Functional Characterization of MdTAC1a Gene Related to Branch Angle in Apple (Malus x domestica Borkh.)
Abstract
1. Introduction
2. Results
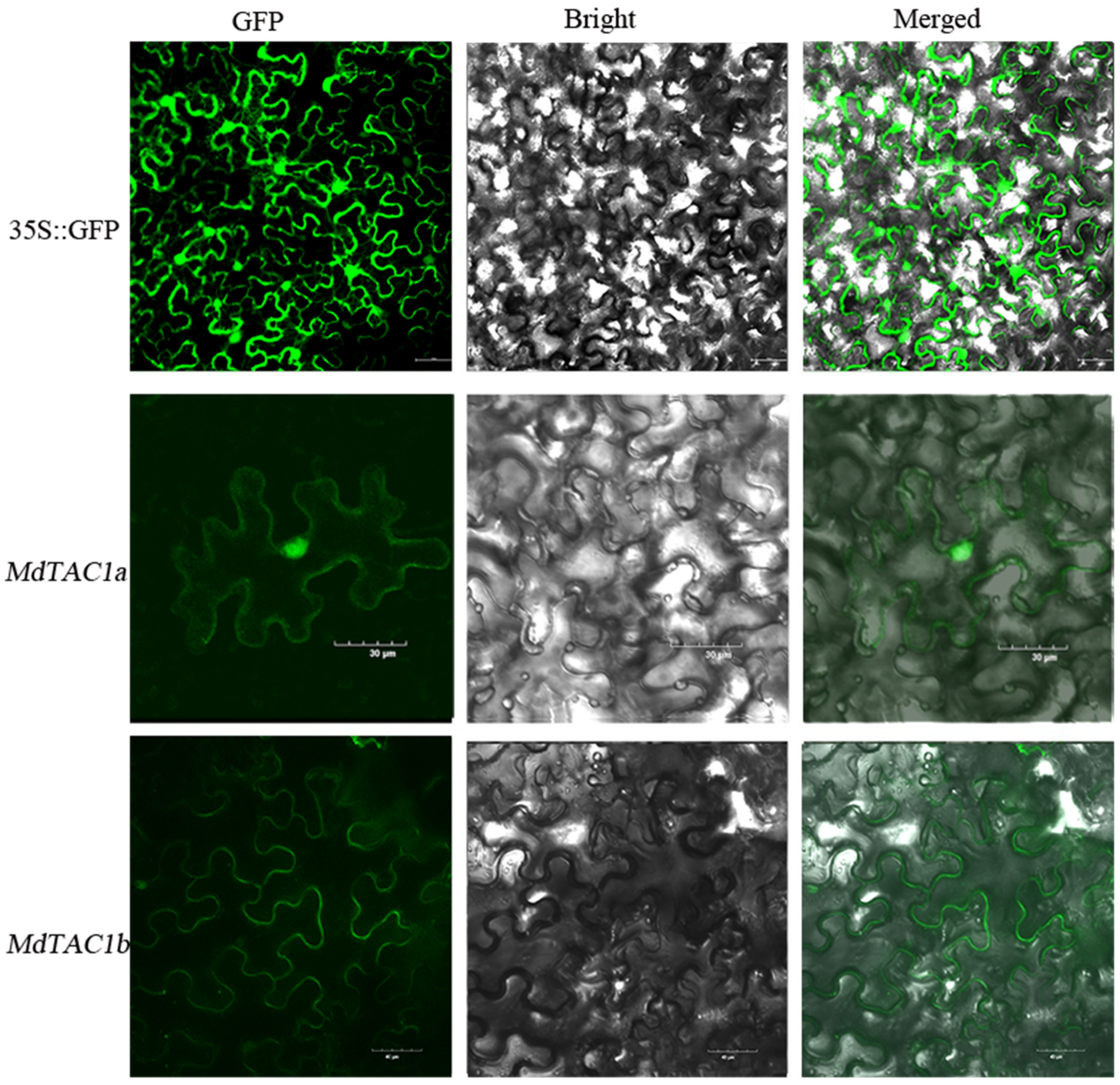
2.1. Subcellular Localization Analysis of MdTAC1a and MdTAC1b
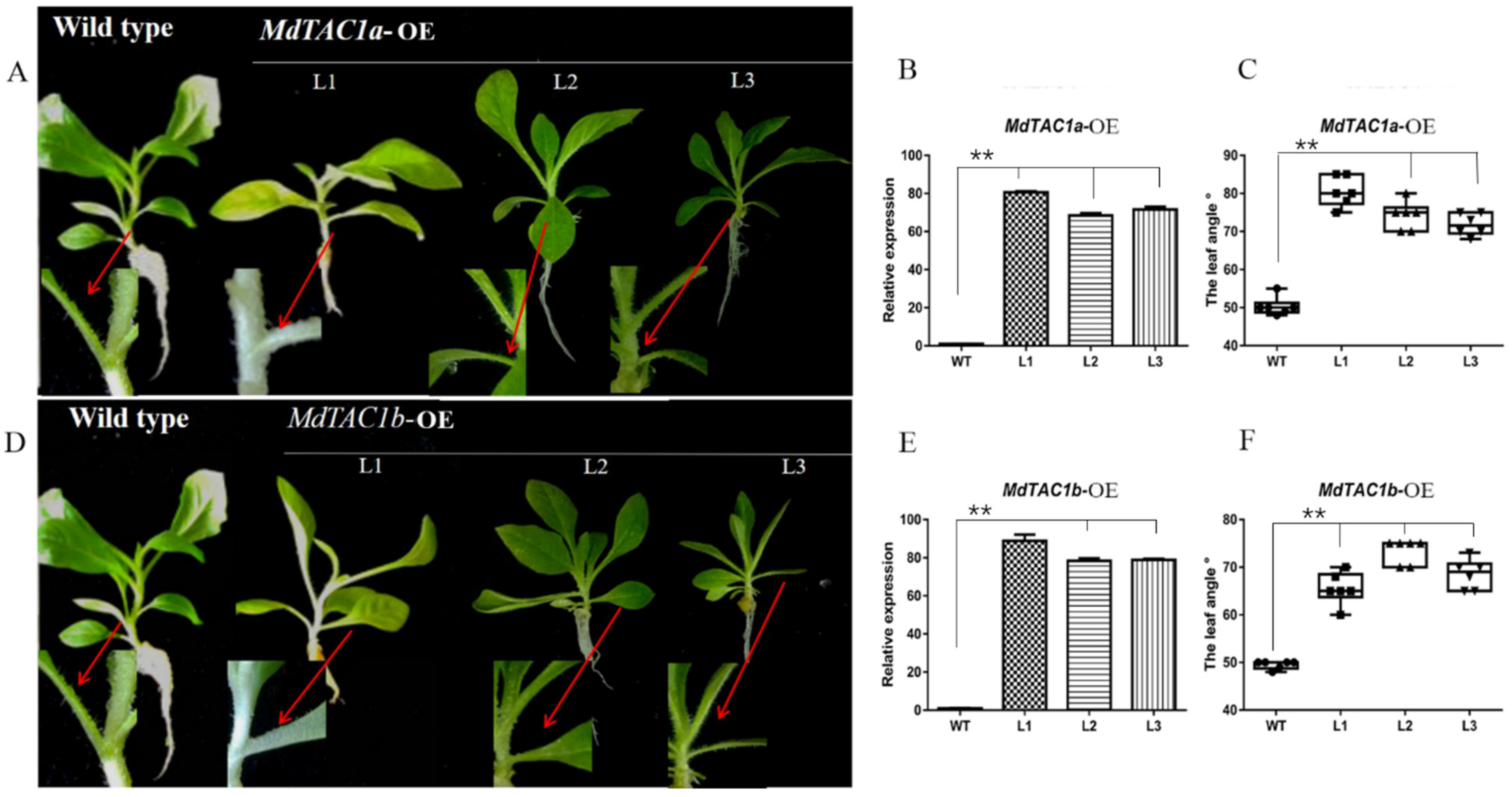
2.2. Phenotypic and Key Gene Analysis of Transgenic MdTAC1a/b in Tobacco
2.3. Phototropism and Gravitropism of MdTAC1-OE Plants
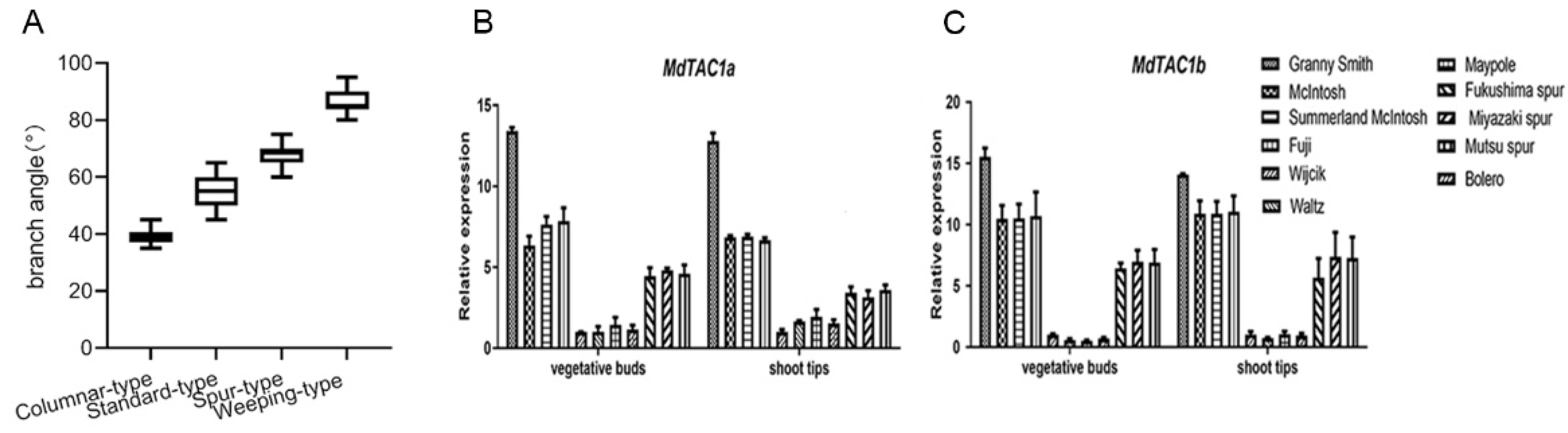
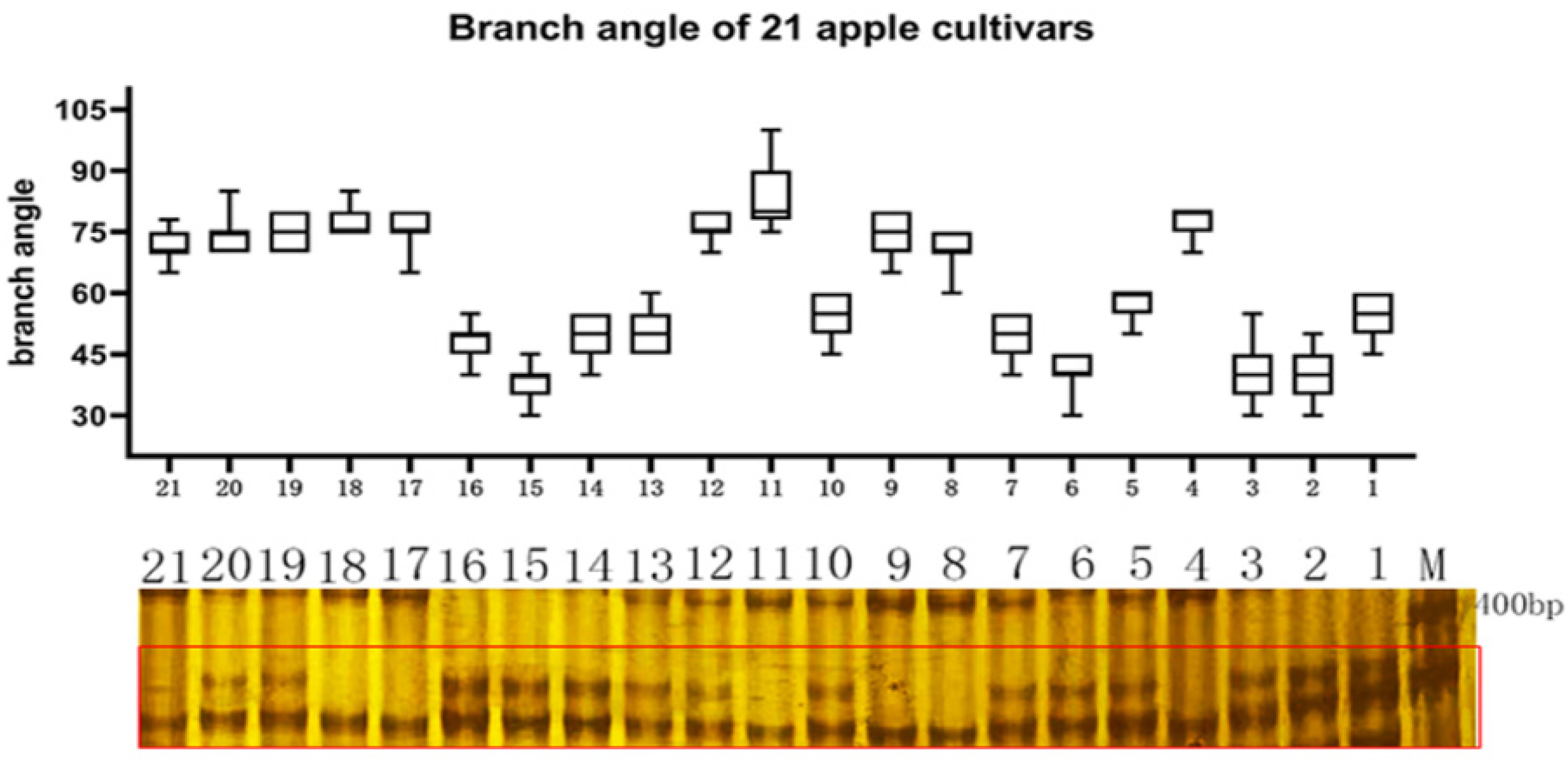
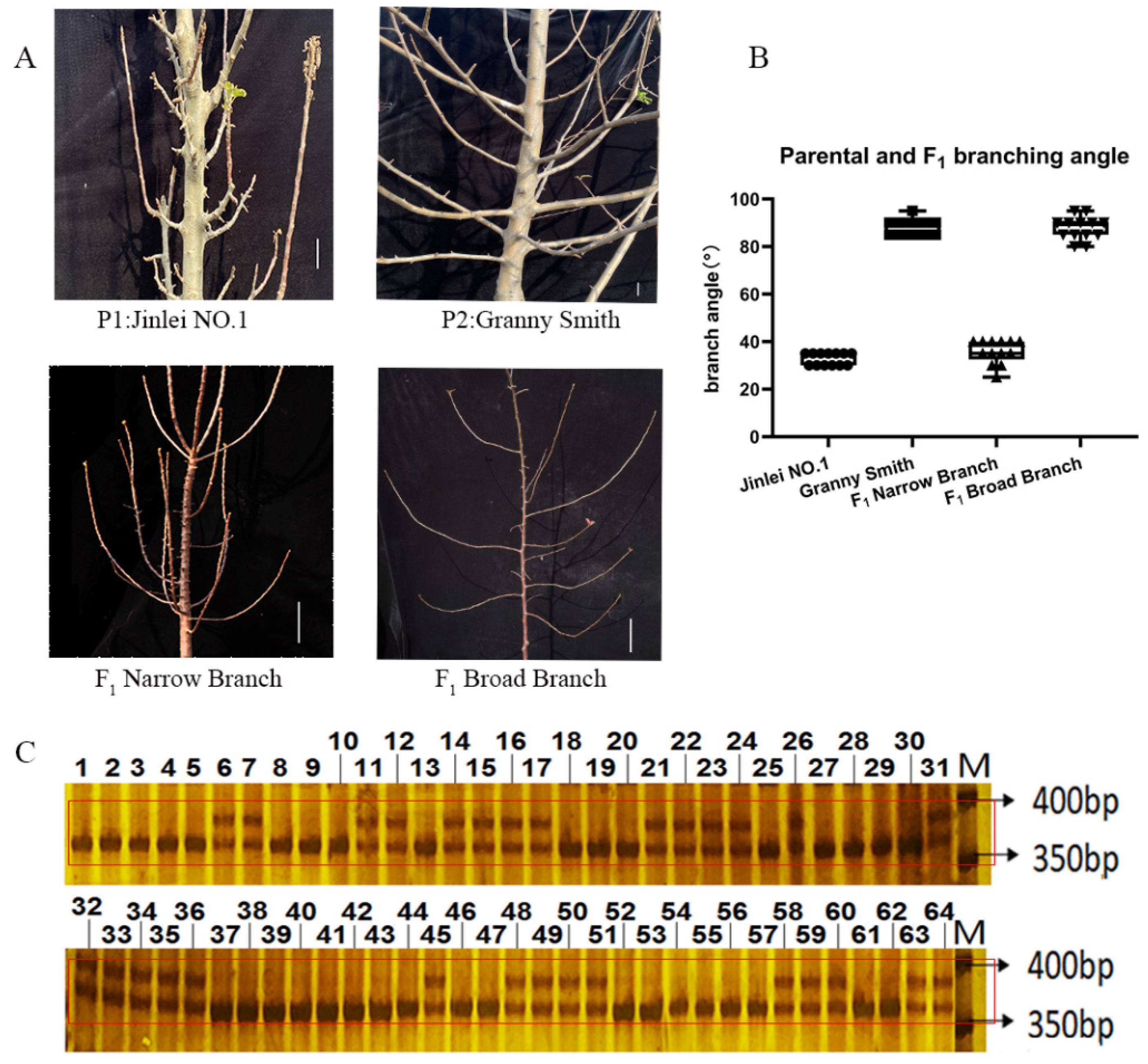
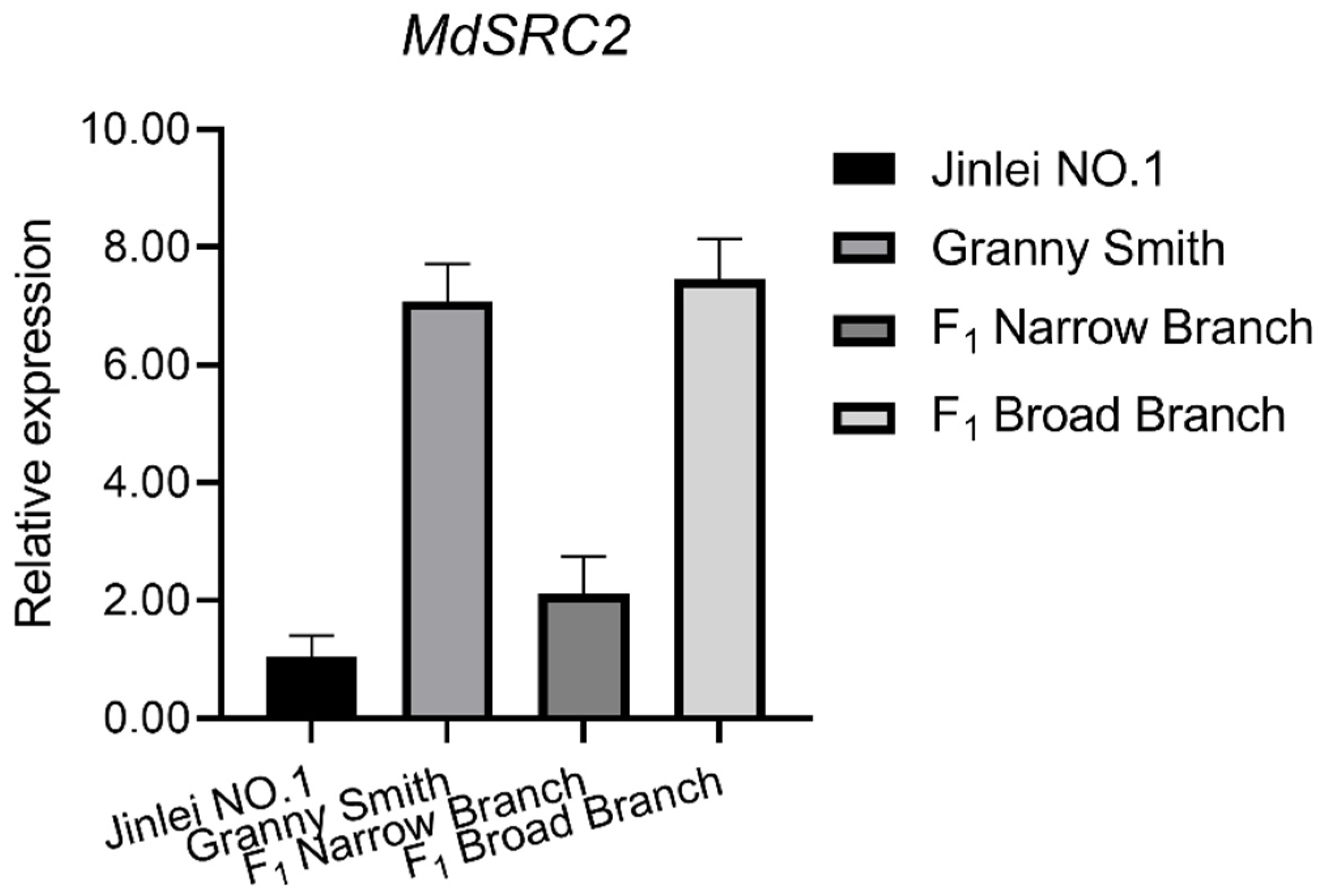
2.4. qRT-PCR Analysis of MdTAC1a/b Expressions in Different Apple Cultivars
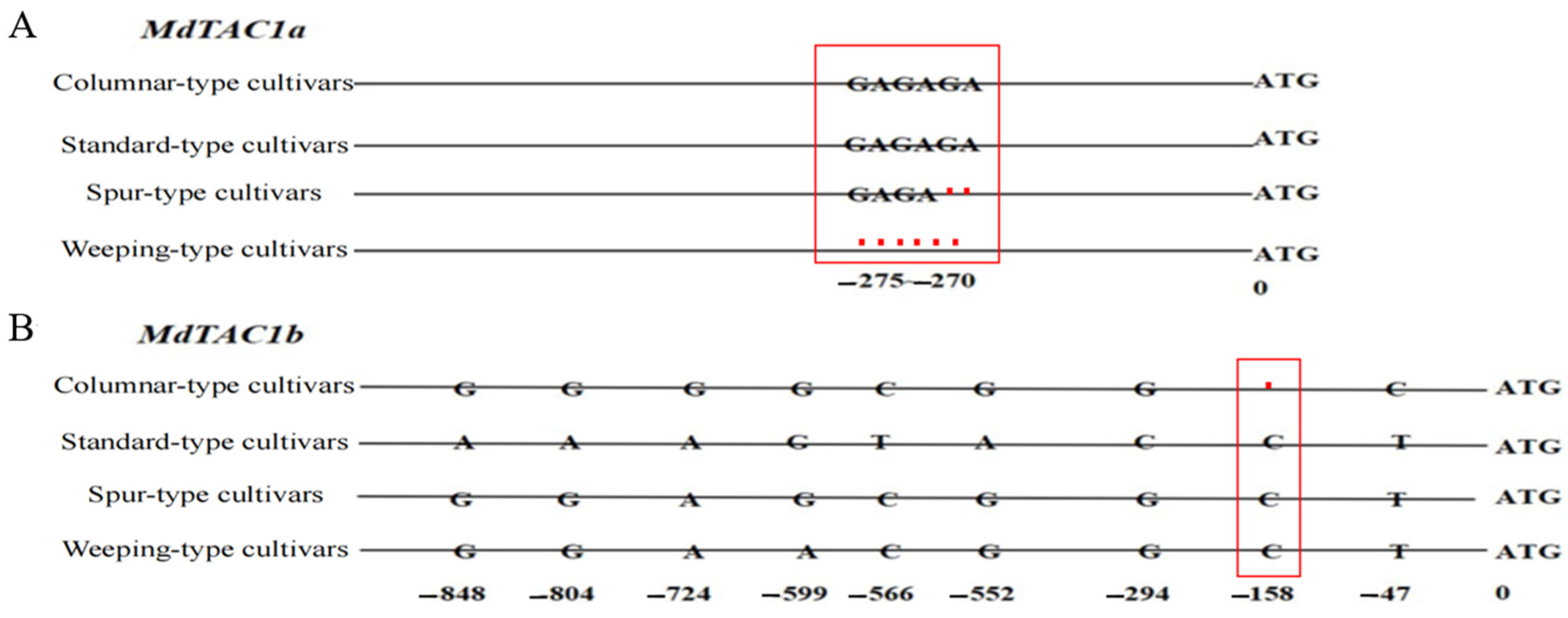
2.5. Development of an InDel Marker and Cosegregation in a Population
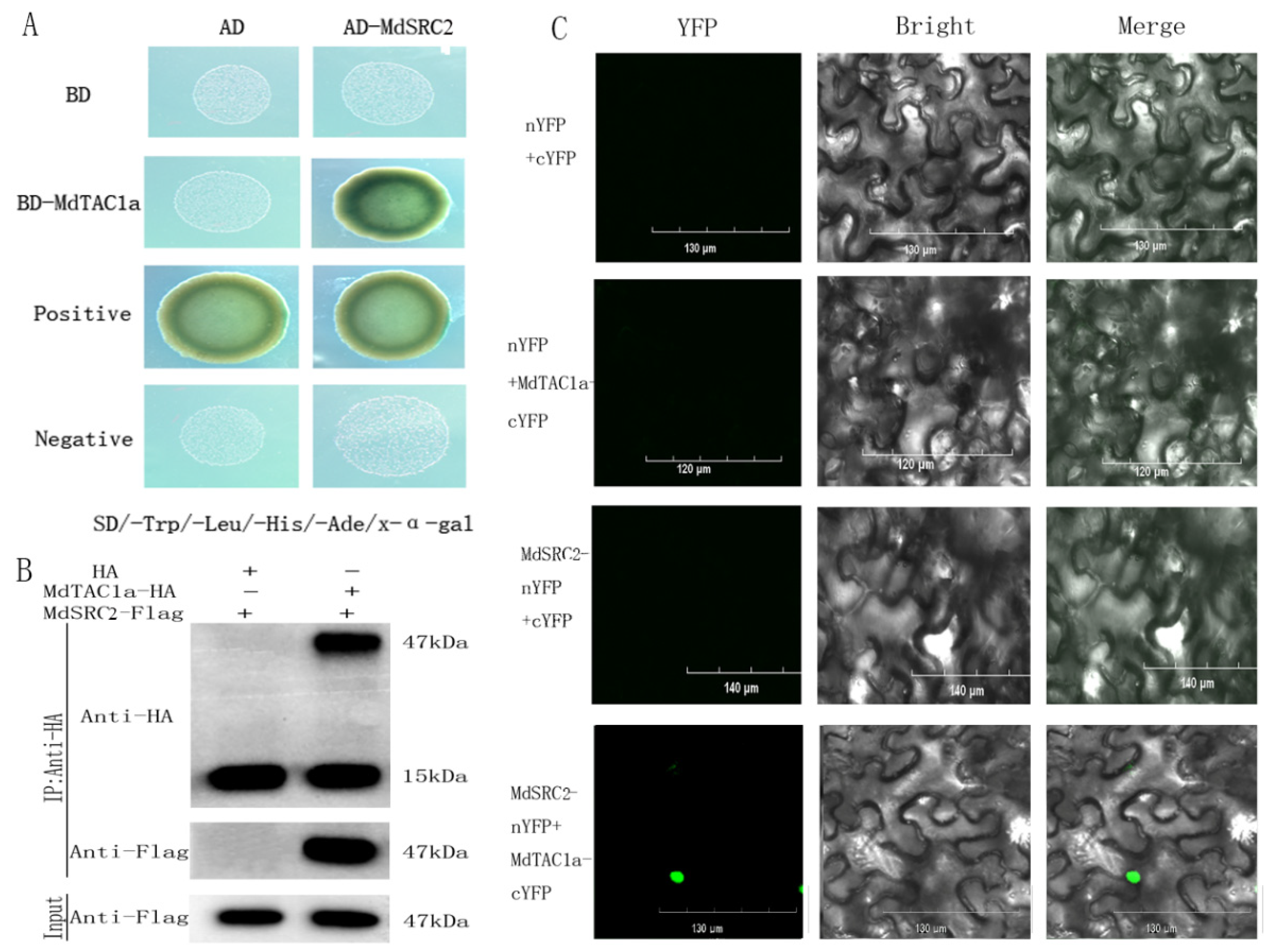
2.6. Interaction of MdTAC1a with MdSRC2
3. Discussion
3.1. Physiological Functions of MdTAC1 and MdTAC1-OE in Tobacco
3.2. Relationship of Growth and Tropic Responses in Transgenic Tobacco
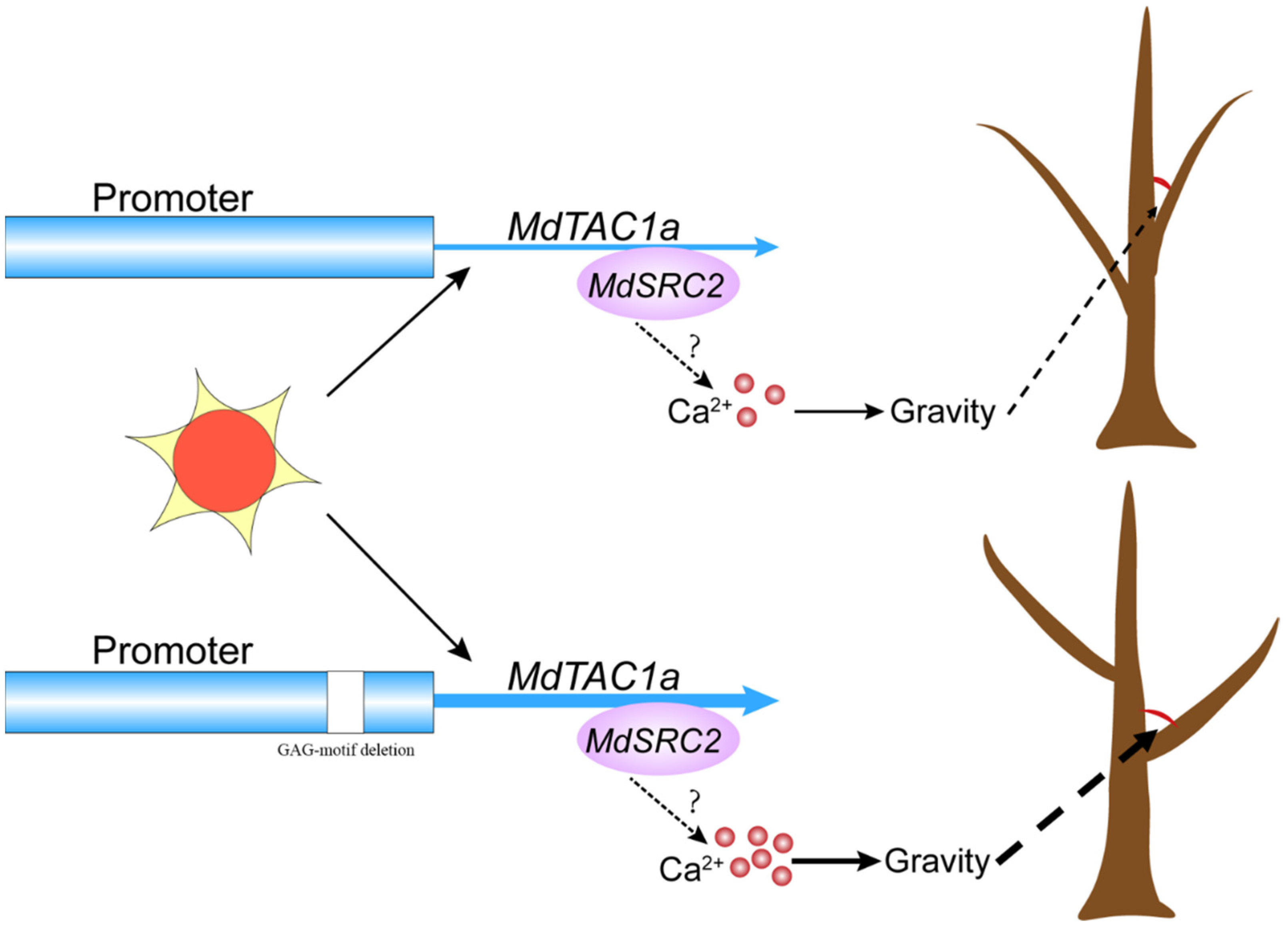
3.3. Variation in the MdTAC1a Promoter among Cultivars and F1 Generation Plants
3.4. The Effect of MdSRC2–MdTAC1a Interaction on Plant Branch Angle
4. Materials and Methods
4.1. Plant Materials
4.2. DNA and RNA Extraction
4.3. Real-Time Quantitative PCR (qRT-PCR) Analysis
4.4. Generation of MdTAC1a/b–GFP Fusion Constructs, Transformation of Tobacco, and Subcellular Localization of MdTAC1a/b Protein
4.5. InDel Marker Development and Genetic Correlation Analysis
4.6. Yeast Two-Hybrid Assay (Y2H), Bimolecular Fluorescence Complementation (BiFC) Analysis, and Co-Immunoprecipitation (Co-IP)
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lespinass, Y. Breeding apple tree: Aims and methods. In Proceedings of the Joint Conference of the EAPR Breeding & Varietal Assessment Section and the EUCARPIA Potato Section, Landerneau, France, 12–17 January 1992; Rousselle-bourgeois, F., Rousselle, P., Eds.; Acdemic Press: Landerneau, France, 1992; pp. 103–110. [Google Scholar]
- Reinhardt, D.; Kuhlemeier, C. Plant architecture. EMBO Rep. 2002, 3, 846–851. [Google Scholar] [CrossRef] [PubMed]
- Costes, E.; Lauri, P.E.; Regnard, J.L. Analyzing fruit tree architecture: Implications for tree management and fruit production. Hortic. Rev. 2010, 32, 1–61. [Google Scholar]
- Bai, F.; Reinheimer, R.; Durantini, D.; Kellogg, E.A.; Schmidt, R.J. TCP transcription factor, BRANCH ANGLE DEFECTIVE 1 (BAD1), is required for normal tassel branch angle formation in maize. Proc. Natl. Acad. Sci. USA 2012, 109, 12225–12230. [Google Scholar] [CrossRef] [PubMed]
- Hollender, C.A.; Dardick, C. Molecular basis of angiosperm tree architecture. New Phytol. 2015, 206, 541–556. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.B.; Jiang, C.; Chen, X.Y.; Zhang, T.; Ding, L.; Song, W.B.; Luo, H.B.; Lai, J.S.; Chen, H.B.; Liu, R.Y.; et al. Maize LAZY1 mediates shoot gravitropism and inflorescence development through regulating auxin transport, auxin signaling, and light response. Plant Physiol. 2013, 163, 1306–1322. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, T.; Iino, M. Identification of the gravitropism-related rice gene LAZY1 and elucidation of LAZY1-dependent and -independent gravity signaling pathways. Plant Cell Physiol. 2007, 48, 678–688. [Google Scholar] [CrossRef]
- Li, X.Y.; Qian, Q.; Fu, Z.M. Control of tillering in rice. Nature 2003, 422, 618–621. [Google Scholar] [CrossRef]
- Ongaro, V.; Leyser, O. Shoot branching in arabidopsis. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2007, 146, S237–S238. [Google Scholar] [CrossRef]
- Sang, D.; Chen, D.; Liu, G.; Liang, Y.; Huang, L.Z.; Meng, X.B.; Chu, J.F.; Sun, X.H.; Dong, G.J.; Yuan, Y.D.; et al. Strigolactones regulate rice tiller angle by attenuating shoot gravitropism through inhibiting auxin biosynthesis. Proc. Natl. Acad. Sci. USA 2014, 111, 11199–11204. [Google Scholar] [CrossRef]
- Li, Y.; Li, J.; Chen, Z.; Wei, Y.; Qi, Y.H.; Wu, C.Y. Osmir167a-targeted auxin response factors modulate tiller angle via fine-tuning auxin distribution in rice. Plant Biotechnol. J. 2020, 18, 2015–2026. [Google Scholar] [CrossRef]
- Busov, V.B.; Strauss, B.S.H. Genes for control of plant stature and form. New Phytol. 2010, 177, 589–607. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.S.; Lin, Z.W.; Li, H.X.; Li, X.J.; Li, J.Y.; Wang, Y.H.; Zhang, X.; Zhu, Z.F.; Zhai, W.X.; Wang, X.K.; et al. TAC1, a major quantitative trait locus controlling tiller angle in rice. Plant J. 2007, 52, 891–898. [Google Scholar] [CrossRef] [PubMed]
- Dardick, C.; Callahan, A.; Horn, R.; Ruiz, K.B.; Zhebentyayeva, T.; Hollender, C.; Whitaker, M.; Abbott, A.; Scorza, R. PpeTAC1 promotes the horizontal growth of branches in peach trees and is a member of a functionally conserved gene family found in diverse plants species. Plant J. 2013, 75, 618–630. [Google Scholar] [CrossRef]
- Ku, L.X.; Wei, X.M.; Zhang, S.F. Cloning and Characterization of a Putative TAC1 Ortholog Associated with Leaf Angle in Maize (Zea mays L.). PLoS ONE 2011, 6, e20621. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.H.; Tan, L.B.; Zhu, Z.F.; Fu, Y.C.; Liu, F.X.; Cai, H.W.; Sun, C.Q. Molecular evolution of the TAC1 gene from rice (Oryza sativa L.). J. Genet. Genom. 2012, 39, 551–560. [Google Scholar] [CrossRef]
- Cao, X.; Deng, M.; Zhang, Z.L.; Liu, Y.J.; Yang, X.L.; Zhou, H.; Liu, Y.X. Molecular Characterization and Expression Analysis of TaTAC1 gene in Triticum aestivum L. J. Plant Genet. Resour. 2017, 18, 125–132. [Google Scholar]
- Hollender, C.A.; Waite, J.M.; Tabb, A.; Raines, D.; Chinnithambi, S.; Dardick, C. Alteration of TAC1 expression in Prunus species leads to pleiotropic shoot phenotypes. Hortic. Res. 2018, 5, 26. [Google Scholar] [CrossRef]
- Xu, D.; Qi, X.; Li, J.H.; Han, X.J.; Wang, J.N.; Jiang, Y.Z.; Tian, Y.T.; Wang, Y.W. PzTAC and PzLAZY from a narrow-crown poplar contribute to regulation of branch angles. Plant Physiol. Biochem. 2017, 118, 571–578. [Google Scholar] [CrossRef]
- Wang, L.M.; Cai, W.; Du, C.H.; Fu, Y.; Xie, X.; Zhu, Y.D. The isolation of the IGT family genes in Malus × domestica and their expressions in four idiotype apple cultivars. Tree Genet. Genomes 2018, 14, 46. [Google Scholar] [CrossRef]
- Takahashi, R.; Shimosaka, E. cDNA sequence analysis and expression of two cold-regulated genes in soybean. Plant Sci. 1997, 123, 93–104. [Google Scholar] [CrossRef]
- Yuan, M.R.; Yang, Y.; Zhang, L.; He, Y.J.; Jin, N.; Yang, H.F. Cloning, Bioinformatics and Tissue-specific Expression Analysis of Sps-TAC2 Gene in Salix psammophila. Mol. Plant Breed. 2019, 17, 2133–2151. [Google Scholar]
- Matthias, F. Targeted CRISPR/Cas9-Based Knock-Out of the Rice Orthologs TILLER ANGLE CONTROL 1 (TAC1) in Poplar Induces Erect Leaf Habit and Shoot Growth. Forests 2021, 12, 1615. [Google Scholar]
- Petersen, R.; Krost, C. Tracing a key player in the regulation of plant architecture: The columnar growth habit of apple trees (Malus × domestica). Planta 2013, 238, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Li, G.; Qi, S.; Liu, X.J.; Chen, X.L.; Ma, J.J.; Zhang, D.; Han, M.Y. Identification and expression analysis of the IPT and CKX gene families during axillary bud outgrowth in apple (Malus domestica Borkh.). Gene 2018, 651, 106–117. [Google Scholar] [CrossRef]
- Cheng, X.; Ruyterspira, C.; Bouwmeester, H. The interaction between strigolactones and other plant hormones in the regulation of plant development. Front. Plant Sci. 2013, 4, 199. [Google Scholar] [CrossRef]
- Dayan, J.; Voronin, N.; Gong, F.; Sun, T.; Hedden, P.; Fromm, H.; Aloni, R. Leaf-induced gibberellin signaling is essential for internode elongation, cambial activity, and fiber differentiation in tobacco stems. Plant Cell 2012, 24, 66–79. [Google Scholar] [CrossRef]
- Hedden, P.; Phillips, A.L. Gibberellin metabolism: New insights revealed by the genes. Trends Plant Sci. 2000, 5, 523–530. [Google Scholar] [CrossRef]
- Yamaguchi, S. Gibberellin Metabolism and its Regulation. Annu. Rev. Plant Biol. 2008, 59, 225–251. [Google Scholar] [CrossRef]
- Nakamura, T.; Saotome, M.; Ishiguro, Y.; Itoh, R.; Higurashi, S.; Hosono, M.; Ishii, Y. The effects of GA3 on weeping of growing shoots of the Japanese cherry, Prunus spachiana. Plant Cell Physiol. 1994, 35, 523–527. [Google Scholar]
- Yoshida, M.; Yamamoto, H.; Okuyama, T.; Nakamura, T. Negative gravitropism and growth stress in GA3-treated branches of Prunus spachiana Kitamura f. spachiana cv. Plenarosea. J. Wood Sci. 1999, 45, 368–372. [Google Scholar] [CrossRef]
- Jessica, M.W.; Chris, D. TILLER ANGLE CONTROL 1 modulates plant architecture in response to photosynthetic signals. J. Exp. Bot. 2018, 69, 4935–4944. [Google Scholar]
- Taniguchi, M.; Nakamura, M.; Tasaka, M.; Morita, M.T. Identification of gravitropic response indicator genes in Arabidopsis inflorescence stems. Plant Signal. Behav. 2014, 9, e29570. [Google Scholar] [CrossRef] [PubMed]
- Espley, R.V.; Brendolise, C.; Chagneé, D.; Kutty-Amma, S.; Green, S.; Volz, R.; Putterill, J.; Schouten, H.J.; Gardiner, S.E.; Hellens, R.P.; et al. Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell 2009, 21, 168–183. [Google Scholar] [CrossRef] [PubMed]
- Liscum, E.; Askinosie, S.K.; Leuchtman, D.L.; Morrow, J.; Willenburg, K.T.; Coats, D.R. Phototropism: Growing Towards an Understanding of Plant Movement. Plant Cell 2014, 26, 38–55. [Google Scholar] [CrossRef]
- Palme, K.; Dovzhenko, A.; Ditengou, F.A. Auxin transport and gravitational research: Perspectives. Protoplasma 2006, 229, 175. [Google Scholar] [CrossRef]
- Bainbridge, K.; Guyomarc’h, S.; Bayer, E.; Swarup, R.; Bennett, M.; Mandel, T.; Kuhlemeier, C. Auxin Influx Carriers Stabilize Phyllotactic Patterning. Genes Dev. 2008, 22, 810–823. [Google Scholar] [CrossRef]
- Rizo, J.; Südhof, T.C. C2-domains, structure and function of a universal Ca2+-binding domain. J. Biol. Chem. 1998, 273, 15879–15882. [Google Scholar] [CrossRef]
- Tomoko, K.; Sachie, K.; Ayako, I.; Shigeru, H.; Hitomi, N.; Masataka, M.; Aya, I.; Mitsutomo, A.; Hidetaka, K.; Kazuyuki, K. A low temperature-inducible protein AtSRC2 enhances the ROS-producing activity of NADPH oxidase AtRbohF. Biochim. Biophys. Acta 2013, 1833, 2775–2780. [Google Scholar]
- Dauwalder, M.; Roux, S.J. Distribution of calmodulin in corn seedlings: Immunocytochemical localization in coleoptiles and root apices. Adv. Space Res. 1986, 6, 67–70. [Google Scholar] [CrossRef]
- Zhang, S.; Chen, S.; Chen, F.; Jiang, B. The Role of Ionic Calcium in the Gravitropic Response of a Creeping Chrysanthemum Cultivar. Russ. J. Plant Physiol. 2011, 58, 696–702. [Google Scholar] [CrossRef]
- Heilmann, I.; Shin, J.; Huang, J.; Perera, I.Y.; Davies, E. Transient dissociation of polyribosomes and concurrent recruitment of calreticulin and calmodulin transcripts in gravistimulated maize pulvini. Plant Physiol. 2001, 127, 1193–1203. [Google Scholar] [PubMed]
- Bushart, T.J.; Cannon, A.; Clark, G.; Roux, S.J. Structure and function of CrACA1, the major PM-type Ca2+-ATPase, expressed at the peak of the gravity-directed trans-cell calcium current in spores of the fern Ceratopteris richardii. Plant Biol. 2014, 16, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Salmi, M.L.; Haque, A.U.; Bushart, T.J.; Stout, S.C.; Roux, S.J.; Porterfield, D.M. Changes in gravity rapidly alter the magnitude and direction of a cellular calcium current. Planta 2011, 233, 911–920. [Google Scholar] [CrossRef] [PubMed]
- Urbina, D.C.; Silva, H.; Meisel, L.A. The Ca2+ pump inhibitor, thapsigargin, inhibits root gravitropism in Arabidopsis thaliana. Biol. Res. 2006, 39, 289–296. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Doyle, J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1986, 19, 11–15. [Google Scholar]
- Asif, M.; Trivedi, P.; Solomos, T.; Tucker, M. Isolation of high-quality RNA from apple (Malus domestica) fruit. J. Agric. Food Chem. 2006, 54, 5227–5229. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Hofgen, R.; Willmitzer, L. Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res. 1988, 16, 9877. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory manual, 3rd ed.; Cold Spring Harbour Laboratory Press: New York, NY, USA, 2001. [Google Scholar]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Tan, X.; Guo, J.; Hu, E.; Pan, Q.; Zhao, Y.; Chu, Y.; Zhu, Y. Functional Characterization of MdTAC1a Gene Related to Branch Angle in Apple (Malus x domestica Borkh.). Int. J. Mol. Sci. 2022, 23, 1870. https://doi.org/10.3390/ijms23031870
Li Y, Tan X, Guo J, Hu E, Pan Q, Zhao Y, Chu Y, Zhu Y. Functional Characterization of MdTAC1a Gene Related to Branch Angle in Apple (Malus x domestica Borkh.). International Journal of Molecular Sciences. 2022; 23(3):1870. https://doi.org/10.3390/ijms23031870
Chicago/Turabian StyleLi, Yongzhou, Xu Tan, Jing Guo, Enyue Hu, Qi Pan, Yuan Zhao, Yu Chu, and Yuandi Zhu. 2022. "Functional Characterization of MdTAC1a Gene Related to Branch Angle in Apple (Malus x domestica Borkh.)" International Journal of Molecular Sciences 23, no. 3: 1870. https://doi.org/10.3390/ijms23031870
APA StyleLi, Y., Tan, X., Guo, J., Hu, E., Pan, Q., Zhao, Y., Chu, Y., & Zhu, Y. (2022). Functional Characterization of MdTAC1a Gene Related to Branch Angle in Apple (Malus x domestica Borkh.). International Journal of Molecular Sciences, 23(3), 1870. https://doi.org/10.3390/ijms23031870





