Overexpression of a Malus baccata MYB Transcription Factor Gene MbMYB4 Increases Cold and Drought Tolerance in Arabidopsis thaliana
Abstract
:1. Introduction
2. Results
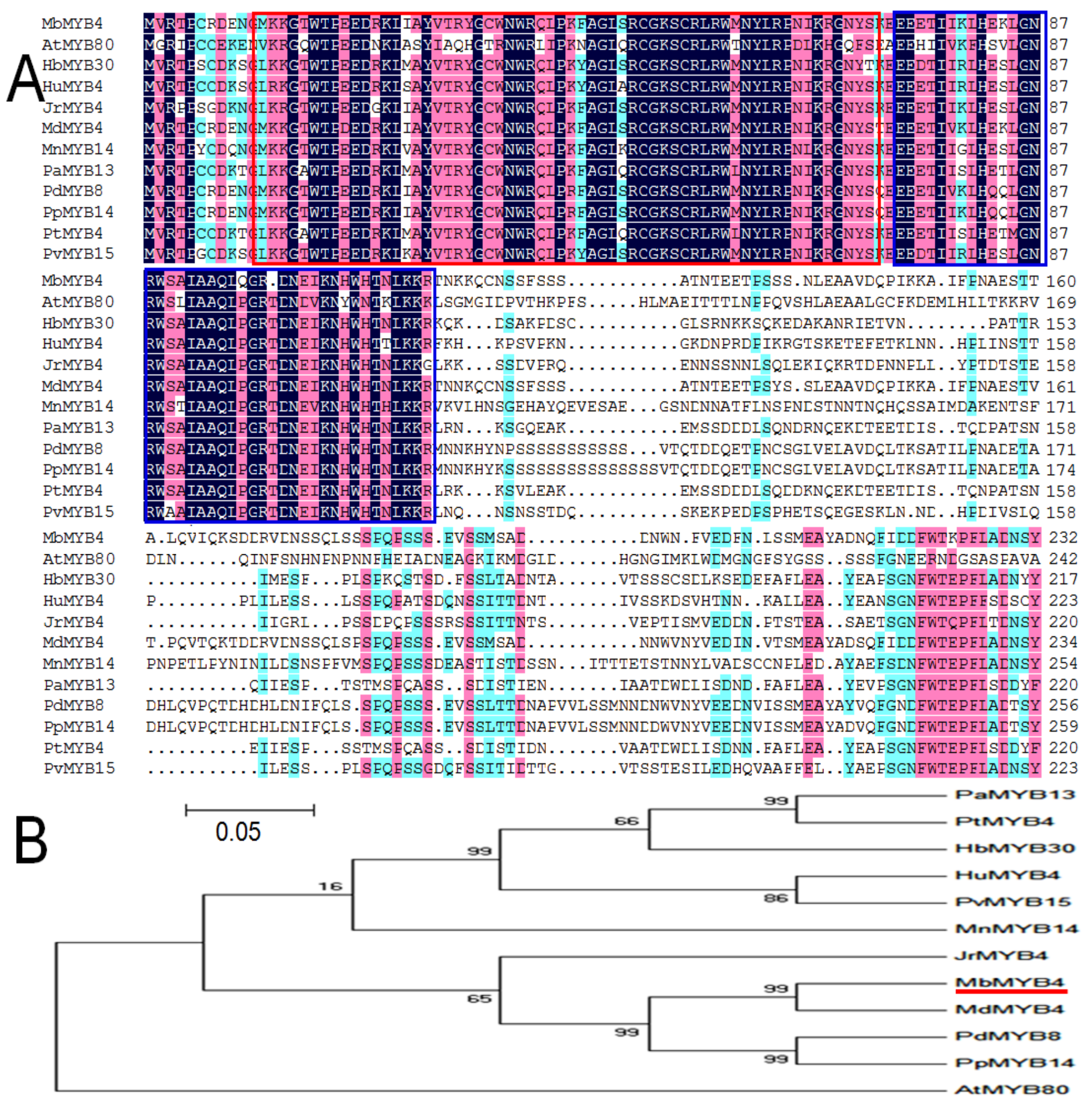
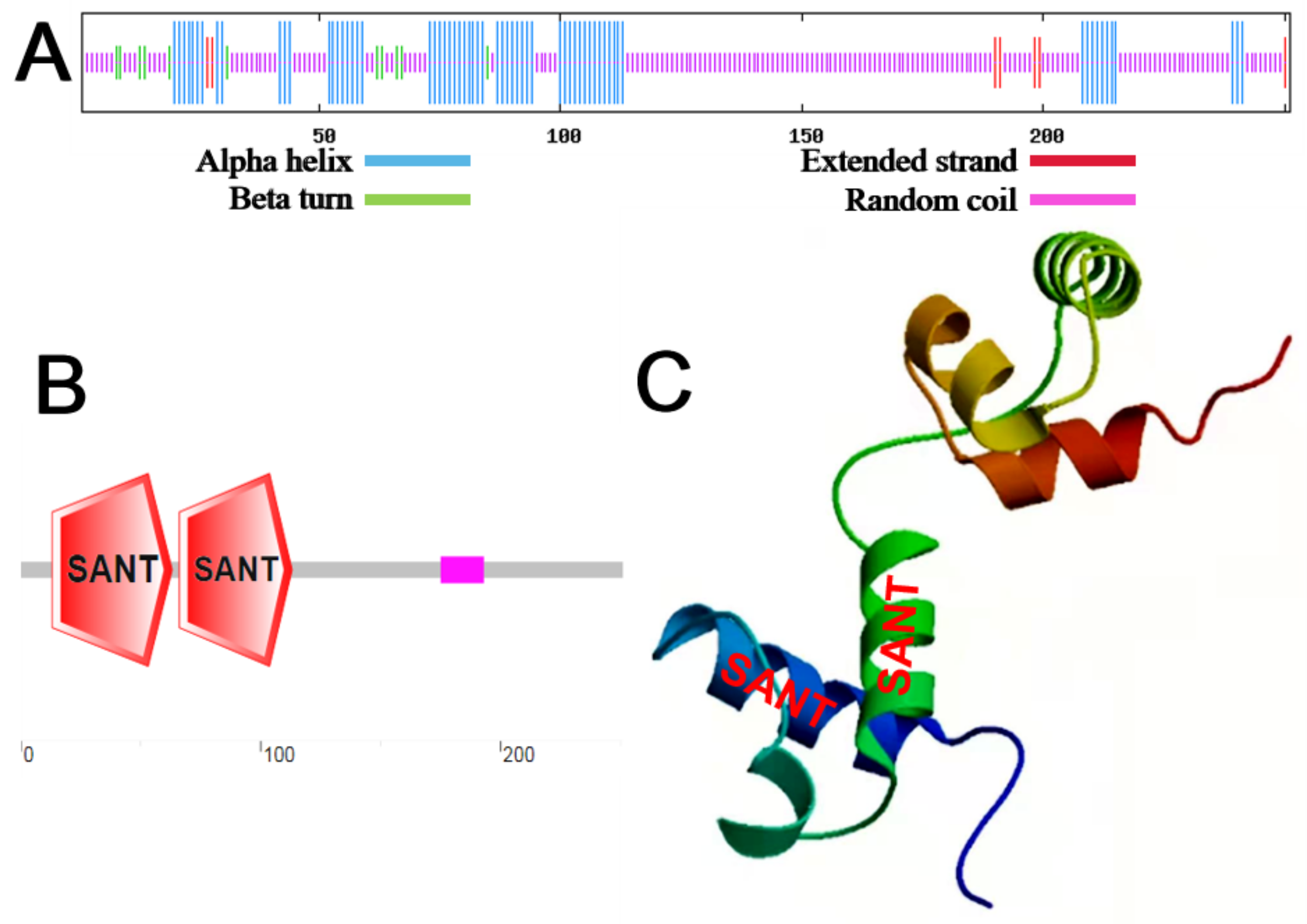
2.1. Cloning and Bioinformatics Analysis of MbMYB4
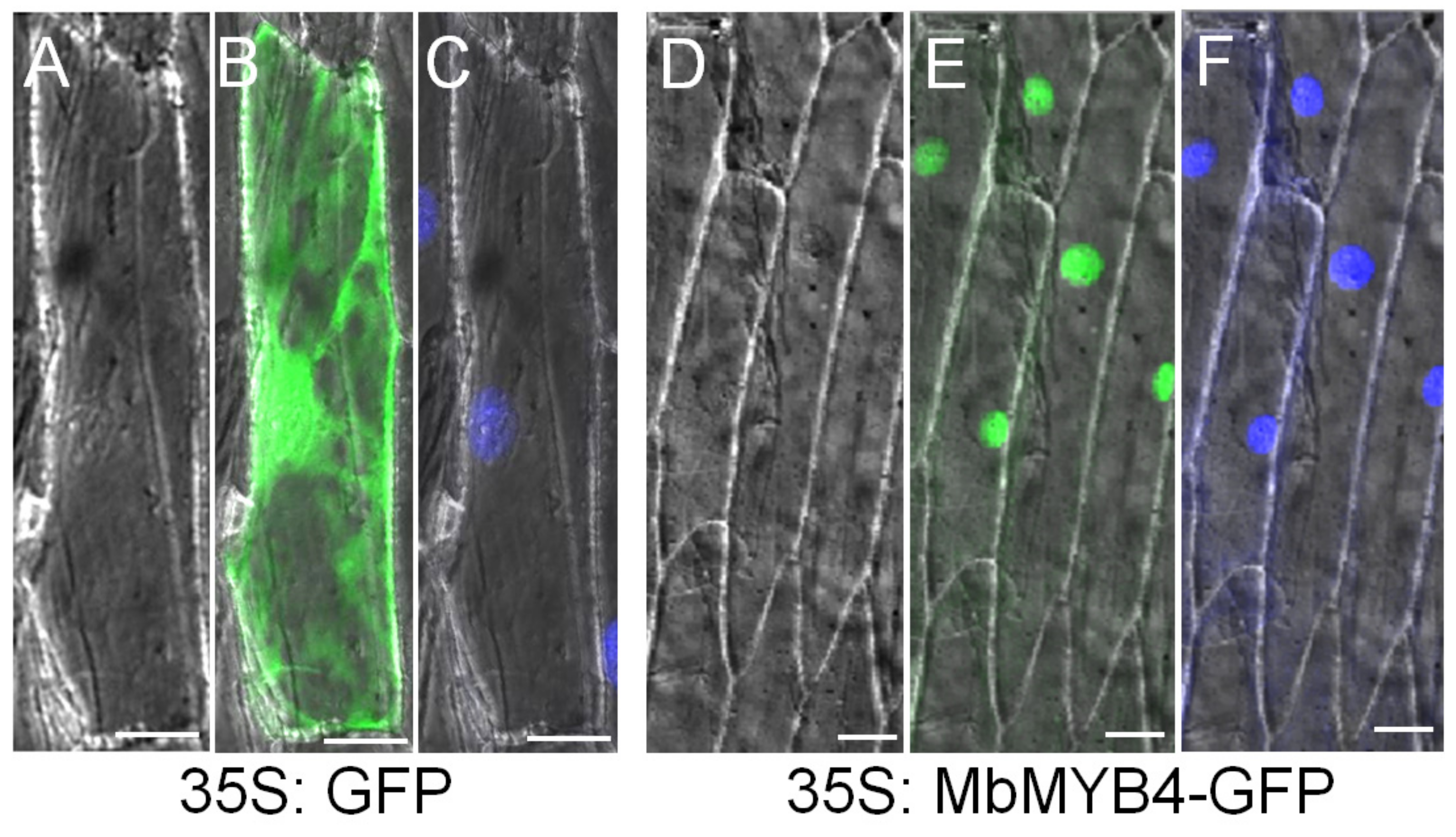
2.2. Subcellular Localization of MbMYB4 Protein
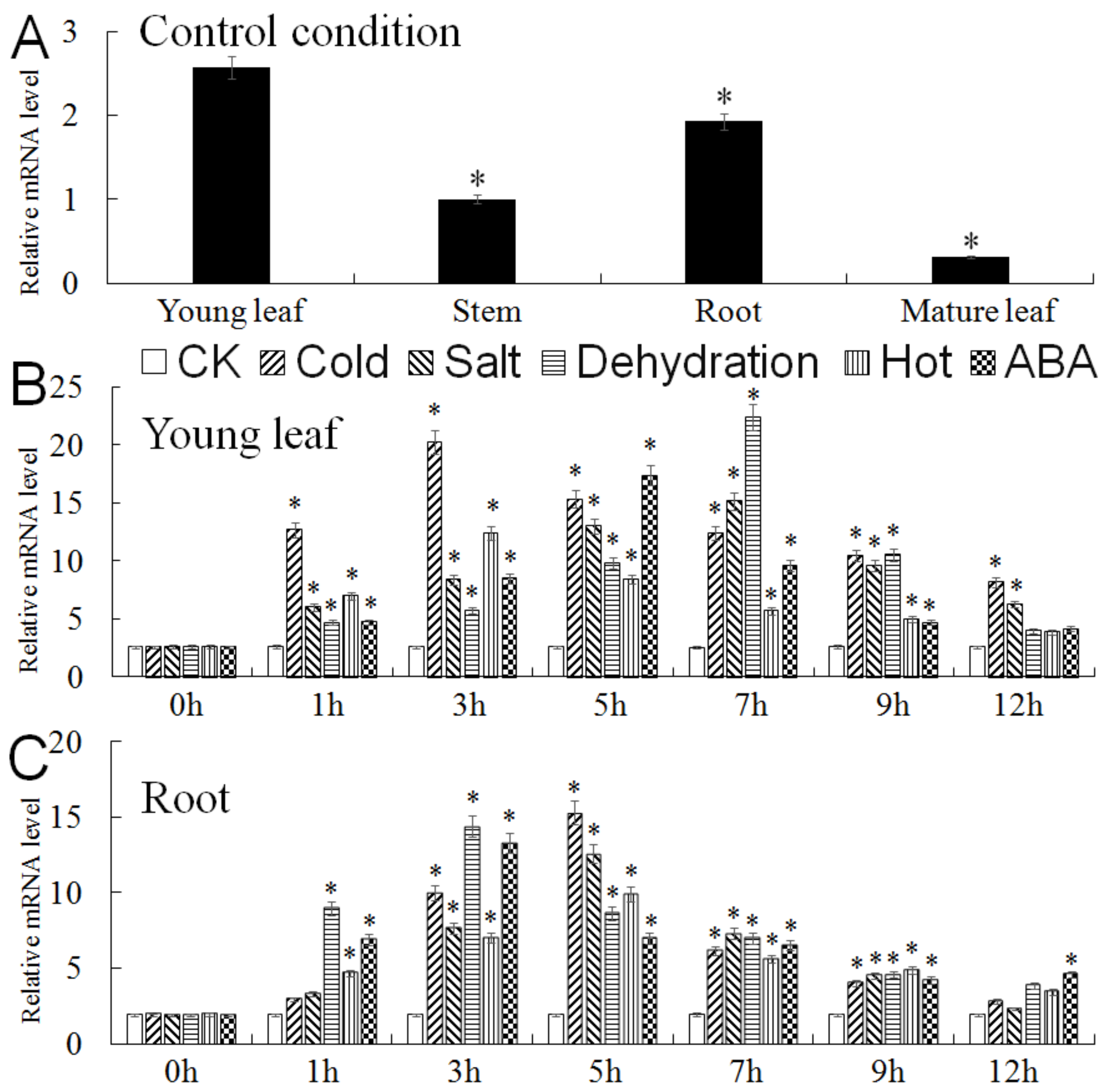
2.3. Expression Analysis of MbMYB4 in Malus baccata
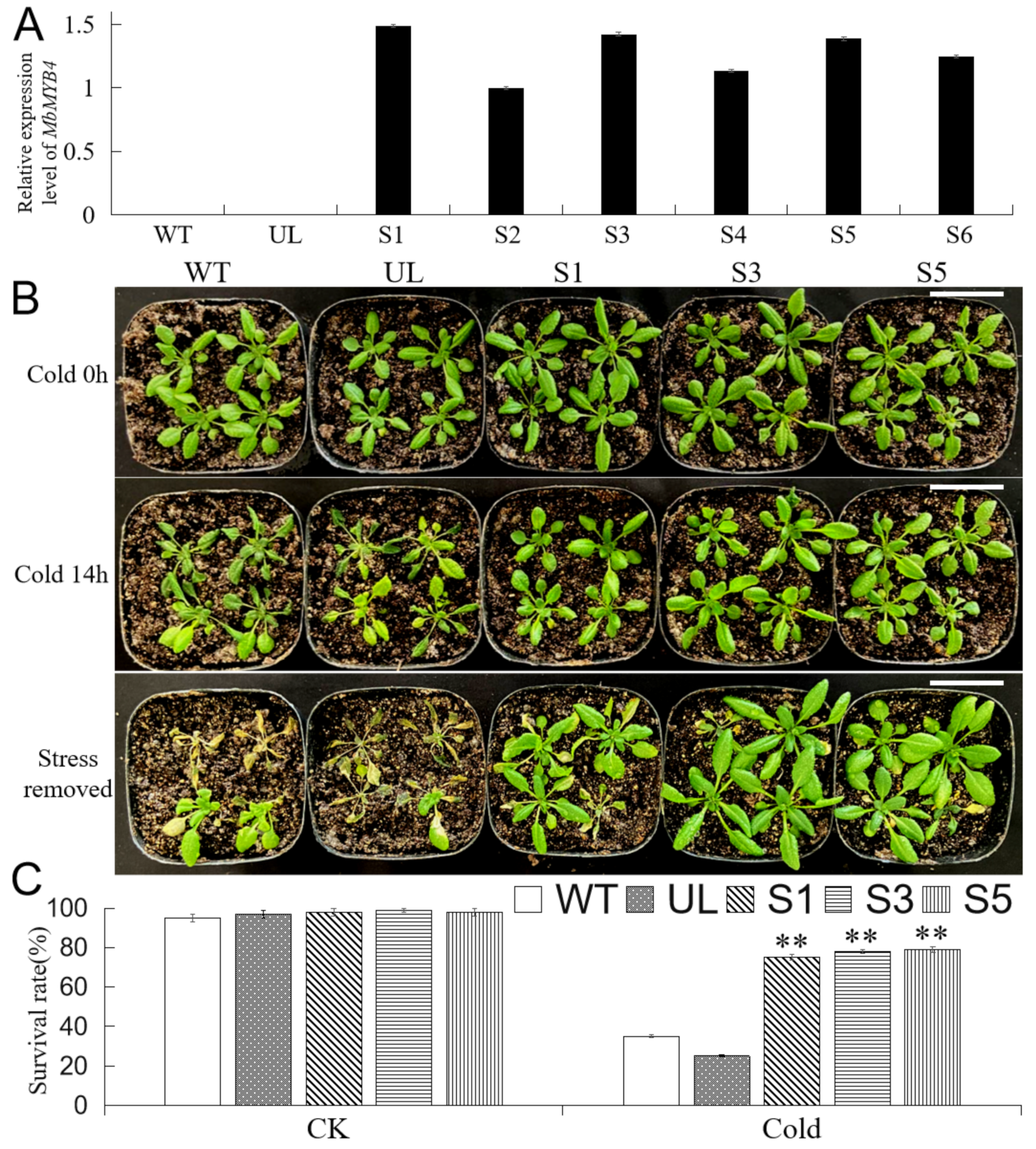
2.4. Overexpression of MbMYB4 in A. thaliana Improved Cold Tolerance
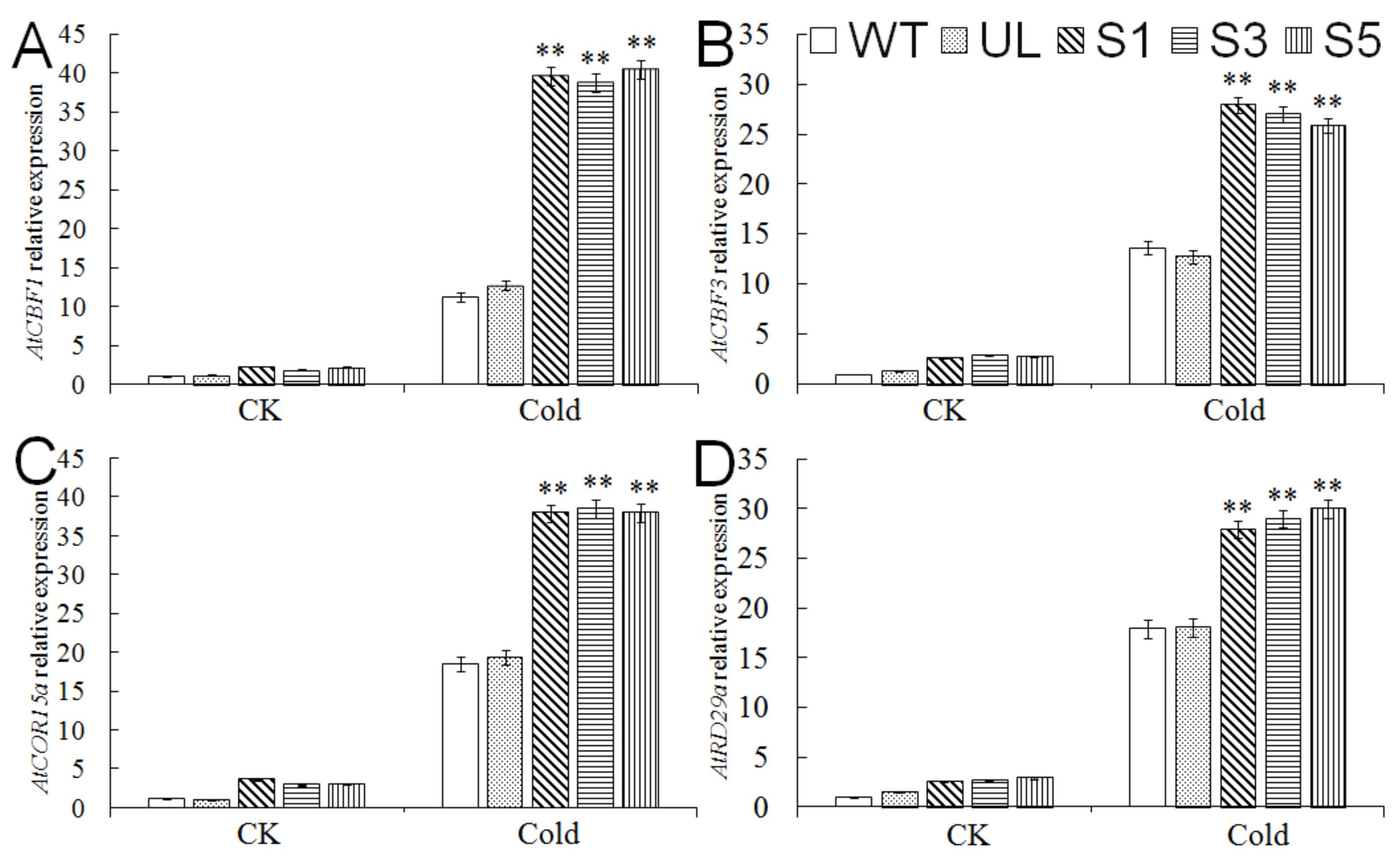
2.5. Expression Analysis of Cold Tolerance-Related Genes in A. thaliana Overexpressing MbMYB4
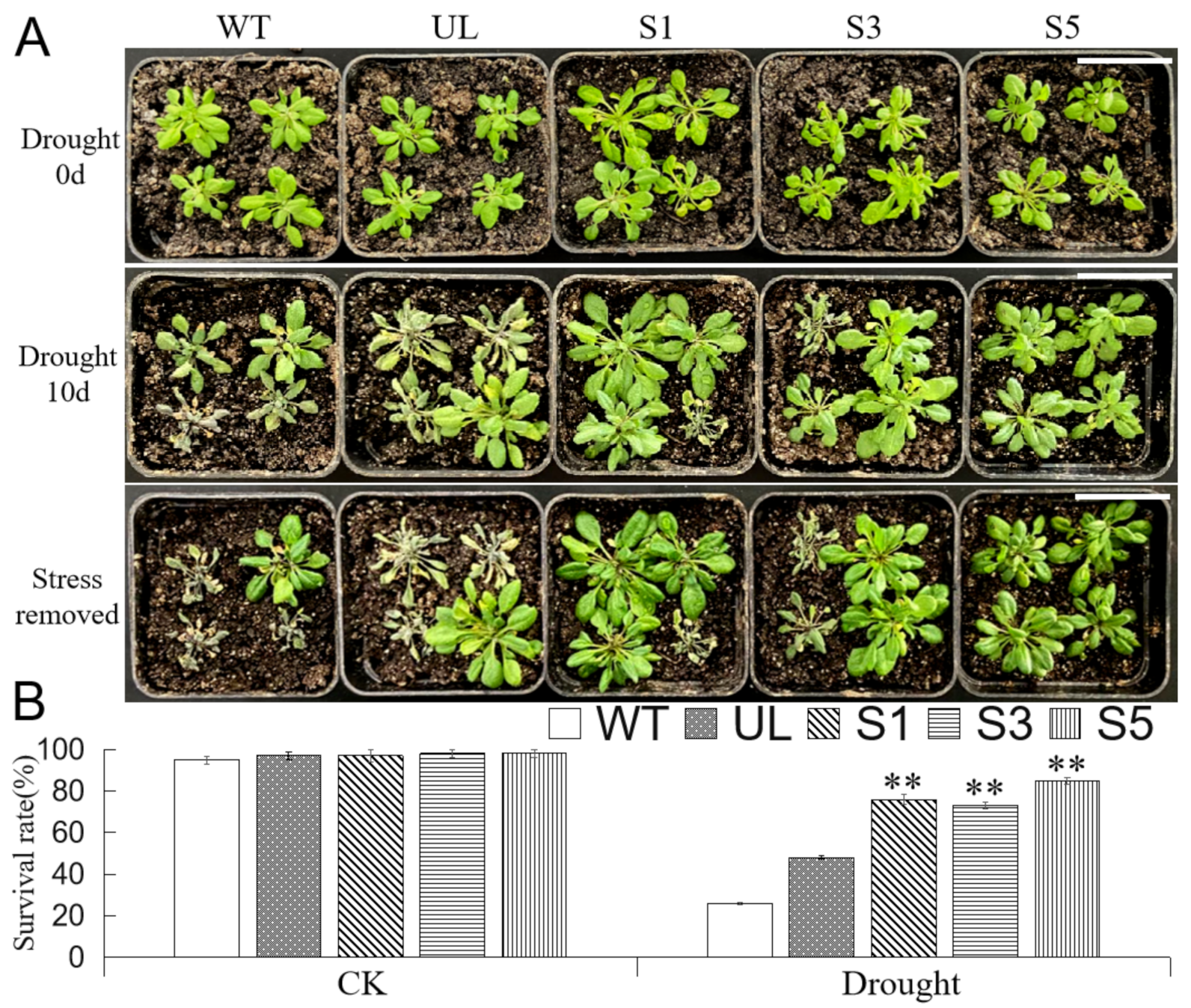
2.6. Overexpression of MbMYB4 in A. thaliana Improved Drought Tolerance
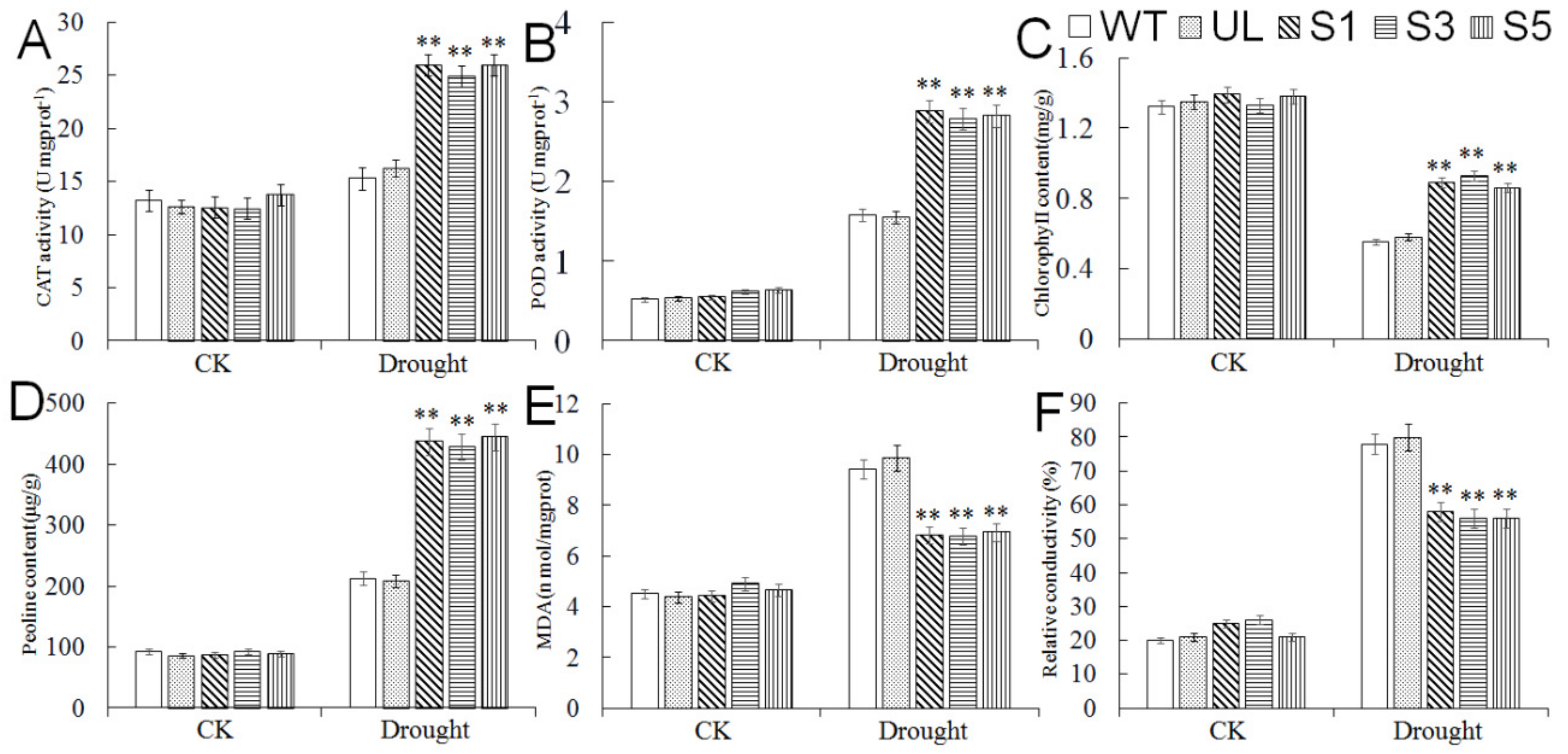
2.7. Expression Analysis of Drought Tolerance-Related Genes in A. thaliana Overexpressing MbMYB4
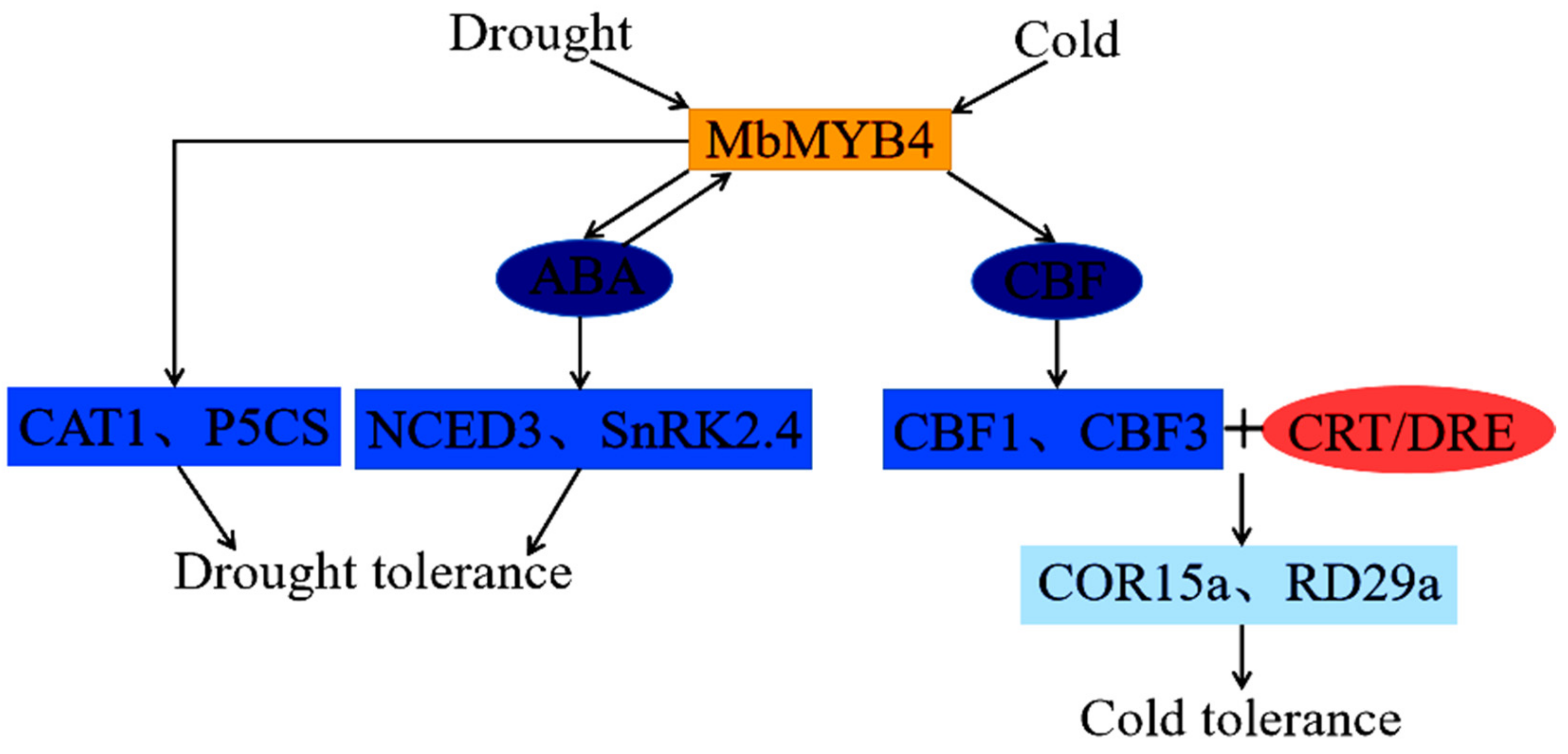
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Isolation and Cloning of MbMYB4
4.3. Bioinformatics Analysis of MbMYB4
4.4. Subcellular Localization Analysis of MbMYB4 Protein
4.5. Quantitative Real-Time PCR (qPCR) Analysis of MbMYB4
4.6. Vector Construction and Agrobacterium-Mediated A. thaliana Transformation
4.7. Determination of Related Physiological Indicators
4.8. Analysis of Downstream Gene Expression of MbMYB4
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, Y.X.; Fu, L.S.; Wang, X.N.; Wang, X.N.; Li, Z.F. Effect of freezing stress on membrane permeability and MDA content in the regrowth plant of winter wheat culticars. J. Northeast Agric. Univ. 2010, 41, 10–16. [Google Scholar]
- Liu, L.; White, M.J.; MacRae, T.H. Transcription factors and their genes in higher plants: Functional domains evolution and regulation. Eur. J. Biochem. 1999, 262, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Guilfoyle, T.J. The structure of plant gene promoters. Genet. Engin. 1997, 19, 15–47. [Google Scholar]
- Grotewold, E.; Chamberlin, M.; Snook, M.; Siame, B.; Butler, L.; Swenson, J.; Maddock, S.; Clair, G.S.; Bowen, B. Engineering secondary metabolism in maize cells by ectopic expression of transcription factors. Plant Cell. 1998, 10, 721–740. [Google Scholar] [CrossRef] [PubMed]
- Romero, I.; Fuertes, A.; Benito, M.J.; Malpical, J.M.; Leyva, A.; Paz-Ares, J. More than 80 R2R3-MYB regulatory genes in the genome of Arabidopsis thaliana. Plant J. 1998, 14, 273–284. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.Z.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis amongeukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Ambawat, S.; Sharma, P.; Yadav, N.R.; Yadav, R.C. MYB transcription factor genes as regulators for plant responses: An overview. Physiol. Mol. Biol. Plants 2013, 19, 307–321. [Google Scholar] [CrossRef] [Green Version]
- Shi, M.Z.; Xie, D.Y. Biosynthesis and metabolic engineering of anthocyanins in Arabidopsis thaliana. Recent Pat. Biotechnol. 2014, 8, 47–60. [Google Scholar] [CrossRef] [Green Version]
- Ogata, K.; Morikawa, S.; Nakamura, H.; Sekikawa, A.; Inoue, T.; Kanai, H.; Sarai, A.; Ishii, S.; Nishimura, Y. Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell 1994, 79, 639–648. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis thaliana. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Jin, H.; Martin, C. Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 1999, 41, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.H.; Jeong, C.Y.; Kang, G.H.; Yoo, S.D.; Hong, S.W.; Lee, H. MYBD employed by HY5 increases anthocyanin accumulation via repression of MYBL2 in Arabidopsis. Plant J. 2015, 84, 1192–1205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, N.; Mittler, R. Reactive oxygen species and temperature stresses: A delicate balance between signaling and destruction. Physiol. Plant. 2006, 126, 45–51. [Google Scholar] [CrossRef]
- Thalhammer, A.; Hincha, D.K. A mechanistic model of COR15 protein function in plant freezing tolerance: Integration of structural and functional characteristics. Plant Signal. Behav. 2014, 9, e977722. [Google Scholar] [CrossRef] [Green Version]
- Rivero, R.M.; Ruiz, J.M.; Garcıa, P.C.; Lopez-Lefebre, L.R.; Sánchez, E.; Romero, L. Resistance to cold and heat stress: Accumulation of phenolic compounds in tomato and watermelon plants. Plant Sci. 2001, 160, 315–321. [Google Scholar] [CrossRef]
- Yang, A.; Dai, X.; Zhang, W. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Ex. Bot. 2012, 63, 2541–2556. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Mao, Z.; Jiang, H.; Zhang, Z.; Chen, X. A feedback loop involving MdMYB108L and MdHY5 controls apple cold tolerance. Biochem. Biophys. Res. Commun. 2019, 512, 381–386. [Google Scholar] [CrossRef]
- An, J.P.; Li, R.; Qu, F.J.; You, C.X.; Wang, X.F.; Hao, Y.J. R2R3-MYB transcription factor MdMYB23 is involved in the cold tolerance and proanthocyanidin accumulation in apple. Plant J. 2018, 96, 562–577. [Google Scholar] [CrossRef] [Green Version]
- Borevitz, J.O.; Xia, Y.J.; Blount, J.; Dixon, R.A.; Lamb, C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell. 2000, 12, 2383–2393. [Google Scholar] [CrossRef] [Green Version]
- Goff, S.A.; Cone, K.C.; Chandler, V.L. Functional analysis of the transcriptional activator encoded by the maize B-gene: Evidence for a direct functional interaction between two classes of regulatory proteins. Genes Dev. 1992, 6, 864–875. [Google Scholar] [CrossRef] [Green Version]
- Rabinowicz, P.; Braun, E.; Wolfe, A.; Bowen, B.; Grotewold, E. Maize R2R3 Myb genes: Sequence analysis reveals amplification in the higher plants. Genetics 1999, 153, 427–444. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Zhang, L.Y.; Yang, L.; Song, J.B.; Yang, Z.M. AtMYB20 is negatively involved in plant adaptive response to drought stress. Plant Soil 2014, 376, 433–443. [Google Scholar] [CrossRef]
- Xiong, H.; Li, J.; Liu, P.; Duan, J.; Zhao, Y.; Guo, X.; Li, Y.; Zhang, H.; Ali, J.; Li, Z. Overexpression of OsMYB48-1, a Novel MYB-Related Transcription Factor, Enhances Drought and Salinity Tolerance in Rice. PLoS ONE 2014, 9, e92913. [Google Scholar]
- Jaglo-Ottosen, K.R.; Gilmour, S.J.; Zarka, D.G.; Schabenberger, O.; Thomashow, M.F. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 1998, 280, 104–106. [Google Scholar] [CrossRef] [Green Version]
- Ogata, K.; Kanei-Ishii, C.; Sasaki, M.; Hatanaka, H.; Nagadoi, A.; Enari, M.; Nakamura, H.; Nishimura, Y.; Ishii, S.; Sarai, A. The cavity in the hydrophobic core of Myb DNA-binding domain is reserved for DNA recognition and trans-activation. Nat. Struct. Biol. 1996, 3, 178–187. [Google Scholar] [CrossRef]
- König, P.; Giraldo, R.; Chapman, L.; Rhodes, D. The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA. Cell 1996, 85, 125–136. [Google Scholar] [CrossRef] [Green Version]
- Ogata, K.; Hojo, H.; Aimoto, S.; Nakai, T.; Nakamura, H.; Sarai, A.; Ishii, S.; Nishimura, Y. Solution structure of a DNA-binding unit of Myb: A helix-turn-helix-related motif with conserved tryptophans forming a hydrophobic core. Proc. Natl. Acad. Sci. USA 1992, 89, 6428–6432. [Google Scholar] [CrossRef] [Green Version]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Han, D.; Du, M.; Zhou, Z.; Wang, S.; Li, T.; Han, J.; Xu, T.; Yang, G. Overexpression of a Malus baccata NAC Transcription Factor Gene MbNAC25 Increases Cold and Salinity Tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2020, 21, 1198. [Google Scholar] [CrossRef] [Green Version]
- Han, D.; Du, M.; Zhou, Z.; Wang, S.; Li, T.; Han, J.; Xu, T.; Yang, G. An NAC transcription factor gene from Malus baccata, MbNAC29, increases cold and high salinity tolerance in Arabidopsis thaliana. Vitr. Cell. Dev. Biol.-Plant 2020, 56, 588–599. [Google Scholar] [CrossRef]
- Lv, B.; Zhang, W.; Li, C.; Ming, F. MYB family transcription factor OsMYB84 participates in salt stress response through ABA signaling pathway. Fudan J. (Nat. Sci. Ed.) 2015, 54, 591–600. [Google Scholar]
- Xie, Y.P. Mechanism of apple MdMYB88 and MdMYB124 transcription factors in low temperature and drought stress. Doctoral Thesis, Northwest A&F University, Yangling, China, 2018; p. 23. [Google Scholar]
- Gao, X.; Zhang, Q.; Zhao, Y.; Yang, J.; He, H.; Jia, G. The lre-miR159a-LrGAMYB pathway mediates resistance to grey mould infection in Lilium regale. Mol. Plant Pathol. 2020, 21, 749–760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, P.; Liu, F.; Zheng, G.; Liu, H. Group 3 late embryogenesis abundant protein in Arabidopsis thaliana: Structure, egulation, and function. Acta Physiol. Plant. 2011, 33, 1063–1073. [Google Scholar] [CrossRef]
- Shen, X.; Wang, Y.; Zhang, Y.; Guo, W.; Jiao, Y.; Zhou, X. Overexpression of the Wild Soybean R2R3-MYB Transcription Factor GsMYB15 Enhances Resistance to Salt Stress and Helicoverpa Armigera in Transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 2018, 19, 3958. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.X. Research Progress on the Mechanism of Increasing Plant Cold Resistance. Acta Ecol. Sin. 2012, 32, 7966–7980. [Google Scholar]
- Ke, X.; He, L.; Su, Z. Relative Chlorophyll Indexes and Distribution of Four Woody Plants in South China. J. Cent. S. Univ. For. Technol. 2010, 30, 82–86. [Google Scholar]
- Yao, H. Physiological and Biochemical Study on Cold Resistance of Four Cupressaceae Plants. Master′s Thesis, Inner Mongolia Agricultural University, Hohhot, China, 2012; pp. 27–29. [Google Scholar]
- Jiao, R.; Liu, H.; Liu, G.; Wang, S.; Hou, N.; Wang, Q.; Liu, Z.; Feng, X.; Hu, X.; Jin, Y. On Proline Accumulation and Plant Resistance to Osmotic Stress. Chin. Agric. Sci. Bull. 2011, 27, 216–221. [Google Scholar]
- Rezayian, M.; Niknam, V.; Ebrahimzadeh, H. Penconazole and calcium ameliorate drought stress in canola by upregulating the antioxidative enzymes. Funct. Plant Biol. 2020, 47, 825–839. [Google Scholar] [CrossRef]
- Lee, H.G.; Seo, P.J. The MYB96-HHP module integrates cold and abscisic acid signaling to activate the CBF-COR pathway in Arabidopsis. Plant J. 2015, 82, 962–977. [Google Scholar] [CrossRef]
- Xie, Y.; Chen, P.; Yan, Y.; Bao, C.; Li, X.; Wang, L.; Wang, L.; Shen, X.; Li, H.; Liu, X.; et al. An atypical R2R3 MYB transcription factor increases cold hardiness by CBF-dependent and CBF-independent pathways in apple. New Phytol. 2018, 218, 201–218. [Google Scholar] [CrossRef]
- Thomashow, M.F. Molecular basis of plant cold acclimation: Insights gained from studying the CBF cold response pathway. Plant Physiol. 2010, 154, 571–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilmour, S.J.; Zarka, D.G.; Stockinger, E.J.; Salazar, M.P.; Houghton, J.M.; Thomashow, M.F. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 1998, 16, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Gilmour, S.J.; Fowler, S.G.; Thomashow, M.F. Arabidopsis transcriptional acti-vators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol. Biol. 2004, 54, 767–781. [Google Scholar] [CrossRef] [PubMed]
- Stockinger, E.J.; Gilmour, S.J.; Thomashow, M.F. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 1997, 94, 1035–1040. [Google Scholar] [CrossRef] [Green Version]
- Medina, J.; Bargues, M.; Terol, J.; Perez-Alonso, M.; Salinas, J. The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol. 1999, 119, 463–470. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Lin, L.; Zhang, Y.; Sui, N. ZmMYB31, a R2R3-MYB transcription factor in maize, positively regulates the expression of CBF genes and enhances resistance to chilling and oxidative stress. Mol. Biol. Rep. 2019, 46, 3937–3944. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Yamaguchi-Shinozaki, K.; Seki, M. Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 2003, 6, 410–417. [Google Scholar] [CrossRef]
- Shan, H.; Chen, S.; Jiang, J.; Chen, F.; Chen, Y.; Gu, C.; Li, P.; Song, A.; Zhu, X.; Gao, H.; et al. Heterologous expression of the Chrysanthemun R2R3-MYB transcription factor CmMYB2 enhances drought and salinity tolerance, increases hypersensitivity to ABA and delays flowering in Arabidopsis thaliana. Mol. Biotechnol. 2012, 51, 160–173. [Google Scholar] [CrossRef]
- Butt, H.I.; Yang, Z.; Gong, Q.; Chen, E.; Wang, X.; Zhao, G.; Ge, X.; Zhang, X.; Li, F. GaMYB85, an R2R3 MYB gene, in transgenic Arabidopsis plays an important role in drought tolerance. BMC Plant Biol. 2017, 17, 1–17. [Google Scholar] [CrossRef]
- Du, Y.; Wang, P.; Chen, J.; Song, C. Comprehensive Functional Analysis of the Catalase Gene Family in Arabidopsis thaliana. J. Integr. Plant Biol. 2008, 50, 1318–1326. [Google Scholar] [CrossRef]
- Han, D.; Han, J.; Yang, G.; Wang, S.; Xu, T.; Li, W. An ERF Transcription Factor Gene from Malus baccata (L.) Borkh, MbERF11, Affects Cold and Salt Stress Tolerance in Arabidopsis. Forests 2020, 11, 514. [Google Scholar] [CrossRef]
- Han, D.; Hou, Y.; Ding, H.; Zhou, Z.; Li, H.; Yang, G. Isolation and preliminary functional analysis of MbWRKY4 gene involved in salt tolerance in transgenic tobacco. Int. J. Agric. Biol. 2018, 20, 2045–2052. [Google Scholar]
- Han, D.; Hou, Y.; Liu, W.; Zhang, Z.; Ding, H.; Li, H.; Yang, G. Isolation and functional analysis of MdNAS1, with functions in improved iron stress tolerance and abnormal flower in transgenic tobacco. J. Plant Interact. 2018, 13, 213–220. [Google Scholar] [CrossRef]
- Jia, M.M. Analysis of the cold resistance function of PtrPRP in Poncirus trifoliata. Master′s Thesis, Huazhong Agricultural University, Wuhan, China, 2014; p. 8. [Google Scholar]
- Han, D.; Shi, Y.; Wang, B.; Liu, W.; Yu, Z.; Lv, B.; Yang, G. Isolation and preliminary functional analysis of MxCS2: A gene encoding a citrate synthase in Malus xiaojinensis. Plant Mol. Biol. Report. 2015, 33, 133–142. [Google Scholar] [CrossRef]
- Tuo, D.C. Detection and Identification of Papaya Malformation Mosaic Virus and Construction and Application of Infectious Clones. Master′s Thesis, Hainan Unitersity, Haikou, China, 2015; pp. 10–12. [Google Scholar]
- Han, D.; Zhang, Z.; Ni, B.; Ding, H.; Liu, W.; Li, W. Isolation and functional analysis of MxNAS3 involved in enhanced iron stress tolerance and abnormal flower in transgenic Arabidopsis thaliana. J. Plant Interact. 2018, 13, 433–441. [Google Scholar] [CrossRef] [Green Version]
- An, G.; Watson, B.D.; Chiang, C.C. Transformation of tobacco, tomato, potato, and Arabidopsis thaliana using a binary ti vector system. Plant Physiol. 1986, 81, 301–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, F.X.; Liu, S.Y.; Liu, Y.F.; Xu, J.; Liu, T.; Dong, S.Z. Effectiveness of lysozyme coatings and 1-MCP treatments on storage and preservation of kiwifruit. Food Chem. 2019, 288, 201–217. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Qian, X.; Jiang, T.; Zheng, X. Effect of eugenol fumigation treatment on chilling injury and CBF gene expression in eggplant fruit during cold storage. Food Chem. 2019, 292, 143–150. [Google Scholar] [CrossRef]
- Sharma, A.; Yuan, H.W.; Kumar, V.; Ramakrishnan, M.; Kohli, S.K.; Kaur, R.; Thukral, A.K.; Bhardwaj, R.; Zheng, B.S. Castasterone attenuates insecticide induced phytotoxicity in mustard. Ecotoxicol. Environ. Saf. 2019, 179, 50–61. [Google Scholar] [CrossRef]
- Wang, X.; Guo, P.; Yuan, X.; Yao, M. Effects of 2,4-D on the antioxidative enzyme activities and lipid peroxidation in Opium poppy (Papaver somniferum L.). Acta Ecol. Sin. 2008, 28, 1098–1103. [Google Scholar]
- Chen, L.; Chen, Y.; Peng, S.; Ma, L.; Wang, R.; Wang, X. Study on physiological and biochemical responses of Camellia oleifera to low phosphorus stress. For. Res. 2010, 23, 782–786. [Google Scholar]
- Jiang, Y.; Lin, L.; Zhong, S.; Cai, Y.; Zhang, F.; Wang, X.; Miao, R.; Zhang, B.; Gao, S.; Hu, X. Overexpression of novel lncRNA NLIPMT inhibits metastasis by reducing phosphorylated glycogen synthase kinase 3 in breast cancer. J. Cell. Physiol. 2019, 234, 10689–10708. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, C.; Li, X.; Li, Y.; Yang, G.; Liu, W.; Shao, B.; Zhong, J.; Huang, P.; Han, D. Overexpression of a Malus baccata MYB Transcription Factor Gene MbMYB4 Increases Cold and Drought Tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 1794. https://doi.org/10.3390/ijms23031794
Yao C, Li X, Li Y, Yang G, Liu W, Shao B, Zhong J, Huang P, Han D. Overexpression of a Malus baccata MYB Transcription Factor Gene MbMYB4 Increases Cold and Drought Tolerance in Arabidopsis thaliana. International Journal of Molecular Sciences. 2022; 23(3):1794. https://doi.org/10.3390/ijms23031794
Chicago/Turabian StyleYao, Chunya, Xingguo Li, Yingmei Li, Guohui Yang, Wanda Liu, Bangtao Shao, Jiliang Zhong, Pengfei Huang, and Deguo Han. 2022. "Overexpression of a Malus baccata MYB Transcription Factor Gene MbMYB4 Increases Cold and Drought Tolerance in Arabidopsis thaliana" International Journal of Molecular Sciences 23, no. 3: 1794. https://doi.org/10.3390/ijms23031794
APA StyleYao, C., Li, X., Li, Y., Yang, G., Liu, W., Shao, B., Zhong, J., Huang, P., & Han, D. (2022). Overexpression of a Malus baccata MYB Transcription Factor Gene MbMYB4 Increases Cold and Drought Tolerance in Arabidopsis thaliana. International Journal of Molecular Sciences, 23(3), 1794. https://doi.org/10.3390/ijms23031794




