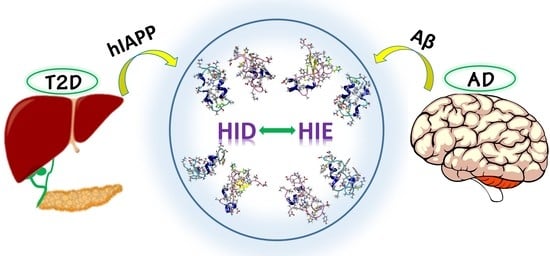
Mechanistic Insights into the Polymorphic Associations and Cross-Seeding of Aβ and hIAPP in the Presence of Histidine Tautomerism: An All-Atom Molecular Dynamic Study
Abstract
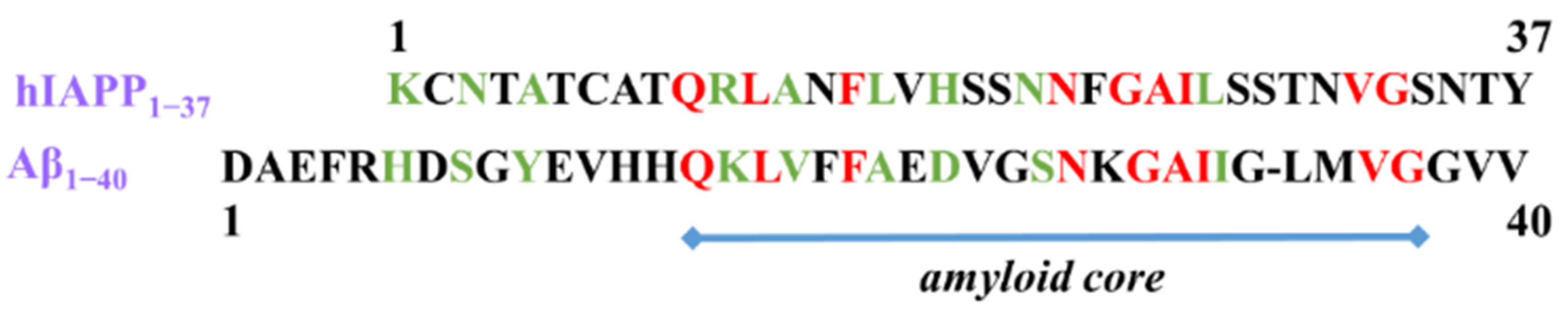
1. Introduction
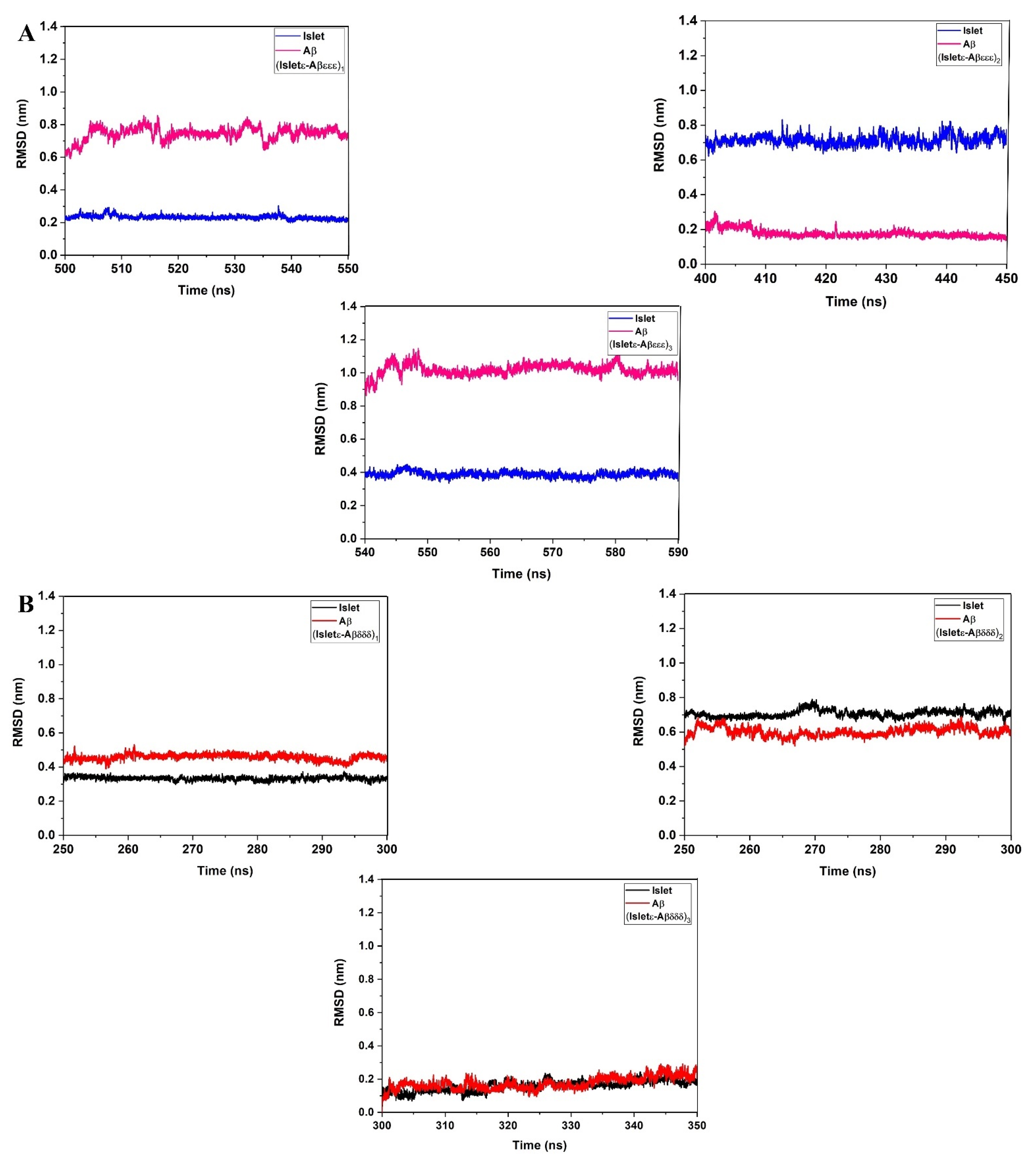
2. Results and Discussion
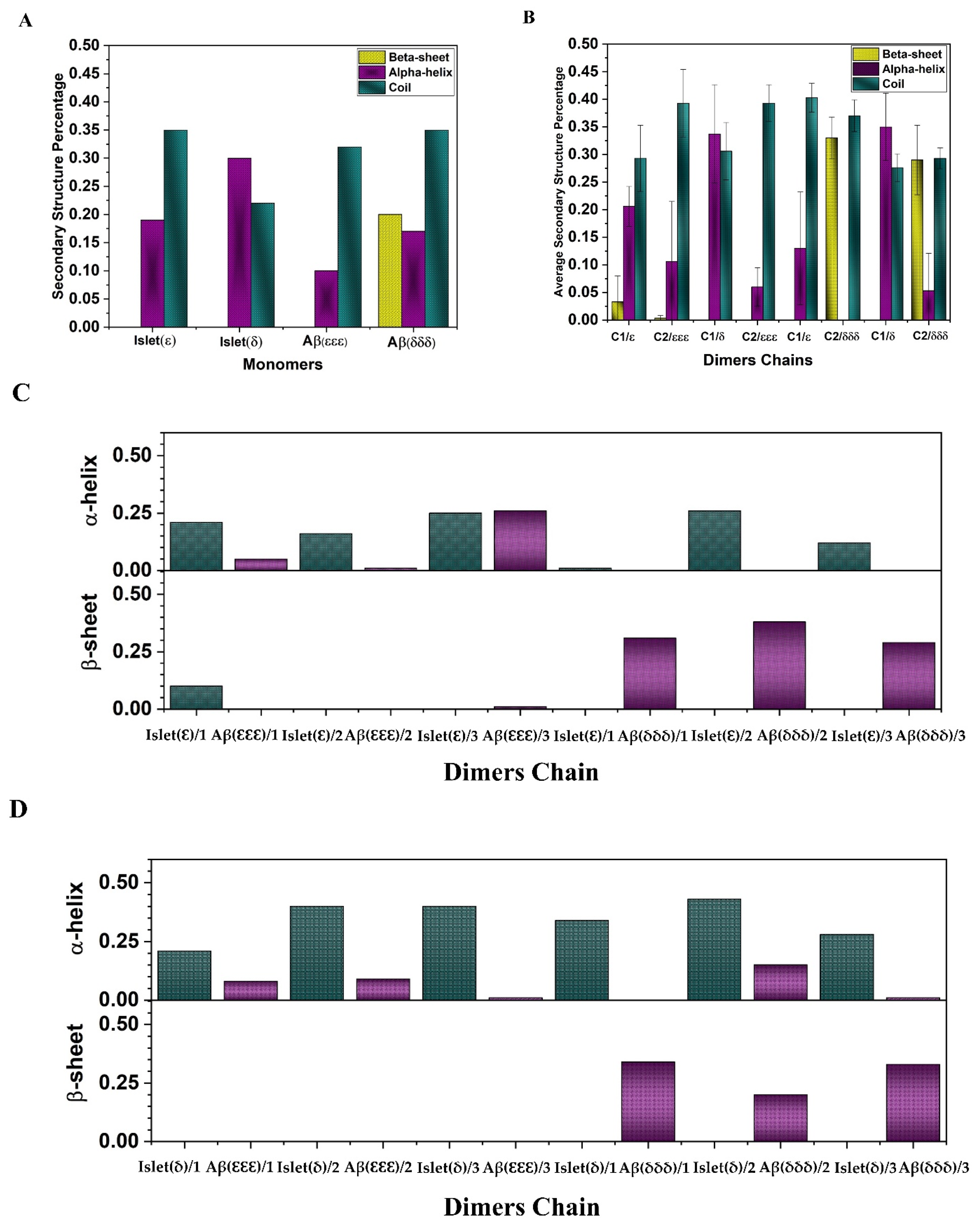
2.1. Secondary Structure
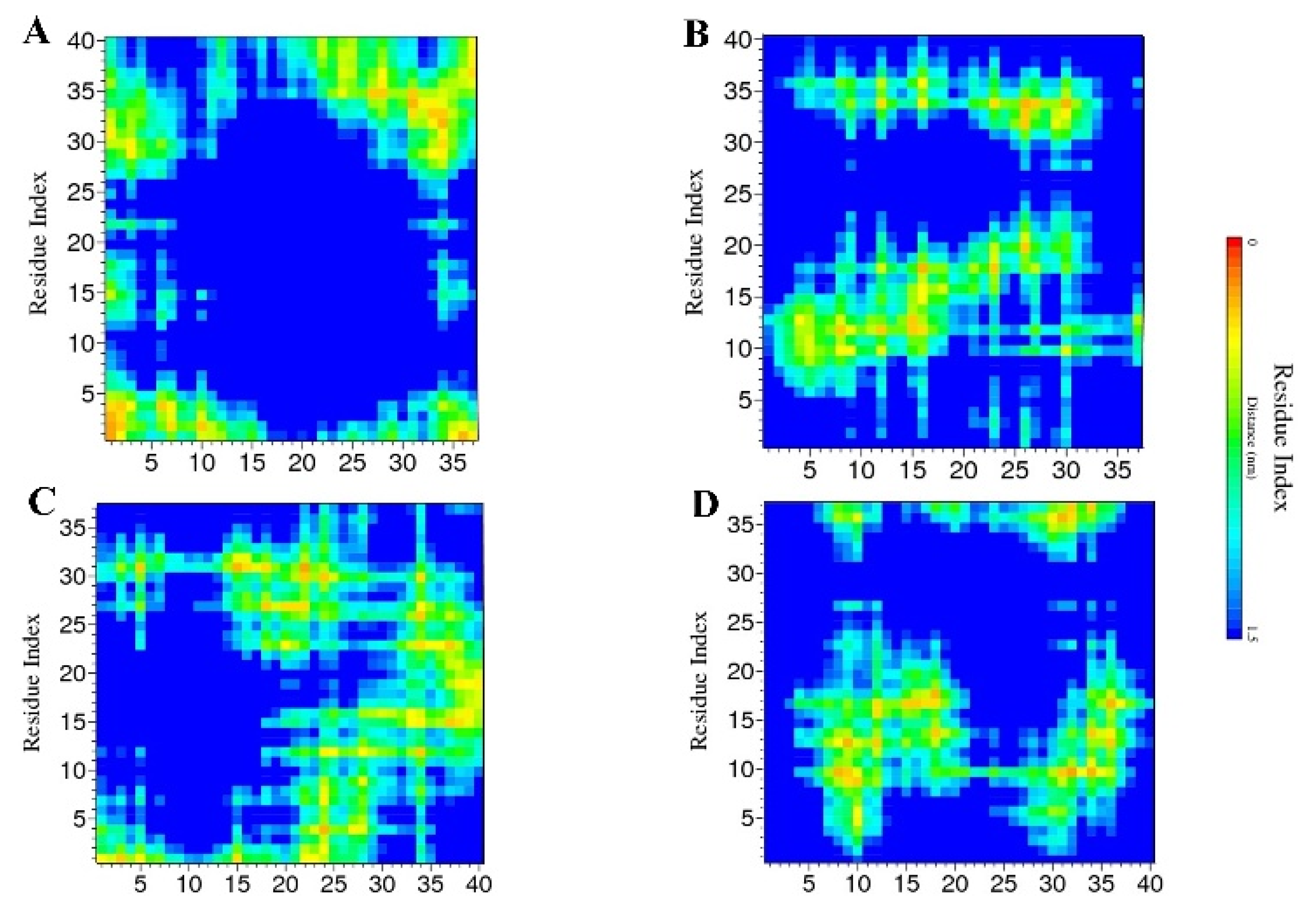
2.2. Contact Maps
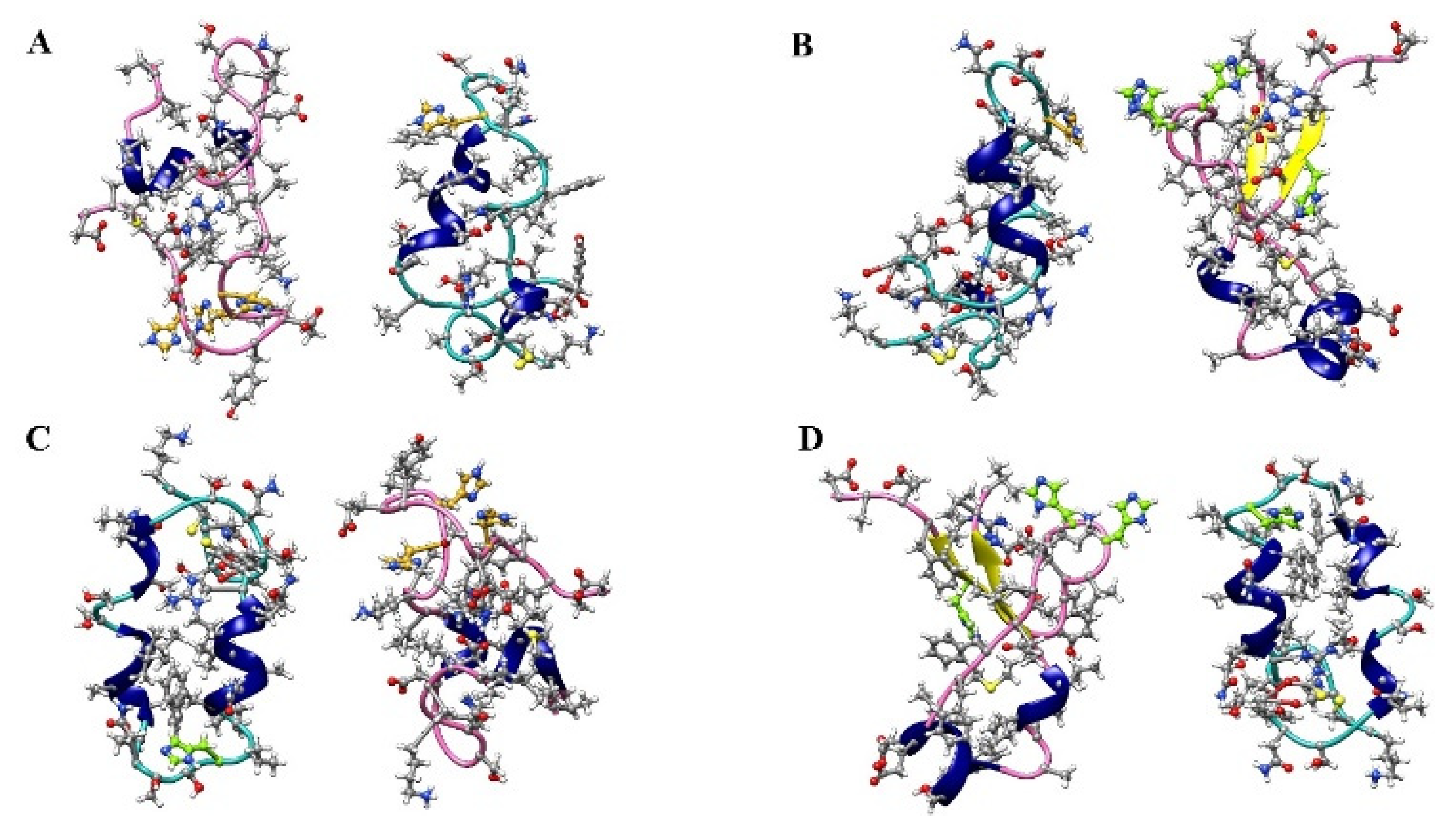
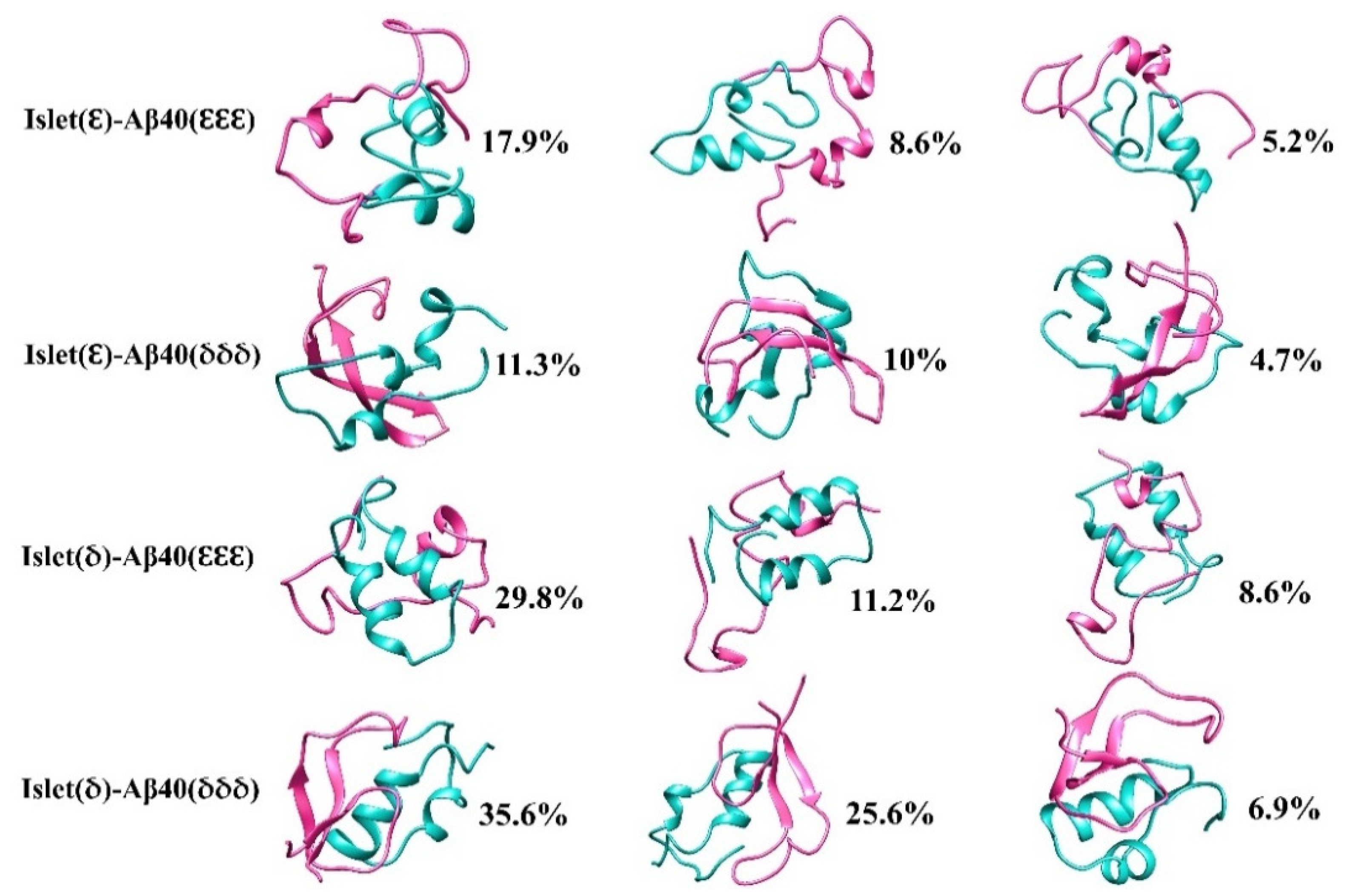
2.3. Cluster Analysis
2.4. Free Energy Landscape (FEL)
2.5. Binding Free Energy
2.6. Hydrogen Bonding
3. Materials and Methods
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Aβ | Aggregated amyloid β-peptide |
| AD | Alzheimer’s disease |
| APP | Amyloid protein precursor |
| hIAPP | Human islet amyloid polypeptide |
| MD | Molecular dynamics |
| RMSD | Root mean square deviation |
| SASA | Solvent accessible surface area |
| PCA | Principal component analysis |
| DSSP | Define Secondary Structure of Proteins |
References
- Zhang, M.; Hu, R.; Ren, B.; Chen, H.; Jiang, B.; Ma, J.; Zheng, J. Molecular Understanding of Aβ-hIAPP Cross-Seeding Assemblies on Lipid Membranes. ACS Chem. Neurosci. 2017, 8, 524–537. [Google Scholar] [CrossRef] [PubMed]
- Qiu, W.Q.; Folstein, M.F. Insulin, insulin-degrading enzyme and amyloid-β peptide in Alzheimer’s disease: Review and hypothesis. Neurobiol. Aging 2006, 27, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Sipe, J.D.; Cohen, A.S. Review: History of the Amyloid Fibril. J. Struct. Biol. 2000, 130, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Chaari, A.; Ladjimi, M. Human islet amyloid polypeptide (hIAPP) aggregation in type 2 diabetes: Correlation between intrinsic physicochemical properties of hIAPP aggregates and their cytotoxicity. Int. J. Biol. Macromol. 2019, 136, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Hölscher, C. Common pathological processes in Alzheimer disease and type 2 diabetes: A review. Brain Res. Rev. 2007, 56, 384–402. [Google Scholar] [CrossRef]
- Jucker, M.; Walker, L.C. Pathogenic protein seeding in alzheimer disease and other neurodegenerative disorders. Ann. Neurol. 2011, 70, 532–540. [Google Scholar] [CrossRef]
- Mark, R.N. The Clinical and Biological Relationship between Type II Diabetes Mellitus and Alzheimers Disease. Curr. Alzheimer Res. 2004, 1, 47–54. [Google Scholar] [CrossRef]
- Oskarsson, M.E.; Paulsson, J.F.; Schultz, S.W.; Ingelsson, M.; Westermark, P.; Westermark, G.T. In Vivo Seeding and Cross-Seeding of Localized Amyloidosis: A Molecular Link between Type 2 Diabetes and Alzheimer Disease. Am. J. Pathol. 2015, 185, 834–846. [Google Scholar] [CrossRef]
- Krishnaswamy, S.; Verdile, G.; Groth, D.; Kanyenda, L.; Martins, R.N. The structure and function of Alzheimer’s gamma secretase enzyme complex. Crit. Rev. Clin. Lab. Sci. 2009, 46, 282–301. [Google Scholar] [CrossRef]
- Cao, P.; Marek, P.; Noor, H.; Patsalo, V.; Tu, L.-H.; Wang, H.; Abedini, A.; Raleigh, D.P. Islet amyloid: From fundamental biophysics to mechanisms of cytotoxicity. FEBS Lett. 2013, 587, 1106–1118. [Google Scholar] [CrossRef]
- Wineman-Fisher, V.; Atsmon-Raz, Y.; Miller, Y. Orientations of Residues along the β-Arch of Self-Assembled Amylin Fibril-Like Structures Lead to Polymorphism. Biomacromolecules 2015, 16, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Brender, J.R.; Lee, E.L.; Hartman, K.; Wong, P.T.; Ramamoorthy, A.; Steel, D.G.; Gafni, A. Biphasic effects of insulin on islet amyloid polypeptide membrane disruption. Biophys. J. 2011, 100, 685–692. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Brender, J.R.; Lee, E.L.; Cavitt, M.A.; Gafni, A.; Steel, D.G.; Ramamoorthy, A. Amyloid Fiber Formation and Membrane Disruption are Separate Processes Localized in Two Distinct Regions of IAPP, the Type-2-Diabetes-Related Peptide. J. Am. Chem. Soc. 2008, 130, 6424–6429. [Google Scholar] [CrossRef] [PubMed]
- Andreetto, E.; Yan, L.-M.; Tatarek-Nossol, M.; Velkova, A.; Frank, R.; Kapurniotu, A. Identification of Hot Regions of the Aβ–IAPP Interaction Interface as High-Affinity Binding Sites in both Cross- and Self-Association. Angew. Chem. Int. Ed. 2010, 49, 3081–3085. [Google Scholar] [CrossRef]
- Reitz, C. Alzheimer’s Disease and the Amyloid Cascade Hypothesis: A Critical Review. Int. J. Alzheimers Dis. 2012, 2012, 369808. [Google Scholar] [CrossRef]
- Hu, R.; Zhang, M.; Chen, H.; Jiang, B.; Zheng, J. Cross-Seeding Interaction between β-Amyloid and Human Islet Amyloid Polypeptide. ACS Chem. Neurosci. 2015, 6, 1759–1768. [Google Scholar] [CrossRef]
- Ren, B.; Zhang, Y.; Zhang, M.; Liu, Y.; Zhang, D.; Gong, X.; Feng, Z.; Tang, J.; Chang, Y.; Zheng, J. Fundamentals of cross-seeding of amyloid proteins: An introduction. J. Mater. Chem. B 2019, 7, 7267–7282. [Google Scholar] [CrossRef]
- O’Nuallain, B.; Williams, A.D.; Westermark, P.; Wetzel, R. Seeding Specificity in Amyloid Growth Induced by Heterologous Fibrils. J. Biol. Chem. 2004, 279, 17490–17499. [Google Scholar] [CrossRef]
- Andreetto, E.; Yan, L.-M.; Caporale, A.; Kapurniotu, A. Dissecting the Role of Single Regions of an IAPP Mimic and IAPP in Inhibition of Aβ40 Amyloid Formation and Cytotoxicity. ChemBioChem 2011, 12, 1313–1322. [Google Scholar] [CrossRef]
- Shi, H.; Kang, B.; Lee, J.Y. Tautomeric Effect of Histidine on the Monomeric Structure of Amyloid β-Peptide(1–40). J. Phys. Chem. B 2016, 120, 11405–11411. [Google Scholar] [CrossRef]
- Shi, H.; Lee, J.Y. Tautomeric Effect of Histidine on the Monomeric Structure of Amyloid β-Peptide(1–42). ACS Chem. Neurosci. 2017, 8, 669–675. [Google Scholar] [CrossRef] [PubMed]
- Salimi, A.; Li, H.; Shi, H.; Lee, J.Y. Intrinsic origin of amyloid aggregation: Behavior of histidine (εεε) and (δδδ) tautomer homodimers of Aβ (1–40). Biochim. Biophys. Acta (BBA)-Gen. Subj. 2019, 1863, 795–801. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Salimi, A.; Lee, J.Y. Intrinsic Origin of Amyloid Aggregation: Collective Effects of the Mutation and Tautomerism of Histidine. ACS Chem. Neurosci. 2019, 10, 4729–4734. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Nam, Y.; Salimi, A.; Lee, J.Y. Impact of A2V Mutation and Histidine Tautomerism on Aβ42 Monomer Structures from Atomistic Simulations. J. Chem. Inf. Model. 2020, 60, 3587–3592. [Google Scholar] [CrossRef]
- Shi, H.; Li, H.; Gong, W.; Gong, R.; Qian, A.; Lee, J.Y.; Guo, W. Structural and Binding Properties on Aβ Mature Fibrils Due to the Histidine Tautomeric Effect. ACS Chem. Neurosci. 2019, 10, 4612–4618. [Google Scholar] [CrossRef]
- Xing, X.; Zhao, W.; Hu, D.; Kang, B.; Shi, H.; Lee, J.Y.; Ai, H. Tautomerization Effect of Histidines on Oligomer Aggregation of β-Amyloid(1–40/42) during the Early Stage: Tautomerism Hypothesis for Misfolding Protein Aggregation. ACS Chem. Neurosci. 2019, 10, 2602–2608. [Google Scholar] [CrossRef]
- Nam, Y.; Kalathingal, M.; Saito, S.; Lee, J.Y. Tautomeric Effect of Histidine on β-Sheet Formation of Amyloid Beta 1–40: 2D-IR Simulations. Biophys. J. 2020, 119, 831–842. [Google Scholar] [CrossRef]
- Chatterjee, S.; Salimi, A.; Lee, J.Y. Intrinsic Origin of Tau Protein Aggregation: Effects of Histidine Tautomerism on Tau267–312 Monomer. ACS Chem. Neurosci. 2020, 11, 3814–3822. [Google Scholar] [CrossRef]
- Chatterjee, S.; Salimi, A.; Lee, J.Y. Molecular mechanism of amyloidogenicity and neurotoxicity of a pro-aggregated tau mutant in the presence of histidine tautomerism via replica-exchange simulation. Phys. Chem. Chem. Phys. 2021, 23, 10475–10486. [Google Scholar] [CrossRef]
- Salimi, A.; Li, H.; Lee, J.Y. Molecular insight into the early stage of amyloid-β(1-42) Homodimers aggregation influenced by histidine tautomerism. Int. J. Biol. Macromol. 2021, 184, 887–897. [Google Scholar] [CrossRef]
- Salimi, A.; Chatterjee, S.; Yong Lee, J. Histidine Tautomerism Driving Human Islet Amyloid Polypeptide Aggregation in the Early Stages of Diabetes Mellitus Progression: Insight at the Atomistic Level. Chem.–Asian J. 2021, 16, 2453–2462. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Salimi, A.; Lee, J.Y. Unraveling the Histidine Tautomerism Effect on the Initial Stages of Prion Misfolding: New Insights from a Computational Perspective. ACS Chem. Neurosci. 2021, 12, 3203–3213. [Google Scholar] [CrossRef] [PubMed]
- Ren, B.; Hu, R.; Zhang, M.; Liu, Y.; Xu, L.; Jiang, B.; Ma, J.; Ma, B.; Nussinov, R.; Zheng, J. Experimental and Computational Protocols for Studies of Cross-Seeding Amyloid Assemblies. Methods Mol. Biol. 2018, 1777, 429–447. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tang, Y.; Zhang, D.; Liu, Y.; He, J.; Chang, Y.; Zheng, J. Amyloid cross-seeding between Aβ and hIAPP in relation to the pathogenesis of Alzheimer and type 2 diabetes. Chin. J. Chem. Eng. 2021, 30, 225–235. [Google Scholar] [CrossRef]
- Morgan, J.E.; Verkhovsky, M.I.; Wikström, M. The histidine cycle: A new model for proton translocation in the respiratory heme-copper oxidases. J. Bioenerg. Biomembr. 1994, 26, 599–608. [Google Scholar] [CrossRef]
- Strothkamp, K.G.; Lippard, S.J. Chemistry of the imidazolate-bridged bimetallic center in the copper-zinc superoxide dismutase and its model compounds. Acc. Chem. Res. 1982, 15, 318–326. [Google Scholar] [CrossRef]
- Ivanov, I.; Klein, M.L. Deprotonation of a Histidine Residue in Aqueous Solution Using Constrained Ab Initio Molecular Dynamics. J. Am. Chem. Soc. 2002, 124, 13380–13381. [Google Scholar] [CrossRef]
- Huang, Z.; Lin, Z.; Song, C. Protonation Processes and Electronic Spectra of Histidine and Related Ions. J. Phys. Chem. A 2007, 111, 4340–4352. [Google Scholar] [CrossRef]
- Vila, J.A.; Arnautova, Y.A.; Vorobjev, Y.; Scheraga, H.A. Assessing the fractions of tautomeric forms of the imidazole ring of histidine in proteins as a function of pH. Proc. Natl. Acad. Sci. USA 2011, 108, 5602. [Google Scholar] [CrossRef]
- Hudáky, P.; Perczel, A. Peptide models XLII. Ab initio study on conformational changes of N-formyl-l-histidinamide caused by protonation or deprotonation of its side chain. J. Mol. Struct. THEOCHEM 2004, 675, 117–127. [Google Scholar] [CrossRef]
- Li, S.; Hong, M. Protonation, Tautomerization, and Rotameric Structure of Histidine: A Comprehensive Study by Magic-Angle-Spinning Solid-State NMR. J. Am. Chem. Soc. 2011, 133, 1534–1544. [Google Scholar] [CrossRef] [PubMed]
- Nydegger, M.W.; Dutta, S.; Cheatum, C.M. Two-dimensional infrared study of 3-azidopyridine as a potential spectroscopic reporter of protonation state. J. Chem. Phys. 2010, 133, 134506. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Hu, R.; Chen, H.; Chang, Y.; Ma, J.; Liang, G.; Mi, J.; Wang, Y.; Zheng, J. Polymorphic cross-seeding amyloid assemblies of amyloid-β and human islet amyloid polypeptide. Phys. Chem. Chem. Phys. 2015, 17, 23245–23256. [Google Scholar] [CrossRef] [PubMed]
- Berhanu, W.M.; Yaşar, F.; Hansmann, U.H.E. In Silico Cross Seeding of Aβ and Amylin Fibril-like Oligomers. ACS Chem. Neurosci. 2013, 4, 1488–1500. [Google Scholar] [CrossRef] [PubMed]
- Baram, M.; Atsmon-Raz, Y.; Ma, B.; Nussinov, R.; Miller, Y. Amylin-Aβ oligomers at atomic resolution using molecular dynamics simulations: A link between Type 2 diabetes and Alzheimer’s disease. Phys. Chem. Chem. Phys. 2016, 18, 2330–2338. [Google Scholar] [CrossRef]
- Brännström, K.; Islam, T.; Sandblad, L.; Olofsson, A. The role of histidines in amyloid β fibril assembly. FEBS Lett. 2017, 591, 1167–1175. [Google Scholar] [CrossRef] [PubMed]
- Huo, G.; Chen, W.; Wang, J.; Chu, X.; Xu, W.; Li, B.; Zhang, Y.; Xu, B.; Zhou, X. His18 promotes reactive oxidative stress production in copper-ion mediated human islet amyloid polypeptide aggregation. RSC Adv. 2020, 10, 5566–5571. [Google Scholar] [CrossRef]
- Jha, S.; Snell, J.M.; Sheftic, S.R.; Patil, S.M.; Daniels, S.B.; Kolling, F.W.; Alexandrescu, A.T. pH Dependence of Amylin Fibrillization. Biochemistry 2014, 53, 300–310. [Google Scholar] [CrossRef]
- Olubiyi, O.O.; Strodel, B. Structures of the Amyloid β-Peptides Aβ1–40 and Aβ1–42 as Influenced by pH and a d-Peptide. J. Phys. Chem. B 2012, 116, 3280–3291. [Google Scholar] [CrossRef]
- Rezaei-Ghaleh, N.; Andreetto, E.; Yan, L.-M.; Kapurniotu, A.; Zweckstetter, M. Interaction between amyloid beta peptide and an aggregation blocker peptide mimicking islet amyloid polypeptide. PLoS ONE 2011, 6, e20289. [Google Scholar] [CrossRef]
- Ge, X.; Yang, Y.; Sun, Y.; Cao, W.; Ding, F. Islet Amyloid Polypeptide Promotes Amyloid-Beta Aggregation by Binding-Induced Helix-Unfolding of the Amyloidogenic Core. ACS Chem. Neurosci. 2018, 9, 967–975. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Hu, R.; Chen, H.; Gong, X.; Zhou, F.; Zhang, L.; Zheng, J. Polymorphic Associations and Structures of the Cross-Seeding of Aβ1–42 and hIAPP1–37 Polypeptides. J. Chem. Inf. Model. 2015, 55, 1628–1639. [Google Scholar] [CrossRef] [PubMed]
- Mawuenyega, K.G.; Sigurdson, W.; Ovod, V.; Munsell, L.; Kasten, T.; Morris, J.C.; Yarasheski, K.E.; Bateman, R.J. Decreased clearance of CNS beta-amyloid in Alzheimer’s disease. Science 2010, 330, 1774. [Google Scholar] [CrossRef] [PubMed]
- González-Alemán, R.; Hernández-Castillo, D.; Caballero, J.; Montero-Cabrera, L.A. Quality Threshold Clustering of Molecular Dynamics: A Word of Caution. J. Chem. Inf. Model. 2020, 60, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Daura, X.; Gademann, K.; Jaun, B.; Seebach, D.; van Gunsteren, W.F.; Mark, A.E. Peptide Folding: When Simulation Meets Experiment. Angew. Chem. Int. Ed. 1999, 38, 236–240. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Amir, M.; Mohammad, T.; Kumar, V.; Alajmi, M.F.; Rehman, M.T.; Hussain, A.; Alam, P.; Dohare, R.; Islam, A.; Ahmad, F.; et al. Structural Analysis and Conformational Dynamics of STN1 Gene Mutations Involved in Coat Plus Syndrome. Front. Mol. Biosci. 2019, 6, 41. [Google Scholar] [CrossRef]
- Basith, S.; Manavalan, B.; Shin, T.H.; Lee, G. A Molecular Dynamics Approach to Explore the Intramolecular Signal Transduction of PPAR-α. Int. J. Mol. Sci. 2019, 20, 1666. [Google Scholar] [CrossRef]
- Sang, P.; Du, X.; Yang, L.-Q.; Meng, Z.-H.; Liu, S.-Q. Molecular motions and free-energy landscape of serine proteinase K in relation to its cold-adaptation: A comparative molecular dynamics simulation study and the underlying mechanisms. RSC Adv. 2017, 7, 28580–28590. [Google Scholar] [CrossRef]
- Kumari, R.; Kumar, R.; Lynn, A. g_mmpbsa—A GROMACS Tool for High-Throughput MM-PBSA Calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [CrossRef]
- Wieczorek, R.; Dannenberg, J.J. H-Bonding Cooperativity and Energetics of α-Helix Formation of Five 17-Amino Acid Peptides. J. Am. Chem. Soc. 2003, 125, 8124–8129. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Ning, L.; Pan, D.; Zhang, Y.; Wang, S.; Liu, H.; Yao, X. Effects of the Pathogenic Mutation A117V and the Protective Mutation H111S on the Folding and Aggregation of PrP106-126: Insights from Replica Exchange Molecular Dynamics Simulations. PLoS ONE 2015, 10, e0125899. [Google Scholar] [CrossRef] [PubMed]
- Fawzi, N.L.; Phillips, A.H.; Ruscio, J.Z.; Doucleff, M.; Wemmer, D.E.; Head-Gordon, T. Structure and Dynamics of the Aβ21–30 Peptide from the Interplay of NMR Experiments and Molecular Simulations. J. Am. Chem. Soc. 2008, 130, 6145–6158. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]



| Dimers | ΔEvdw | ΔEelec | ΔGpolar | ΔGsasa | ΔEbinding |
|---|---|---|---|---|---|
| Islet(ε)-Aβ(εεε) (1) | −151.013 (1.868) | −508.965 (5.505) | 969.206 (10.696) | −19.514 (0.244) | 289.111 (2.990) |
| Islet(ε)-Aβ(εεε) (2) | −300.080 (0.576) | −284.750 (0.705) | 686.494 (1.496) | −35.159 (0.050) | 66.520 (0.913) |
| Islet(ε)-Aβ(εεε) (3) | −348.255 (0.580) | −506.954 (0.983) | 1021.628 (1.707) | −38.343 (0.051) | 128.096 (1.065) |
| Islet(δ)-Aβ(εεε) (1) | −361.515 (0.752) | −449.112 (1.370) | 1055.068 (2.821) | −40.605 (0.085) | 203.830 (1.572) |
| Islet(δ)-Aβ(εεε) (2) | −302.575 (0.641) | −574.749 (1.589) | 1249.936 (2.726) | −38.848 (0.059) | 333.757 (1.397) |
| Islet(δ)-Aβ(εεε) (3) | −160.475 (0.343) | −269.774 (0.785) | 596.982 (1.471) | −20.898 (0.041) | 145.758 (0.951) |
| Islet(ε)-Aβ(δδδ) (1) | −274.357 (0.439) | −477.366 (0.575) | 961.720 (1.081) | −29.931 (0.034) | 180.096 (0.925) |
| Islet(ε)-Aβ(δδδ) (2) | −226.746 (0.532) | −243.161 (0.987) | 577.932 (1.928) | −25.630 (0.051) | 82.379 (1.186) |
| Islet(ε)-Aβ(δδδ) (3) | −274.619 (0.441) | −474.310 (0.677) | 919.789 (1.201) | −28.932 (0.042) | 141.888 (0.945) |
| Islet(δ)-Aβ(δδδ) (1) | −209.193 (0.407) | −224.150 (0.453) | 503.763 (1.132) | −23.461 (0.039) | 46.955 (0.858) |
| Islet(δ)-Aβ(δδδ) (2) | −258.789 (0.399) | −370.938 (0.879) | 872.423 (1.706) | −26.770 (0.035) | 215.987 (1.035) |
| Islet(δ)-Aβ(δδδ) (3) | −193.606 (0.467) | −247.322 (0.705) | 572.019 (1.560) | −23.159 (0.048) | 107.935 (1.245) |
| Dimers | Average H-Bond | Std |
|---|---|---|
| Islet(ε)-Aβ(εεε) (1) | 6.8 | 1.9 |
| Islet(ε)-Aβ(εεε) (2) | 5.7 | 1.6 |
| Islet(ε)-Aβ(εεε) (3) | 6.2 | 1.3 |
| Islet(δ)-Aβ(εεε) (1) | 5.5 | 1.7 |
| Islet(δ)-Aβ(εεε) (2) | 4.8 | 1.6 |
| Islet(δ)-Aβ(εεε) (3) | 1.8 | 0.9 |
| Islet(ε)-Aβ(δδδ) (1) | 5.8 | 1.1 |
| Islet(ε)-Aβ(δδδ) (2) | 1.1 | 0.8 |
| Islet(ε)-Aβ(δδδ) (3) | 6.8 | 1.5 |
| Islet(δ)-Aβ(δδδ) (1) | 3.2 | 1.0 |
| Islet(δ)-Aβ(δδδ) (2) | 2.9 | 1 |
| Islet(δ)-Aβ(δδδ) (3) | 4.22 | 1.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salimi, A.; Chatterjee, S.; Lee, J.Y. Mechanistic Insights into the Polymorphic Associations and Cross-Seeding of Aβ and hIAPP in the Presence of Histidine Tautomerism: An All-Atom Molecular Dynamic Study. Int. J. Mol. Sci. 2022, 23, 1930. https://doi.org/10.3390/ijms23041930
Salimi A, Chatterjee S, Lee JY. Mechanistic Insights into the Polymorphic Associations and Cross-Seeding of Aβ and hIAPP in the Presence of Histidine Tautomerism: An All-Atom Molecular Dynamic Study. International Journal of Molecular Sciences. 2022; 23(4):1930. https://doi.org/10.3390/ijms23041930
Chicago/Turabian StyleSalimi, Abbas, Sompriya Chatterjee, and Jin Yong Lee. 2022. "Mechanistic Insights into the Polymorphic Associations and Cross-Seeding of Aβ and hIAPP in the Presence of Histidine Tautomerism: An All-Atom Molecular Dynamic Study" International Journal of Molecular Sciences 23, no. 4: 1930. https://doi.org/10.3390/ijms23041930
APA StyleSalimi, A., Chatterjee, S., & Lee, J. Y. (2022). Mechanistic Insights into the Polymorphic Associations and Cross-Seeding of Aβ and hIAPP in the Presence of Histidine Tautomerism: An All-Atom Molecular Dynamic Study. International Journal of Molecular Sciences, 23(4), 1930. https://doi.org/10.3390/ijms23041930