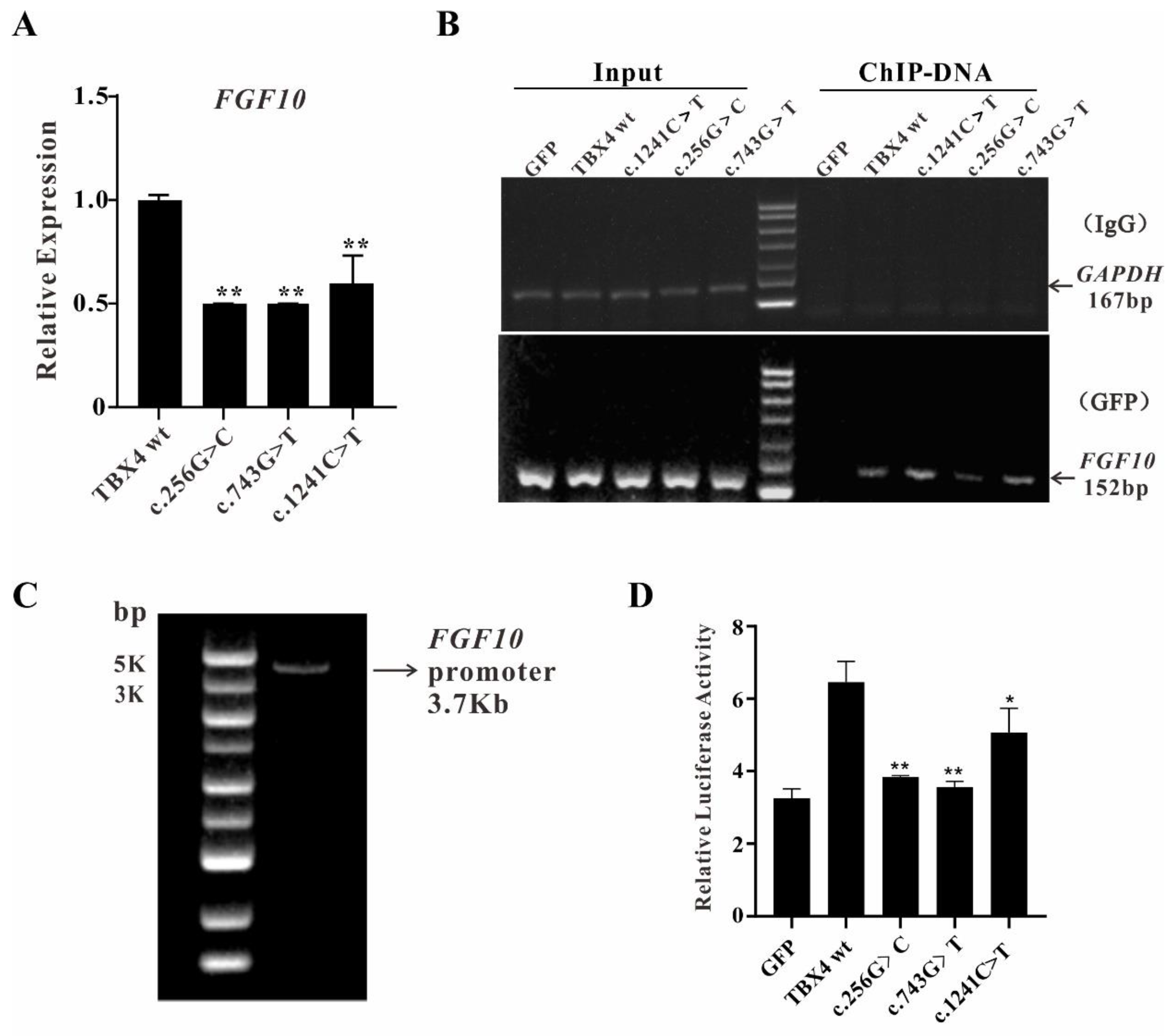
2.4. The Reduced Binding Efficiences of TBX4 Mutants to FGF10 Promoter
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Whole-Exome Sequencing and Sanger Sequencing
4.3. Plasmids and Stable Cell Lines Construction
4.4. Immunoflurorescence
4.5. Osteogenic Differentiation of MSCs
4.6. Senescence Associated β-Galactosidase Assay
4.7. qRT-PCR
4.8. Chromatin Immunoprecipitation (ChIP)
4.9. Dual Luciferase Reporter Assay
4.10. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Takeuchi, J.K.; Koshiba-Takeuchi, K.; Suzuki, T.; Kamimura, M.; Ogura, K.; Ogura, T. Tbx5 and Tbx4 trigger limb initiation through activation of the Wnt/Fgf signaling cascade. Development 2003, 130, 2729–2739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasson, P.; DeLaurier, A.; Bennett, M.; Grigorieva, E.; Naiche, L.A.; Papaioannou, V.E.; Mohun, T.J.; Logan, M.P. Tbx4 and tbx5 acting in connective tissue are required for limb muscle and tendon patterning. Dev. Cell 2010, 18, 148–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, W.; Bacino, C.A.; Richards, B.S.; Alvarez, C.; VanderMeer, J.E.; Vella, M.; Ahituv, N.; Sikka, N.; Dietz, F.R.; Blanton, S.H.; et al. Studies of TBX4 and chromosome 17q23.1q23.2: An uncommon cause of nonsyndromic clubfoot. Am. J. Med. Genet. A 2012, 158, 1620–1627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez-Esteban, C.; Tsukui, T.; Yonei, S.; Magallon, J.; Tamura, K.; Izpisua Belmonte, J.C. The T-box genes Tbx4 and Tbx5 regulate limb outgrowth and identity. Nature 1999, 398, 814–818. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, J.K.; Koshiba-Takeuchi, K.; Matsumoto, K.; Vogel-Hopker, A.; Naitoh-Matsuo, M.; Ogura, K.; Takahashi, N.; Yasuda, K.; Ogura, T. Tbx5 and Tbx4 genes determine the wing/leg identity of limb buds. Nature 1999, 398, 810–814. [Google Scholar] [CrossRef] [PubMed]
- Sheeba, C.J.; Logan, M.P. The Roles of T-Box Genes in Vertebrate Limb Development. Curr. Top. Dev. Biol. 2017, 122, 355–381. [Google Scholar] [CrossRef]
- Bongers, E.M.; Duijf, P.H.; van Beersum, S.E.; Schoots, J.; Van Kampen, A.; Burckhardt, A.; Hamel, B.C.; Losan, F.; Hoefsloot, L.H.; Yntema, H.G.; et al. Mutations in the human TBX4 gene cause small patella syndrome. Am. J. Hum. Genet. 2004, 74, 1239–1248. [Google Scholar] [CrossRef] [Green Version]
- Hajduk, P.; Murphy, P.; Puri, P. Mesenchymal expression of Tbx4 gene is not altered in Adriamycin mouse model. Pediatr. Surg. Int. 2010, 26, 407–411. [Google Scholar] [CrossRef]
- Suhrie, K.; Pajor, N.M.; Ahlfeld, S.K.; Dawson, D.B.; Dufendach, K.R.; Kitzmiller, J.A.; Leino, D.; Lombardo, R.C.; Smolarek, T.A.; Rathbun, P.A.; et al. Neonatal Lung Disease Associated with TBX4 Mutations. J. Pediatr. 2019, 206, 286–292 e281. [Google Scholar] [CrossRef]
- Don, E.K.; de Jong-Curtain, T.A.; Doggett, K.; Hall, T.E.; Heng, B.; Badrock, A.P.; Winnick, C.; Nicholson, G.A.; Guillemin, G.J.; Currie, P.D.; et al. Genetic basis of hindlimb loss in a naturally occurring vertebrate model. Biol. Open 2016, 5, 359–366. [Google Scholar] [CrossRef] [Green Version]
- Isaac, A.; Rodriguez-Esteban, C.; Ryan, A.; Altabef, M.; Tsukui, T.; Patel, K.; Tickle, C.; Izpisua-Belmonte, J.C. Tbx genes and limb identity in chick embryo development. Development 1998, 125, 1867–1875. [Google Scholar] [CrossRef] [PubMed]
- Douglas, N.C.; Arora, R.; Chen, C.Y.; Sauer, M.V.; Papaioannou, V.E. Investigating the role of tbx4 in the female germline in mice. Biol. Reprod. 2013, 89, 148. [Google Scholar] [CrossRef] [PubMed]
- Hart, A.; Papadopoulou, S.; Edlund, H. Fgf10 maintains notch activation, stimulates proliferation, and blocks differentiation of pancreatic epithelial cells. Dev. Dyn. 2003, 228, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Sahara, S.; O’Leary, D.D. Fgf10 regulates transition period of cortical stem cell differentiation to radial glia controlling generation of neurons and basal progenitors. Neuron 2009, 63, 48–62. [Google Scholar] [CrossRef] [Green Version]
- Sakiyama, J.; Yamagishi, A.; Kuroiwa, A. Tbx4-Fgf10 system controls lung bud formation during chicken embryonic development. Development 2003, 130, 1225–1234. [Google Scholar] [CrossRef] [Green Version]
- Naiche, L.A.; Papaioannou, V.E. Loss of Tbx4 blocks hindlimb development and affects vascularization and fusion of the allantois. Development 2003, 130, 2681–2693. [Google Scholar] [CrossRef] [Green Version]
- Haarman, M.G.; Kerstjens-Frederikse, W.S.; Berger, R.M.F. The ever-expanding phenotypical spectrum of human TBX4 mutations: From toe to lung. Eur. Respir. J. 2019, 54, 1901504. [Google Scholar] [CrossRef]
- Vincent, M.; Karolak, J.A.; Deutsch, G.; Gambin, T.; Popek, E.; Isidor, B.; Szafranski, P.; Le Caignec, C.; Stankiewicz, P. Clinical, Histopathological, and Molecular Diagnostics in Lethal Lung Developmental Disorders. Am. J. Respir. Crit. Care Med. 2019, 200, 1093–1101. [Google Scholar] [CrossRef]
- Karolak, J.A.; Vincent, M.; Deutsch, G.; Gambin, T.; Cogne, B.; Pichon, O.; Vetrini, F.; Mefford, H.C.; Dines, J.N.; Golden-Grant, K.; et al. Complex Compound Inheritance of Lethal Lung Developmental Disorders Due to Disruption of the TBX-FGF Pathway. Am. J. Hum. Genet. 2019, 104, 213–228. [Google Scholar] [CrossRef] [Green Version]
- Szafranski, P.; Coban-Akdemir, Z.H.; Rupps, R.; Grazioli, S.; Wensley, D.; Jhangiani, S.N.; Popek, E.; Lee, A.F.; Lupski, J.R.; Boerkoel, C.F.; et al. Phenotypic expansion of TBX4 mutations to include acinar dysplasia of the lungs. Am. J. Med. Genet. A 2016, 170, 2440–2444. [Google Scholar] [CrossRef]
- Zhang, X.; Li, J.; Nie, J.; Jiang, K.; Zhen, Z.; Wang, J.; Shen, L. Differentiation character of adult mesenchymal stem cells and transfection of MSCs with lentiviral vectors. J. Huazhong Univ. Sci. Technol. 2010, 30, 687–693. [Google Scholar] [CrossRef] [PubMed]
- Nishimoto, S.; Wilde, S.M.; Wood, S.; Logan, M.P. RA Acts in a Coherent Feed-Forward Mechanism with Tbx5 to Control Limb Bud Induction and Initiation. Cell Rep. 2015, 12, 879–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oda, T.; Matsushita, M.; Ono, Y.; Kitoh, H.; Sakai, T. A Novel Heterozygous Mutation in the T-box Protein 4 Gene in an Adult Case of Small Patella Syndrome. J. Orthop. Case Rep. 2018, 8, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Tickle, C. How the embryo makes a limb: Determination, polarity and identity. J. Anat. 2015, 227, 418–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawakami, Y.; Capdevila, J.; Buscher, D.; Itoh, T.; Rodriguez Esteban, C.; Izpisua Belmonte, J.C. WNT signals control FGF-dependent limb initiation and AER induction in the chick embryo. Cell 2001, 104, 891–900. [Google Scholar] [CrossRef]
- Almalki, S.G.; Agrawal, D.K. Key transcription factors in the differentiation of mesenchymal stem cells. Differentiation 2016, 92, 41–51. [Google Scholar] [CrossRef] [Green Version]
- Ohuchi, H.; Takeuchi, J.; Yoshioka, H.; Ishimaru, Y.; Ogura, K.; Takahashi, N.; Ogura, T.; Noji, S. Correlation of wing-leg identity in ectopic FGF-induced chimeric limbs with the differential expression of chick Tbx5 and Tbx4. Development 1998, 125, 51–60. [Google Scholar] [CrossRef]
- Isaac, A.; Cohn, M.J.; Ashby, P.; Ataliotis, P.; Spicer, D.B.; Cooke, J.; Tickle, C. FGF and genes encoding transcription factors in early limb specification. Mech. Dev. 2000, 93, 41–48. [Google Scholar] [CrossRef]
- Prince, L.S. FGF10 and Human Lung Disease Across the Life Spectrum. Front. Genet. 2018, 9, 517. [Google Scholar] [CrossRef]
- Kerstjens-Frederikse, W.S.; Bongers, E.M.; Roofthooft, M.T.; Leter, E.M.; Douwes, J.M.; Van Dijk, A.; Vonk-Noordegraaf, A.; Dijk-Bos, K.K.; Hoefsloot, L.H.; Hoendermis, E.S.; et al. TBX4 mutations (small patella syndrome) are associated with childhood-onset pulmonary arterial hypertension. J. Med. Genet. 2013, 50, 500–506. [Google Scholar] [CrossRef] [Green Version]
- Sekine, K.; Ohuchi, H.; Fujiwara, M.; Yamasaki, M.; Yoshizawa, T.; Sato, T.; Yagishita, N.; Matsui, D.; Koga, Y.; Itoh, N.; et al. Publisher Correction: Fgf10 is essential for limb and lung formation. Nat. Genet. 2019, 51, 921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibson-Brown, J.J.; Agulnik, S.I.; Chapman, D.L.; Alexiou, M.; Garvey, N.; Silver, L.M.; Papaioannou, V.E. Evidence of a role for T-box genes in the evolution of limb morphogenesis and the specification of forelimb/hindlimb identity. Mech. Dev. 1996, 56, 93–101. [Google Scholar] [CrossRef]
- Barriscale, K.A.; O’Sullivan, S.A.; McCarthy, T.V. A single secreted luciferase-based gene reporter assay. Anal. Biochem. 2014, 453, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Shigehisa, M.; Amaba, N.; Arai, S.; Higashi, C.; Kawanabe, R.; Matsunaga, A.; Laksmi, F.A.; Tokunaga, M.; Ishibashi, M. Stabilization of luciferase from Renilla reniformis using random mutations. Protein Eng. Des. Sel. PEDS 2017, 30, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Pavesi, G. ChIP-Seq Data Analysis to Define Transcriptional Regulatory Networks. Adv. Biochem. Eng. Biotechnol. 2017, 160, 1–14. [Google Scholar] [CrossRef]
- Vaquerizas, J.M.; Kummerfeld, S.K.; Teichmann, S.A.; Luscombe, N.M. A census of human transcription factors: Function, expression and evolution. Nat. Rev. Genet. 2009, 10, 252–263. [Google Scholar] [CrossRef]
- Slattery, M.; Zhou, T.; Yang, L.; Dantas Machado, A.C.; Gordan, R.; Rohs, R. Absence of a simple code: How transcription factors read the genome. Trends Biochem. Sci. 2014, 39, 381–399. [Google Scholar] [CrossRef] [Green Version]
- Glaser, A.; Arora, R.; Hoffmann, S.; Li, L.; Gretz, N.; Papaioannou, V.E.; Rappold, G.A. Tbx4 interacts with the short stature homeobox gene Shox2 in limb development. Dev. Dyn. 2014, 243, 629–639. [Google Scholar] [CrossRef] [Green Version]
- Duboc, V.; Sulaiman, F.A.; Feneck, E.; Kucharska, A.; Bell, D.; Holder-Espinasse, M.; Logan, M.P.O. Tbx4 function during hindlimb development reveals a mechanism that explains the origins of proximal limb defects. Development 2021, 148. [Google Scholar] [CrossRef]
- Solberg, N.; Krauss, S. Luciferase assay to study the activity of a cloned promoter DNA fragment. Methods Mol. Biol. 2013, 977, 65–78. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, P.; Lan, W.; Li, J.; Zhang, Y.; Xiong, Q.; Ye, J.; Wu, C.; Xiao, H. Identification and Functional Evaluation of a Novel TBX4 Mutation Underlies Small Patella Syndrome. Int. J. Mol. Sci. 2022, 23, 2075. https://doi.org/10.3390/ijms23042075
Li P, Lan W, Li J, Zhang Y, Xiong Q, Ye J, Wu C, Xiao H. Identification and Functional Evaluation of a Novel TBX4 Mutation Underlies Small Patella Syndrome. International Journal of Molecular Sciences. 2022; 23(4):2075. https://doi.org/10.3390/ijms23042075
Chicago/Turabian StyleLi, Ping, Wenli Lan, Jiaying Li, Yanping Zhang, Qiuhong Xiong, Jinpei Ye, Changxin Wu, and Han Xiao. 2022. "Identification and Functional Evaluation of a Novel TBX4 Mutation Underlies Small Patella Syndrome" International Journal of Molecular Sciences 23, no. 4: 2075. https://doi.org/10.3390/ijms23042075
APA StyleLi, P., Lan, W., Li, J., Zhang, Y., Xiong, Q., Ye, J., Wu, C., & Xiao, H. (2022). Identification and Functional Evaluation of a Novel TBX4 Mutation Underlies Small Patella Syndrome. International Journal of Molecular Sciences, 23(4), 2075. https://doi.org/10.3390/ijms23042075






