Profiling of the Bacterial Microbiota along the Murine Alimentary Tract
Abstract
:1. Introduction
2. Results
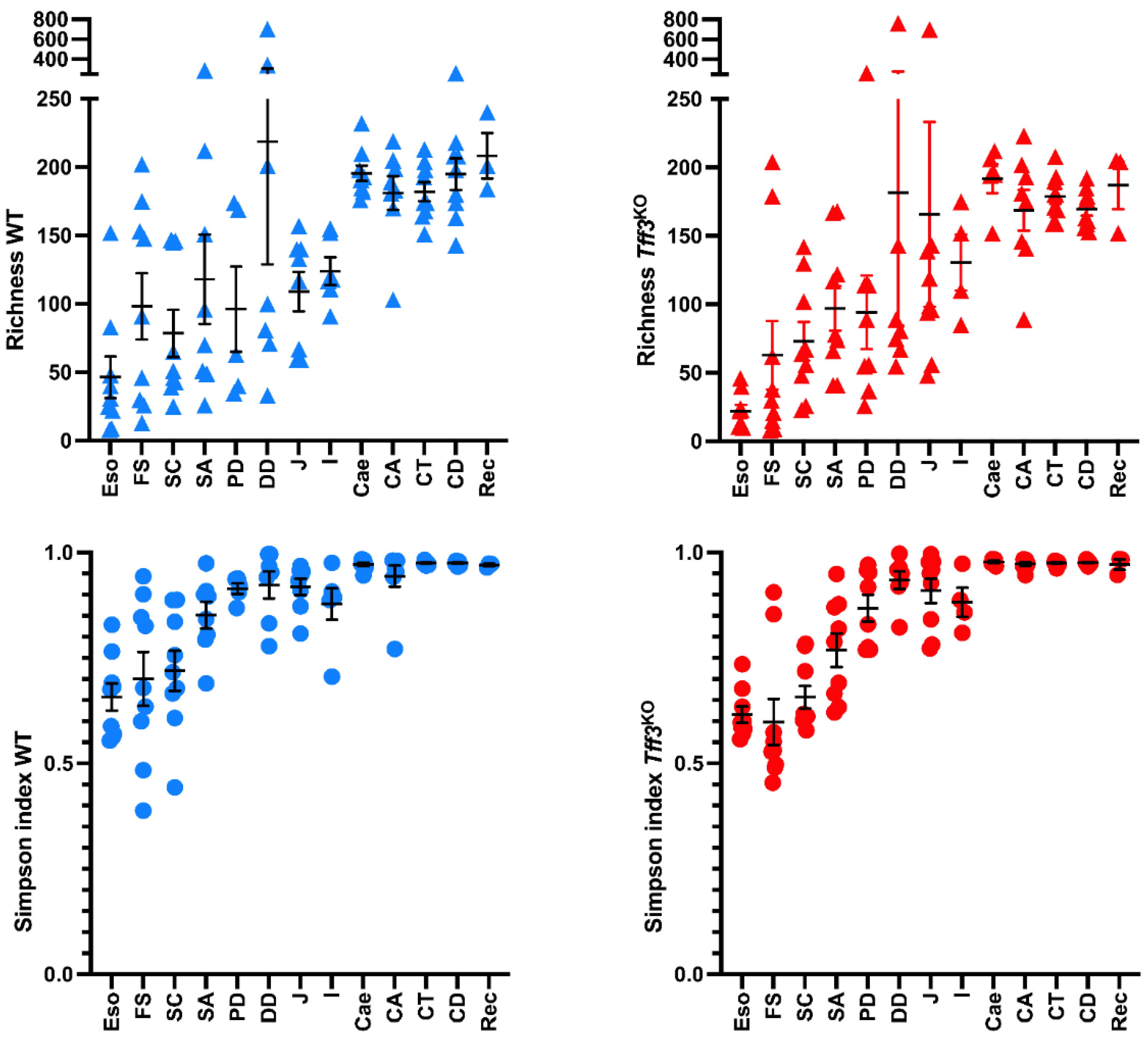
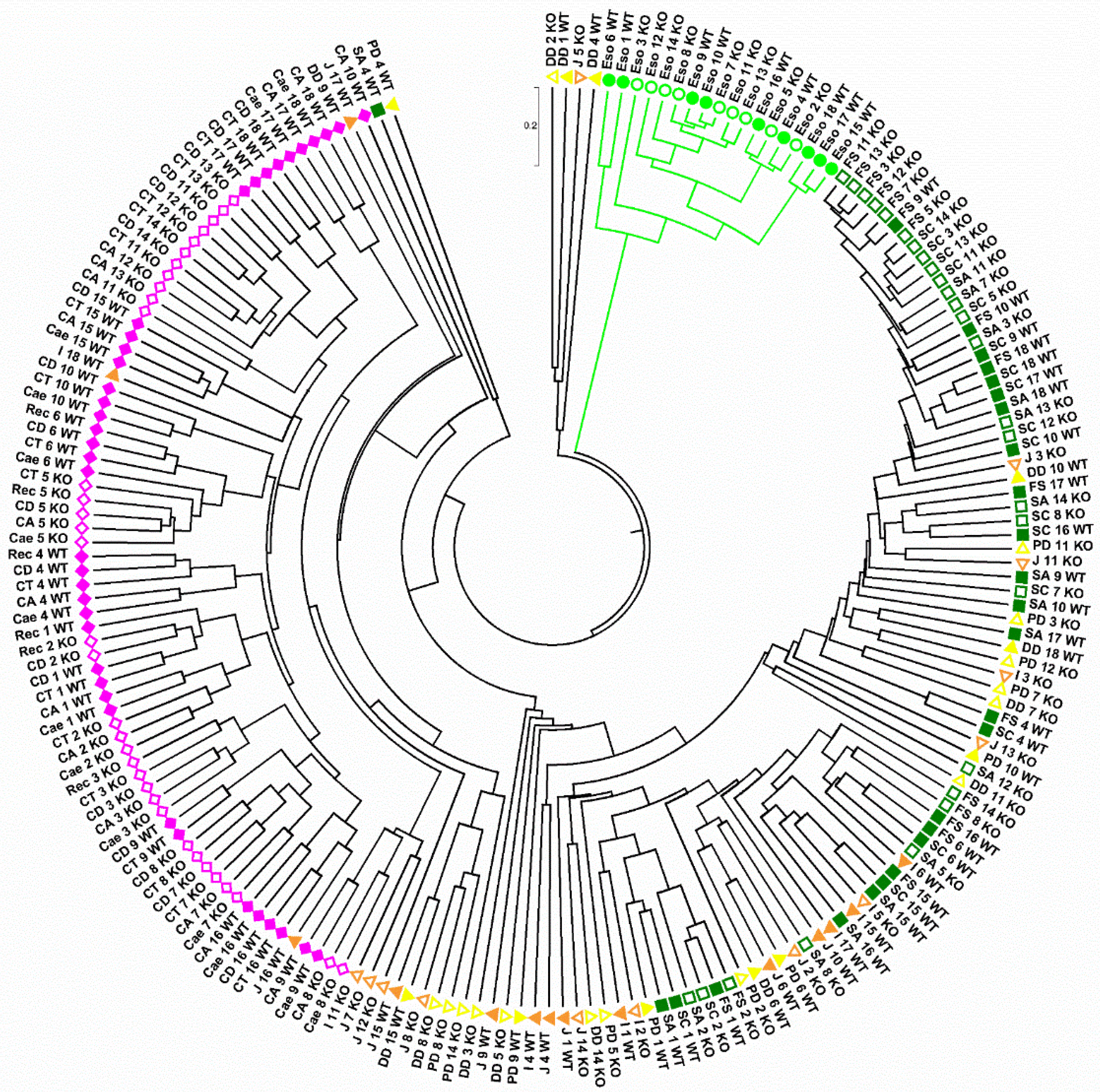
2.1. Overall Bacterial Communities in Wild-Type and Tff3KO Mice
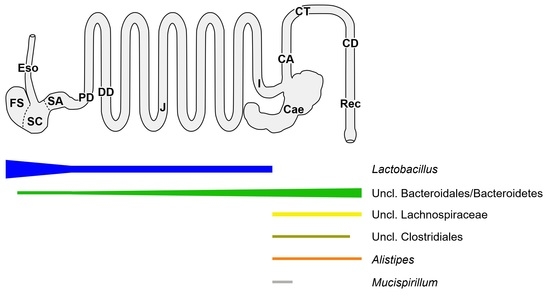
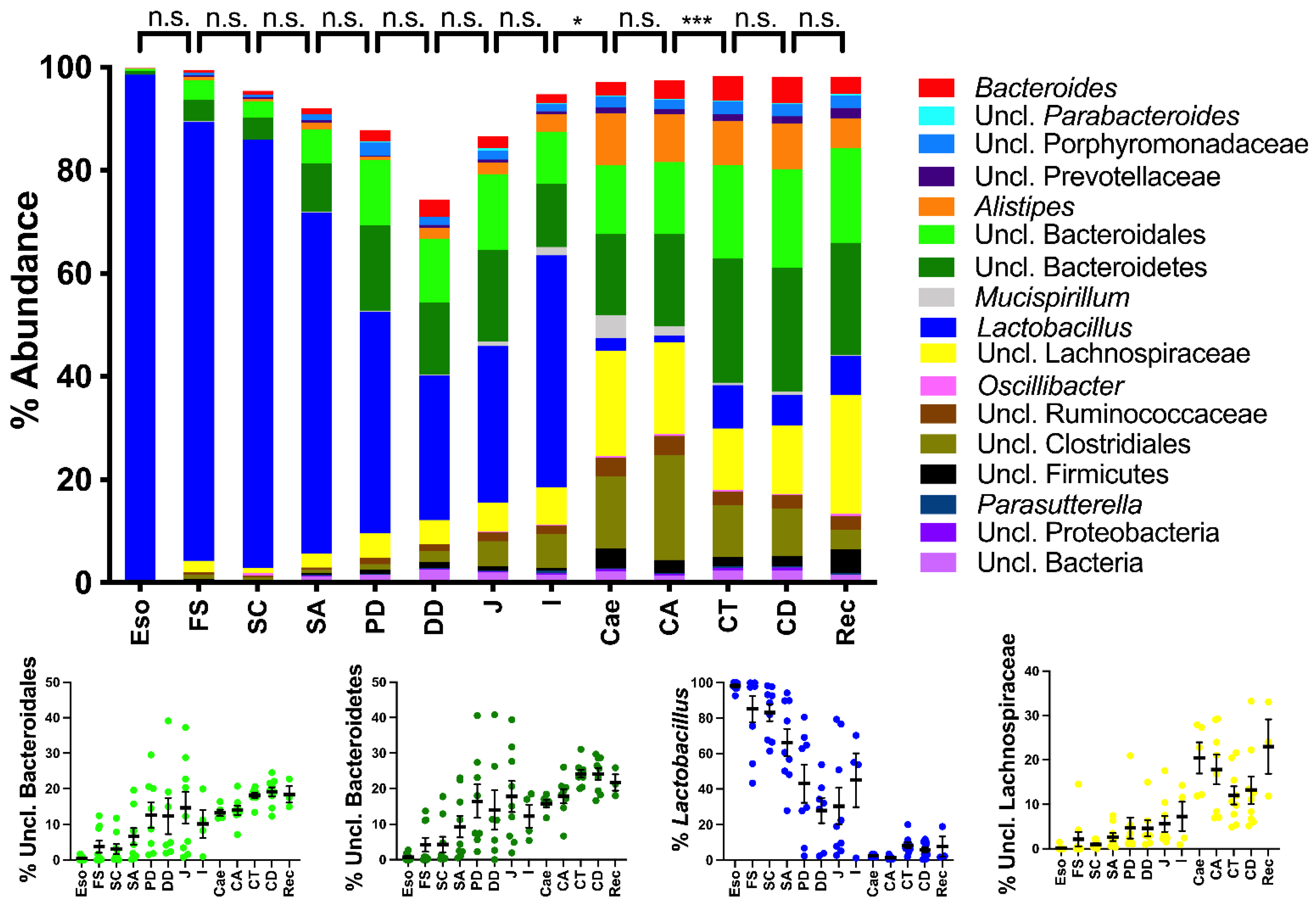
2.2. Spatial Analysis of the Bacterial Communities at the Genus Level
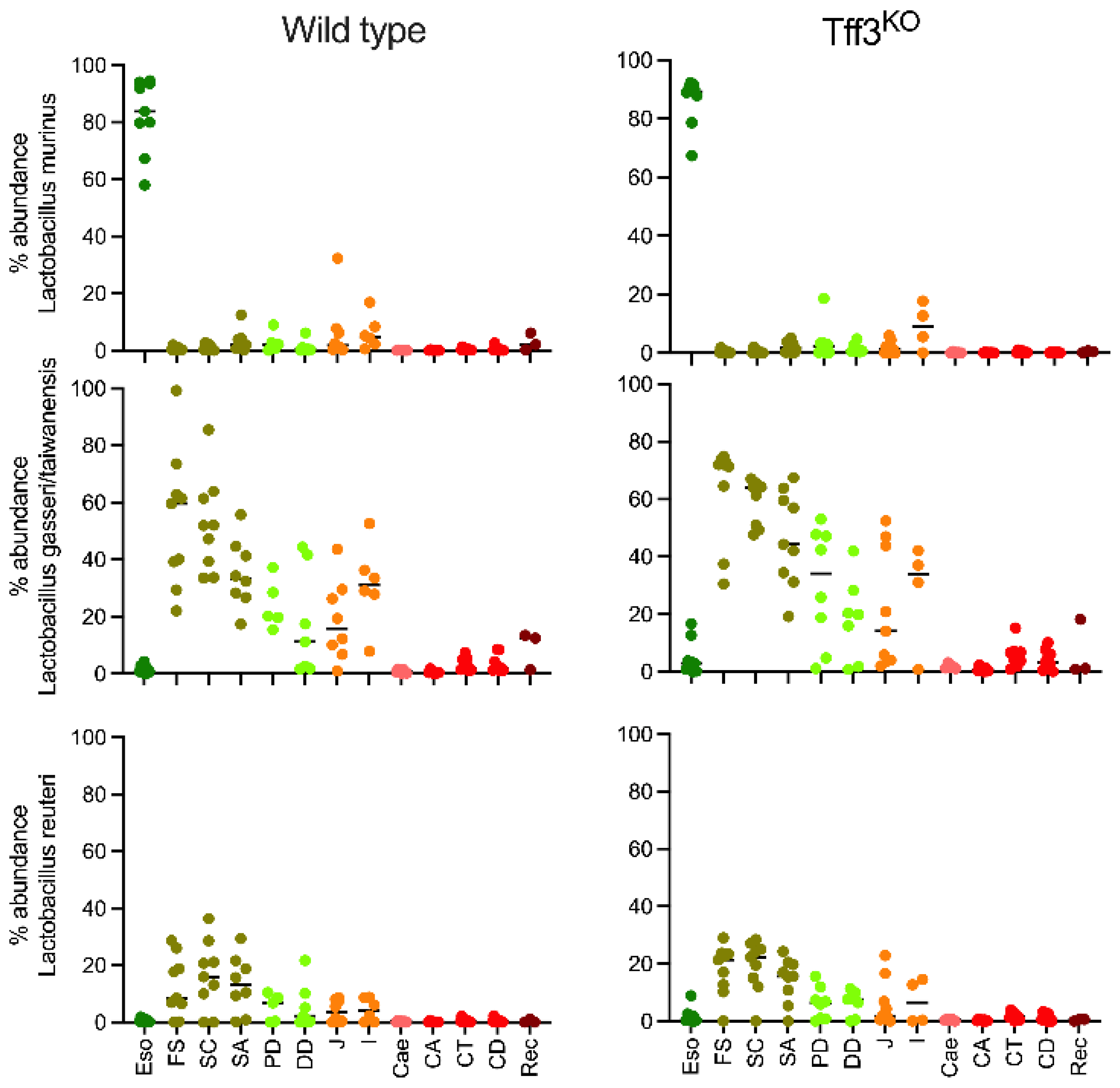
2.3. Spatial Analysis of Lactobacillus Phylotypes
3. Discussion
3.1. The Bacterial Communities Differ Significantly along the Murine Alimentary Tract
3.1.1. The Upper Alimentary Tract
3.1.2. The Lower Alimentary Tract
3.2. Comparison of Wild-Type Animals and Tff3KO Mice
3.3. Comparison of Gastrointestinal Bacterial Communities in Humans vs. Mice
4. Materials and Methods
4.1. Murine Tissue
4.2. DNA Extraction and Library Construction
4.3. Bioinformatic and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DSS | Dextran sulfate sodium |
| FCGBP | IgG Fc binding protein |
| GC | Goblet cell |
| GI | gastrointestinal |
| icGC | Intercrypt goblet cell |
| senGC | Sentinel goblet cell |
| TFF | Trefoil factor family |
| TLR | Toll-like receptor |
References
- Spor, A.; Koren, O.; Ley, R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 2011, 9, 279–290. [Google Scholar] [CrossRef]
- Integrative, H.M.P.; Proctor, L.M.; Creasy, H.H.; Fettweis, J.M.; Lloyd-Price, J.; Mahurkar, A.; Zhou, W.; Buck, G.A.; Snyder, M.P.; Strauss, J.F., III; et al. The Integrative Human Microbiome Project. Nature 2019, 569, 641–648. [Google Scholar] [CrossRef] [Green Version]
- Kau, A.L.; Ahern, P.P.; Griffin, N.W.; Goodman, A.L.; Gordon, J.I. Human nutrition, the gut microbiome and the immune system. Nature 2011, 474, 327–336. [Google Scholar] [CrossRef] [Green Version]
- Tilg, H.; Moschen, A.R. Food, immunity, and the microbiome. Gastroenterology 2015, 148, 1107–1119. [Google Scholar] [CrossRef]
- Gentile, C.L.; Weir, T.L. The gut microbiota at the intersection of diet and human health. Science 2018, 362, 776–780. [Google Scholar] [CrossRef] [Green Version]
- Pothuraju, R.; Chaudhary, S.; Rachagani, S.; Kaur, S.; Roy, H.K.; Bouvet, M.; Batra, S.K. Mucins, gut microbiota, and postbiotics role in colorectal cancer. Gut Microbes 2021, 13, 1974795. [Google Scholar] [CrossRef]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Microbial ecology: Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef]
- Tilg, H.; Kaser, A. Gut microbiome, obesity, and metabolic dysfunction. J. Clin. Investig. 2011, 121, 2126–2132. [Google Scholar] [CrossRef]
- Hirschberg, S.; Gisevius, B.; Duscha, A.; Haghikia, A. Implications of Diet and The Gut Microbiome in Neuroinflammatory and Neurodegenerative Diseases. Int. J. Mol. Sci. 2019, 20, 3109. [Google Scholar] [CrossRef] [Green Version]
- Arthur, J.C.; Perez-Chanona, E.; Mühlbauer, M.; Tomkovich, S.; Uronis, J.M.; Fan, T.-J.; Campbell, B.J.; Abujamel, T.; Dogan, B.; Rogers, A.B.; et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 2012, 338, 120–123. [Google Scholar] [CrossRef] [Green Version]
- Boccellato, F.; Meyer, T.F. Bacteria Moving into Focus of Human Cancer. Cell Host Microbe 2015, 17, 728–730. [Google Scholar] [CrossRef] [Green Version]
- Wargo, J.A. Modulating gut microbes. Science 2020, 369, 1302–1303. [Google Scholar] [CrossRef]
- Yang, I.; Nell, S.; Suerbaum, S. Survival in hostile territory: The microbiota of the stomach. FEMS Microbiol. Rev. 2013, 37, 736–761. [Google Scholar] [CrossRef]
- Nardone, G.; Compare, D. The human gastric microbiota: Is it time to rethink the pathogenesis of stomach diseases? United Eur. Gastroenterol. J. 2015, 3, 255–260. [Google Scholar] [CrossRef] [Green Version]
- Ley, R.E.; Peterson, D.A.; Gordon, J.I. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 2006, 124, 837–848. [Google Scholar] [CrossRef] [Green Version]
- Mahowald, M.A.; Rey, F.E.; Seedorf, H.; Turnbaugh, P.J.; Fulton, R.S.; Wollam, A.; Shah, N.; Wang, C.; Magrini, V.; Wilson, R.K.; et al. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc. Natl. Acad. Sci. USA 2009, 106, 5859–5864. [Google Scholar] [CrossRef] [Green Version]
- McGuckin, M.A.; Lindén, S.K.; Sutton, P.; Florin, T.H. Mucin dynamics and enteric pathogens. Nat. Rev. Microbiol. 2011, 9, 265–278. [Google Scholar] [CrossRef]
- Johansson, M.E.V.; Sjövall, H.; Hansson, G.C. The gastrointestinal mucus system in health and disease. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 352–361. [Google Scholar] [CrossRef] [Green Version]
- Duncan, K.; Carey-Ewend, K.; Vaishnava, S. Spatial analysis of gut microbiome reveals a distinct ecological niche associated with the mucus layer. Gut Microbes 2021, 13, 1874815. [Google Scholar] [CrossRef]
- Hoffmann, W. TFF2, a MUC6-binding lectin stabilizing the gastric mucus barrier and more. Int. J. Oncol. 2015, 47, 806–816. [Google Scholar] [CrossRef] [Green Version]
- Johansson, M.E.V.; Larsson, J.M.H.; Hansson, G.C. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc. Natl. Acad. Sci. USA 2011, 108, 4659–4665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pelaseyed, T.; Bergström, J.H.; Gustafsson, J.K.; Ermund, A.; Birchenough, G.M.H.; Schütte, A.; van der Post, S.; Svensson, F.; Rodríguez-Piñeiro, A.M.; Nyström, E.E.L.; et al. The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol. Rev. 2014, 260, 8–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergstrom, K.; Shan, X.; Casero, D.; Batushansky, A.; Lagishetty, V.; Jacobs, J.P.; Hoover, C.; Kondo, Y.; Shao, B.; Gao, L.; et al. Proximal colon-derived O-glycosylated mucus encapsulates and modulates the microbiota. Science 2020, 370, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Birchenough, G.M.H.; Johansson, M.E.V. Forming a mucus barrier along the colon. Science 2020, 370, 402–403. [Google Scholar] [CrossRef]
- Bäckhed, F.; Ding, H.; Wang, T.; Hooper, L.V.; Koh, G.Y.; Nagy, A.; Semenkovich, C.F.; Gordon, J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA 2004, 101, 15718–15723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, C.; Murdock, M.H.; Jing, D.; Won, T.H.; Chung, H.; Kressel, A.M.; Tsaava, T.; Addorisio, M.E.; Putzel, G.G.; Zhou, L.; et al. The microbiota regulate neuronal function and fear extinction learning. Nature 2019, 574, 543–548. [Google Scholar] [CrossRef]
- Muller, P.A.; Matheis, F.; Schneeberger, M.; Kerner, Z.; Jové, V.; Mucida, D. Microbiota-modulated CART(+) enteric neurons autonomously regulate blood glucose. Science 2020, 370, 314–321. [Google Scholar] [CrossRef]
- Vasapolli, R.; Schütte, K.; Schulz, C.; Vital, M.; Schomburg, D.; Pieper, D.H.; Vilchez-Vargas, R.; Malfertheiner, P. Analysis of Transcriptionally Active Bacteria Throughout the Gastrointestinal Tract of Healthy Individuals. Gastroenterology 2019, 157, 1081–1092.e3. [Google Scholar] [CrossRef] [Green Version]
- Mashimo, H.; Wu, D.C.; Podolsky, D.K.; Fishman, M.C. Impaired defense of intestinal mucosa in mice lacking intestinal trefoil factor. Science 1996, 274, 262–265. [Google Scholar] [CrossRef]
- Hoffmann, W. Trefoil Factor Family (TFF) Peptides and Their Links to Inflammation: A Re-evaluation and New Medical Perspectives. Int. J. Mol. Sci. 2021, 22, 4909. [Google Scholar] [CrossRef]
- Johansson, M.E.V.; Gustafsson, J.K.; Sjöberg, K.E.; Petersson, J.; Holm, L.; Sjövall, H.; Hansson, G.C. Bacteria penetrate the inner mucus layer before inflammation in the dextran sulfate colitis model. PLoS ONE 2010, 5, e12238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, W. Trefoil Factor Family (TFF) Peptides and Their Diverse Molecular Functions in Mucus Barrier Protection and More: Changing the Paradigm. Int. J. Mol. Sci. 2020, 21, 4535. [Google Scholar] [CrossRef]
- Hoffmann, W. Trefoil Factor Family (TFF) Peptides. Encyclopedia 2021, 1, 74. [Google Scholar] [CrossRef]
- Albert, T.K.; Laubinger, W.; Müller, S.; Hanisch, F.-G.; Kalinski, T.; Meyer, F.; Hoffmann, W. Human intestinal TFF3 forms disulfide-linked heteromers with the mucus-associated FCGBP protein and is released by hydrogen sulfide. J. Proteome Res. 2010, 9, 3108–3117. [Google Scholar] [CrossRef] [PubMed]
- Houben, T.; Harder, S.; Schlüter, H.; Kalbacher, H.; Hoffmann, W. Different Forms of TFF3 in the Human Saliva: Heterodimerization with IgG Fc Binding Protein (FCGBP). Int. J. Mol. Sci. 2019, 20, 5000. [Google Scholar] [CrossRef] [Green Version]
- Hauser, F.; Poulsom, R.; Chinery, R.; Rogers, L.A.; Hanby, A.M.; Wright, N.A.; Hoffmann, W. hP1.B, a human P-domain peptide homologous with rat intestinal trefoil factor, is expressed also in the ulcer-associated cell lineage and the uterus. Proc. Natl. Acad. Sci. USA 1993, 90, 6961–6965. [Google Scholar] [CrossRef] [Green Version]
- Harada, N.; Iijima, S.; Kobayashi, K.; Yoshida, T.; Brown, W.R.; Hibi, T.; Oshima, A.; Morikawa, M. Human IgGFc binding protein (FcgammaBP) in colonic epithelial cells exhibits mucin-like structure. J. Biol. Chem. 1997, 272, 15232–15241. [Google Scholar] [CrossRef] [Green Version]
- Jagla, W.; Wiede, A.; Hinz, M.; Dietzmann, K.; Gülicher, D.; Gerlach, K.L.; Hoffmann, W. Secretion of TFF-peptides by human salivary glands. Cell Tissue Res. 1999, 298, 161–166. [Google Scholar] [CrossRef]
- Kouznetsova, I.; Gerlach, K.L.; Zahl, C.; Hoffmann, W. Expression analysis of human salivary glands by laser microdissection: Differences between submandibular and labial glands. Cell. Physiol. Biochem. 2010, 26, 375–382. [Google Scholar] [CrossRef]
- Kouznetsova, I.; Kalinski, T.; Peitz, U.; Mönkemüller, K.E.; Kalbacher, H.; Vieth, M.; Meyer, F.; Roessner, A.; Malfertheiner, P.; Lippert, H.; et al. Localization of TFF3 peptide in human esophageal submucosal glands and gastric cardia: Differentiation of two types of gastric pit cells along the rostro-caudal axis. Cell Tissue Res. 2007, 328, 365–374. [Google Scholar] [CrossRef]
- Kobayashi, K.; Ogata, H.; Morikawa, M.; Iijima, S.; Harada, N.; Yoshida, T.; Brown, W.R.; Inoue, N.; Hamada, Y.; Ishii, H.; et al. Distribution and partial characterisation of IgG Fc binding protein in various mucin producing cells and body fluids. Gut 2002, 51, 169–176. [Google Scholar] [CrossRef] [Green Version]
- Kouznetsova, I.; Peitz, U.; Vieth, M.; Meyer, F.; Vestergaard, E.M.; Malfertheiner, P.; Roessner, A.; Lippert, H.; Hoffmann, W. A gradient of TFF3 (trefoil factor family 3) peptide synthesis within the normal human gastric mucosa. Cell Tissue Res. 2004, 316, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Znalesniak, E.B.; Salm, F.; Hoffmann, W. Molecular Alterations in the Stomach of Tff1-Deficient Mice: Early Steps in Antral Carcinogenesis. Int. J. Mol. Sci. 2020, 21, 644. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Wang, R.; Su, B.; Luo, Y.; Terhune, J.; Beck, B.; Peatman, E. Evasion of mucosal defenses during Aeromonas hydrophila infection of channel catfish (Ictalurus punctatus) skin. Dev. Comp. Immunol. 2013, 39, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, W. Trefoil Factor Family (TFF) Peptides and their Different Roles in the Mucosal Innate Immune Defense and More: An Update. Curr. Med. Chem. 2021, 28, 7387–7399. [Google Scholar] [CrossRef]
- Madsen, J.; Sorensen, G.L.; Nielsen, O.; Tornøe, I.; Thim, L.; Fenger, C.; Mollenhauer, J.; Holmskov, U. A variant form of the human deleted in malignant brain tumor 1 (DMBT1) gene shows increased expression in inflammatory bowel diseases and interacts with dimeric trefoil factor 3 (TFF3). PLoS ONE 2013, 8, e64441. [Google Scholar] [CrossRef] [Green Version]
- Nyström, E.E.L.; Martinez-Abad, B.; Arike, L.; Birchenough, G.M.H.; Nonnecke, E.B.; Castillo, P.A.; Svensson, F.; Bevins, C.L.; Hansson, G.C.; Johansson, M.E.V. An intercrypt subpopulation of goblet cells is essential for colonic mucus barrier function. Science 2021, 372, eabb1590. [Google Scholar] [CrossRef] [PubMed]
- Bathum Nexoe, A.; Pedersen, A.A.; von Huth, S.; Detlefsen, S.; Hansen, P.L.; Holmskov, U. Immunohistochemical Localization of Deleted in Malignant Brain Tumors 1 in Normal Human Tissues. J. Histochem. Cytochem. 2020, 68, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Prakobphol, A.; Xu, F.; Hoang, V.M.; Larsson, T.; Bergstrom, J.; Johansson, I.; Frängsmyr, L.; Holmskov, U.; Leffler, H.; Nilsson, C.; et al. Salivary agglutinin, which binds Streptococcus mutans and Helicobacter pylori, is the lung scavenger receptor cysteine-rich protein gp-340. J. Biol. Chem. 2000, 275, 39860–39866. [Google Scholar] [CrossRef] [Green Version]
- Khoder, G.; Al-Yassir, F.; Al Menhali, A.; Saseedharan, P.; Sugathan, S.; Tomasetto, C.; Karam, S.M. Probiotics Upregulate Trefoil Factors and Downregulate Pepsinogen in the Mouse Stomach. Int. J. Mol. Sci. 2019, 20, 3901. [Google Scholar] [CrossRef] [Green Version]
- Bergstrom, K.S.B.; Guttman, J.A.; Rumi, M.; Ma, C.; Bouzari, S.; Khan, M.A.; Gibson, D.L.; Vogl, A.W.; Vallance, B.A. Modulation of intestinal goblet cell function during infection by an attaching and effacing bacterial pathogen. Infect. Immun. 2008, 76, 796–811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shalapour, S.; Deiser, K.; Kühl, A.A.; Glauben, R.; Krug, S.M.; Fischer, A.; Sercan, O.; Chappaz, S.; Bereswill, S.; Heimesaat, M.M.; et al. Interleukin-7 links T lymphocyte and intestinal epithelial cell homeostasis. PLoS ONE 2012, 7, e31939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Podolsky, D.K.; Gerken, G.; Eyking, A.; Cario, E. Colitis-associated variant of TLR2 causes impaired mucosal repair because of TFF3 deficiency. Gastroenterology 2009, 137, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Frese, S.A.; Benson, A.K.; Tannock, G.W.; Loach, D.M.; Kim, J.; Zhang, M.; Oh, P.L.; Heng, N.C.K.; Patil, P.B.; Juge, N.; et al. The evolution of host specialization in the vertebrate gut symbiont Lactobacillus reuteri. PLoS Genet. 2011, 7, e1001314. [Google Scholar] [CrossRef] [Green Version]
- Frese, S.A.; Mackenzie, D.A.; Peterson, D.A.; Schmaltz, R.; Fangman, T.; Zhou, Y.; Zhang, C.; Benson, A.K.; Cody, L.A.; Mulholland, F.; et al. Molecular characterization of host-specific biofilm formation in a vertebrate gut symbiont. PLoS Genet. 2013, 9, e1004057. [Google Scholar] [CrossRef]
- Sarma-Rupavtarm, R.B.; Ge, Z.; Schauer, D.B.; Fox, J.G.; Polz, M.F. Spatial distribution and stability of the eight microbial species of the altered schaedler flora in the mouse gastrointestinal tract. Appl. Environ. Microbiol. 2004, 70, 2791–2800. [Google Scholar] [CrossRef] [Green Version]
- Swidsinski, A.; Loening-Baucke, V.; Lochs, H.; Hale, L.-P. Spatial organization of bacterial flora in normal and inflamed intestine: A fluorescence in situ hybridization study in mice. World J. Gastroenterol. 2005, 11, 1131–1140. [Google Scholar] [CrossRef]
- El Kaoutari, A.; Armougom, F.; Gordon, J.I.; Raoult, D.; Henrissat, B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. Microbiol. 2013, 11, 497–504. [Google Scholar] [CrossRef]
- Kamphuis, J.B.J.; Mercier-Bonin, M.; Eutamène, H.; Theodorou, V. Mucus organisation is shaped by colonic content; a new view. Sci. Rep. 2017, 7, 8527. [Google Scholar] [CrossRef] [Green Version]
- Parikh, K.; Antanaviciute, A.; Fawkner-Corbett, D.; Jagielowicz, M.; Aulicino, A.; Lagerholm, C.; Davis, S.; Kinchen, J.; Chen, H.H.; Alham, N.K.; et al. Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature 2019, 567, 49–55. [Google Scholar] [CrossRef]
- Nava, G.M.; Stappenbeck, T.S. Diversity of the autochthonous colonic microbiota. Gut Microbes 2011, 2, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Ermund, A.; Schütte, A.; Johansson, M.E.V.; Gustafsson, J.K.; Hansson, G.C. Studies of mucus in mouse stomach, small intestine, and colon. I. Gastrointestinal mucus layers have different properties depending on location as well as over the Peyer’s patches. Am. J. Physiol. Gastrointest. Liver Physiol. 2013, 305, G341–G347. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.E.V.; Phillipson, M.; Petersson, J.; Velcich, A.; Holm, L.; Hansson, G.C. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl. Acad. Sci. USA 2008, 105, 15064–15069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swidsinski, A.; Sydora, B.C.; Doerffel, Y.; Loening-Baucke, V.; Vaneechoutte, M.; Lupicki, M.; Scholze, J.; Lochs, H.; Dieleman, L.A. Viscosity gradient within the mucus layer determines the mucosal barrier function and the spatial organization of the intestinal microbiota. Inflamm. Bowel Dis. 2007, 13, 963–970. [Google Scholar] [CrossRef]
- Birchenough, G.M.H.; Nyström, E.E.L.; Johansson, M.E.V.; Hansson, G.C. A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion. Science 2016, 352, 1535–1542. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Devkota, S.; Musch, M.W.; Jabri, B.; Nagler, C.; Antonopoulos, D.A.; Chervonsky, A.; Chang, E.B. Regional mucosa-associated microbiota determine physiological expression of TLR2 and TLR4 in murine colon. PLoS ONE 2010, 5, e13607. [Google Scholar] [CrossRef]
- Li, H.; Limenitakis, J.P.; Fuhrer, T.; Geuking, M.B.; Lawson, M.A.; Wyss, M.; Brugiroux, S.; Keller, I.; Macpherson, J.A.; Rupp, S.; et al. The outer mucus layer hosts a distinct intestinal microbial niche. Nat. Commun. 2015, 6, 8292. [Google Scholar] [CrossRef]
- Herp, S.; Brugiroux, S.; Garzetti, D.; Ring, D.; Jochum, L.M.; Beutler, M.; Eberl, C.; Hussain, S.; Walter, S.; Gerlach, R.G.; et al. Mucispirillum schaedleri Antagonizes Salmonella Virulence to Protect Mice against Colitis. Cell Host Microbe 2019, 25, 681–694.e8. [Google Scholar] [CrossRef]
- Loy, A.; Pfann, C.; Steinberger, M.; Hanson, B.; Herp, S.; Brugiroux, S.; Gomes Neto, J.C.; Boekschoten, M.V.; Schwab, C.; Urich, T.; et al. Lifestyle and Horizontal Gene Transfer-Mediated Evolution of Mucispirillum schaedleri, a Core Member of the Murine Gut Microbiota. mSystems 2017, 2, e00171-16. [Google Scholar] [CrossRef] [Green Version]
- Nava, G.M.; Friedrichsen, H.J.; Stappenbeck, T.S. Spatial organization of intestinal microbiota in the mouse ascending colon. ISME J. 2011, 5, 627–638. [Google Scholar] [CrossRef] [Green Version]
- Ringel, Y.; Maharshak, N.; Ringel-Kulka, T.; Wolber, E.A.; Sartor, R.B.; Carroll, I.M. High throughput sequencing reveals distinct microbial populations within the mucosal and luminal niches in healthy individuals. Gut Microbes 2015, 6, 173–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bastholm, S.K.; Samson, M.H.; Becher, N.; Hansen, L.K.; Stubbe, P.R.; Chronakis, I.S.; Nexo, E.; Uldbjerg, N. Trefoil factor peptide 3 is positively correlated with the viscoelastic properties of the cervical mucus plug. Acta Obstet. Gynecol. Scand. 2017, 96, 47–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, T.; Znalesniak, E.B.; Kalinski, T.; Möhle, L.; Biswas, A.; Salm, F.; Dunay, I.R.; Hoffmann, W. TFF Peptides Play a Role in the Immune Response Following Oral Infection of Mice with Toxoplasma gondii. Eur. J. Microbiol. Immunol. 2015, 5, 221–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vilchez-Vargas, R.; Geffers, R.; Suárez-Diez, M.; Conte, I.; Waliczek, A.; Kaser, V.S.; Kralova, M.; Junca, H.; Pieper, D.H. Analysis of the microbial gene landscape and transcriptome for aromatic pollutants and alkane degradation using a novel internally calibrated microarray system. Environ. Microbiol. 2013, 15, 1016–1039. [Google Scholar] [CrossRef] [PubMed]
- Rath, S.; Heidrich, B.; Pieper, D.H.; Vital, M. Uncovering the trimethylamine-producing bacteria of the human gut microbiota. Microbiome 2017, 5, 54. [Google Scholar] [CrossRef] [Green Version]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vilchez-Vargas, R.; Salm, F.; Znalesniak, E.B.; Haupenthal, K.; Schanze, D.; Zenker, M.; Link, A.; Hoffmann, W. Profiling of the Bacterial Microbiota along the Murine Alimentary Tract. Int. J. Mol. Sci. 2022, 23, 1783. https://doi.org/10.3390/ijms23031783
Vilchez-Vargas R, Salm F, Znalesniak EB, Haupenthal K, Schanze D, Zenker M, Link A, Hoffmann W. Profiling of the Bacterial Microbiota along the Murine Alimentary Tract. International Journal of Molecular Sciences. 2022; 23(3):1783. https://doi.org/10.3390/ijms23031783
Chicago/Turabian StyleVilchez-Vargas, Ramiro, Franz Salm, Eva B. Znalesniak, Katharina Haupenthal, Denny Schanze, Martin Zenker, Alexander Link, and Werner Hoffmann. 2022. "Profiling of the Bacterial Microbiota along the Murine Alimentary Tract" International Journal of Molecular Sciences 23, no. 3: 1783. https://doi.org/10.3390/ijms23031783
APA StyleVilchez-Vargas, R., Salm, F., Znalesniak, E. B., Haupenthal, K., Schanze, D., Zenker, M., Link, A., & Hoffmann, W. (2022). Profiling of the Bacterial Microbiota along the Murine Alimentary Tract. International Journal of Molecular Sciences, 23(3), 1783. https://doi.org/10.3390/ijms23031783