Molecular Docking and Molecular Dynamics Simulations Discover Curcumin Analogue as a Plausible Dual Inhibitor for SARS-CoV-2
Abstract
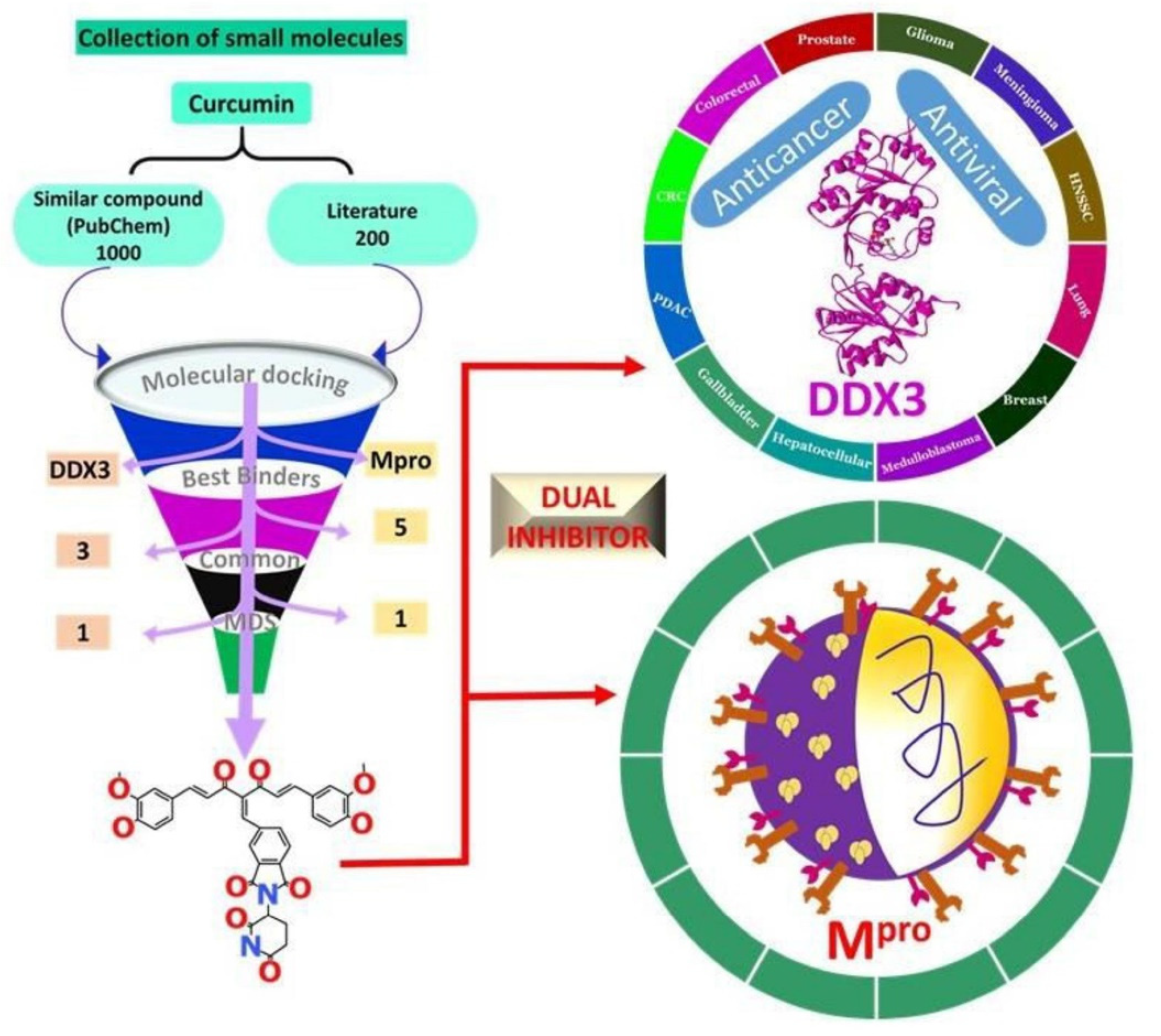
1. Introduction
2. Results
2.1. Effect of Natural Compound Analogues on Mpro
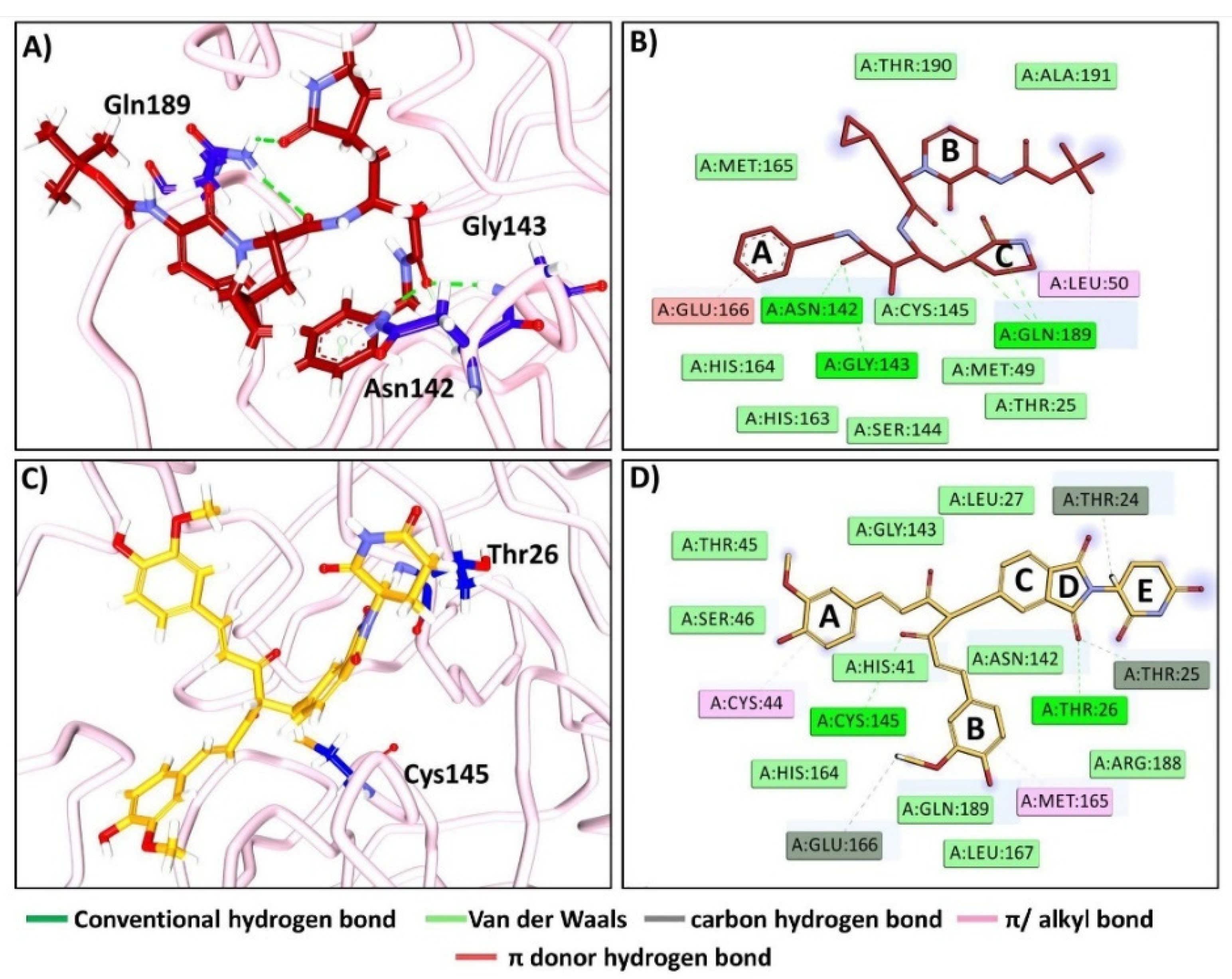
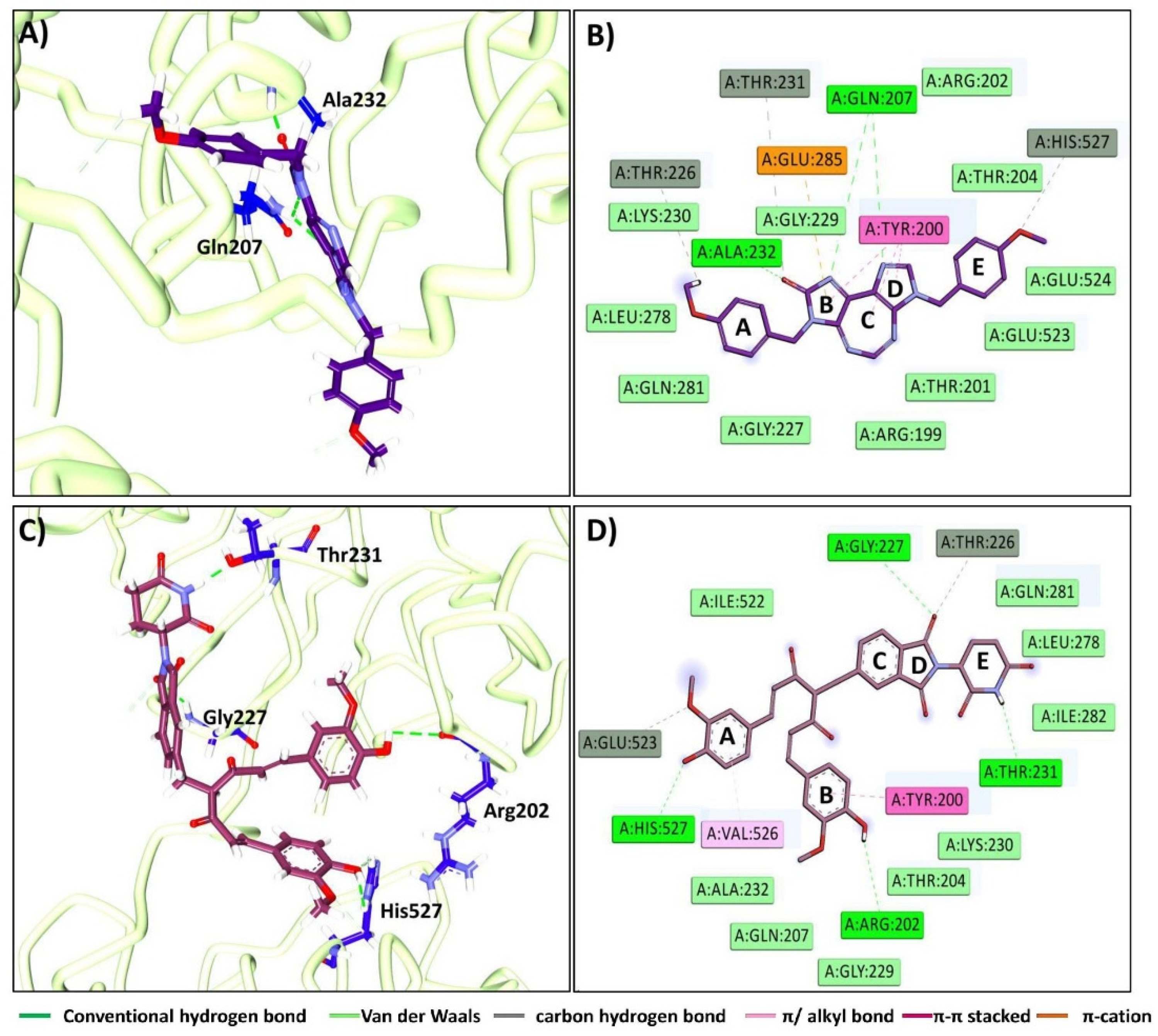
2.1.1. Binding Affinity Studies and the Section of the Pose
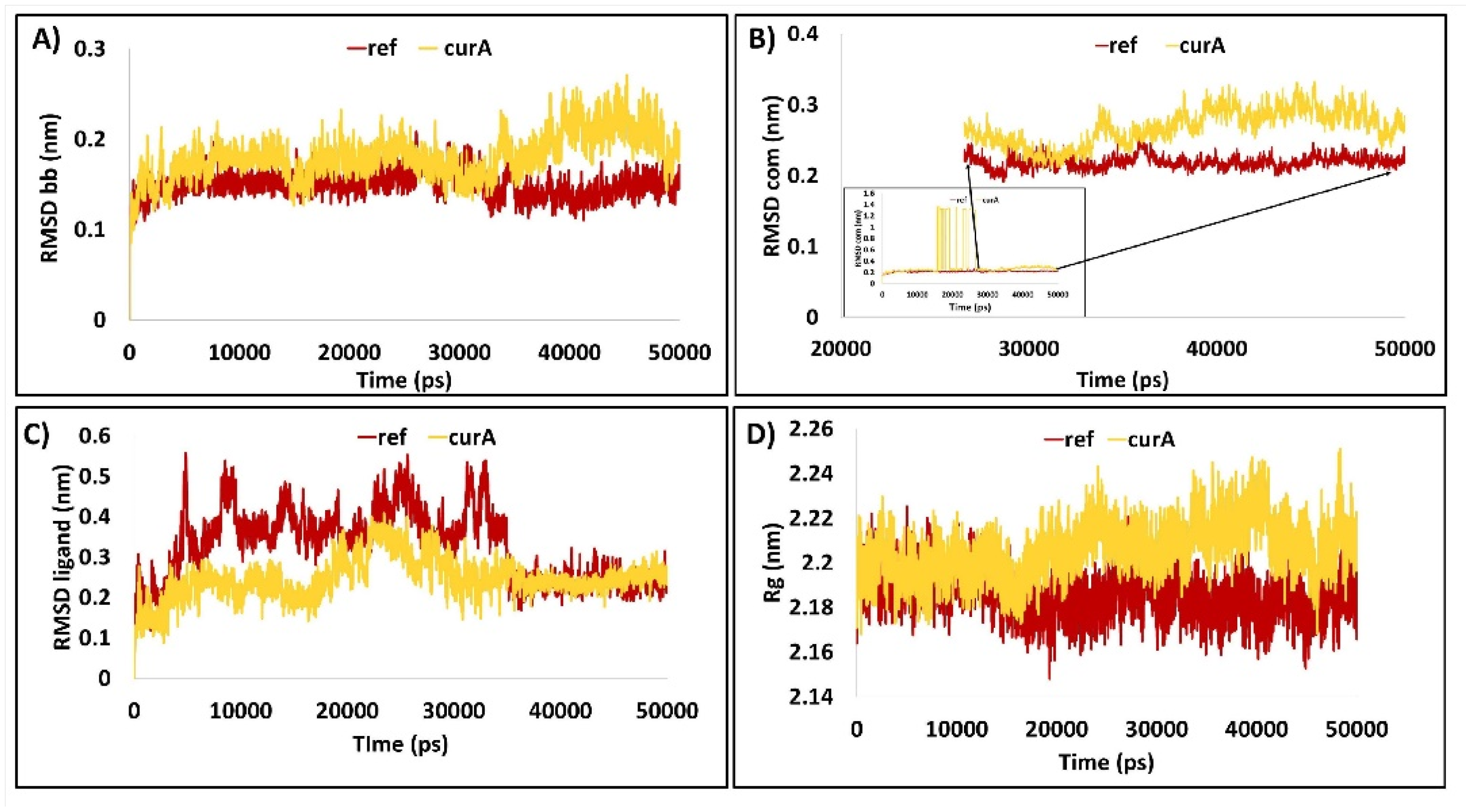
2.1.2. Molecular Dynamics Simulation Studies
2.1.3. The Compound Demonstrates a Stable RMSD
2.1.4. Protein Backbone Is Compact during the Simulation
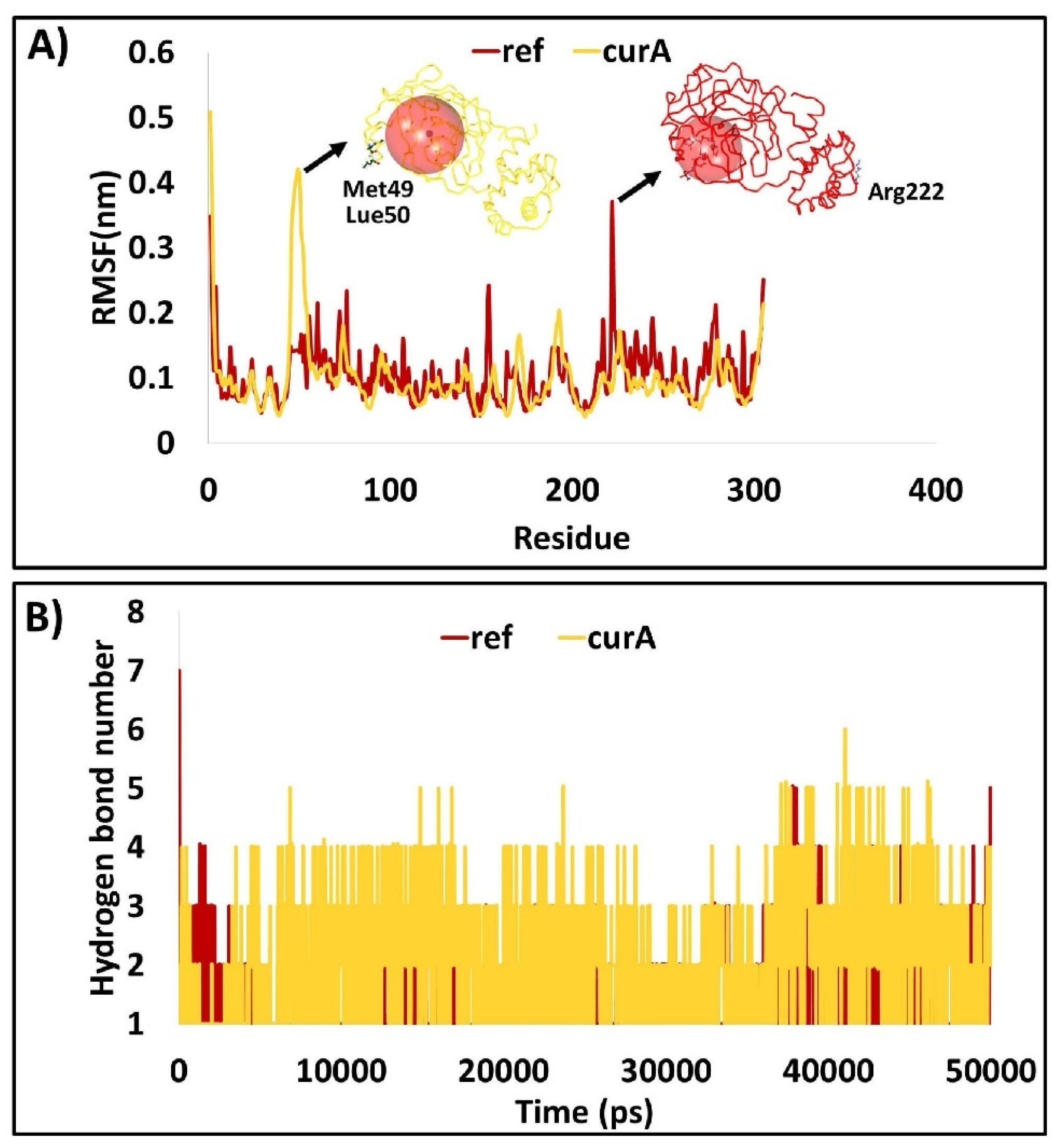
2.1.5. The Protein Backbone Has Demonstrated Negligible Fluctuations
2.1.6. Ligand Is Accommodated in the Binding Pocket throughout the Simulation Run
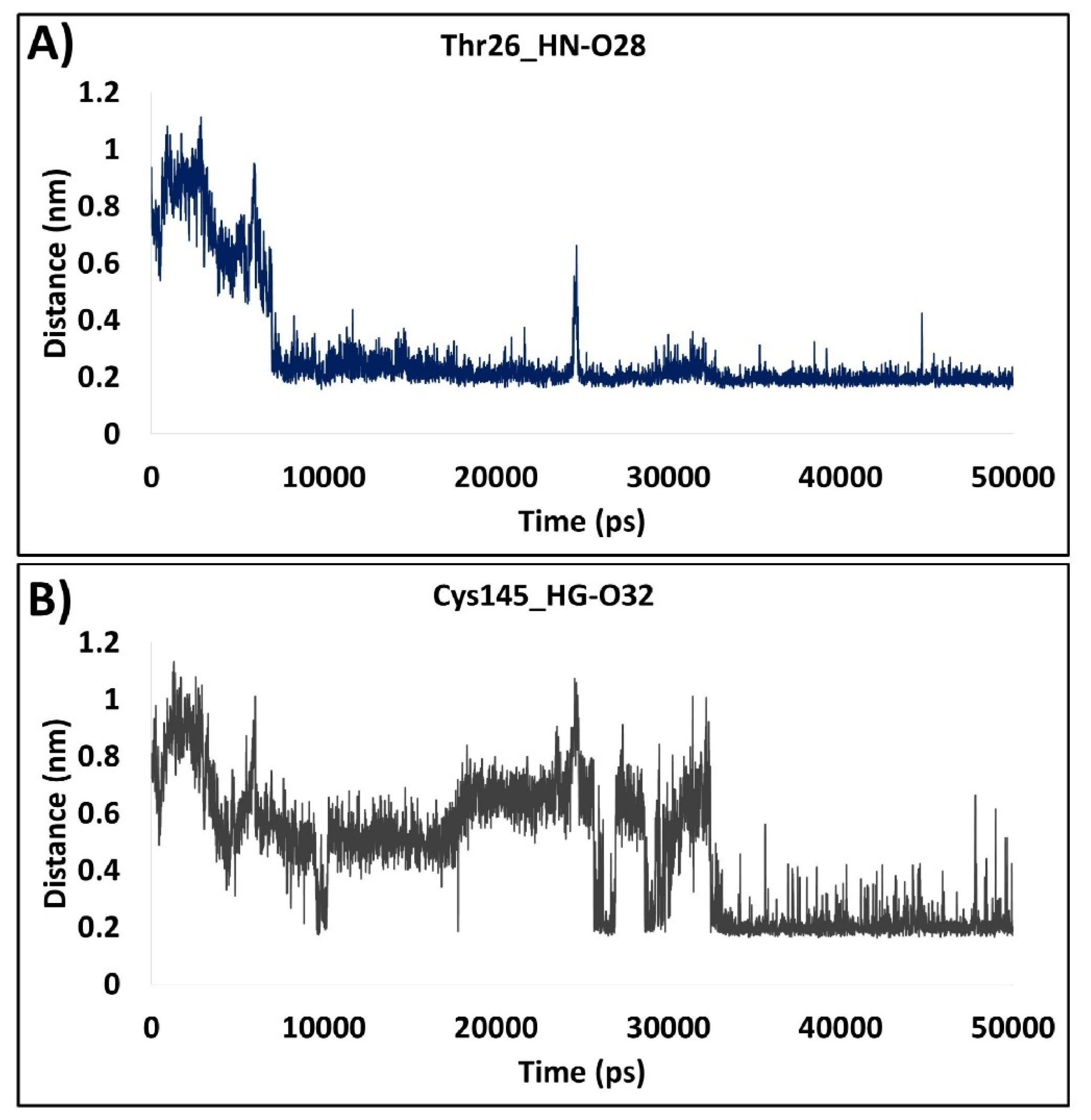
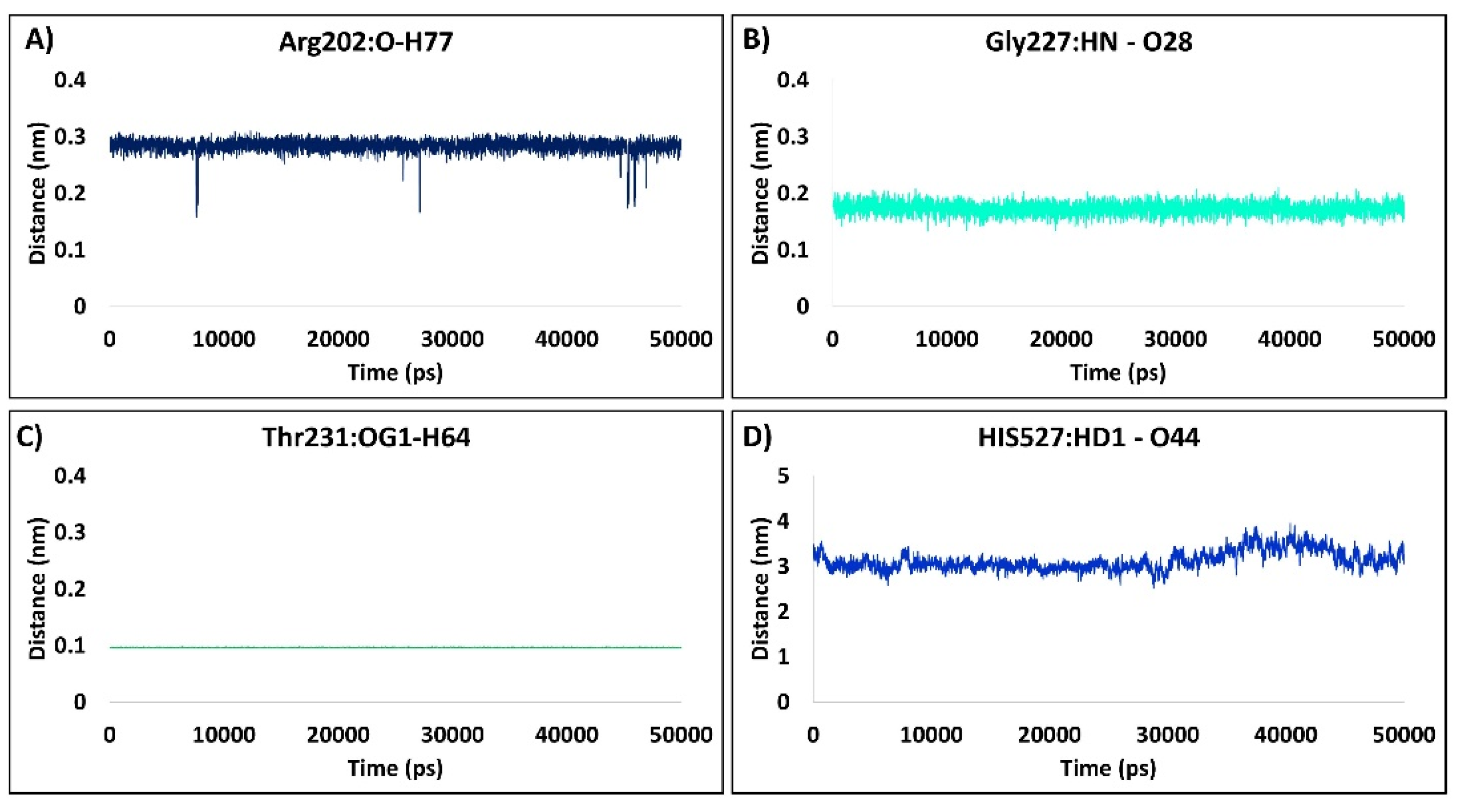
2.1.7. Distance Measure between the Interacting Residues and the Ligand Atoms
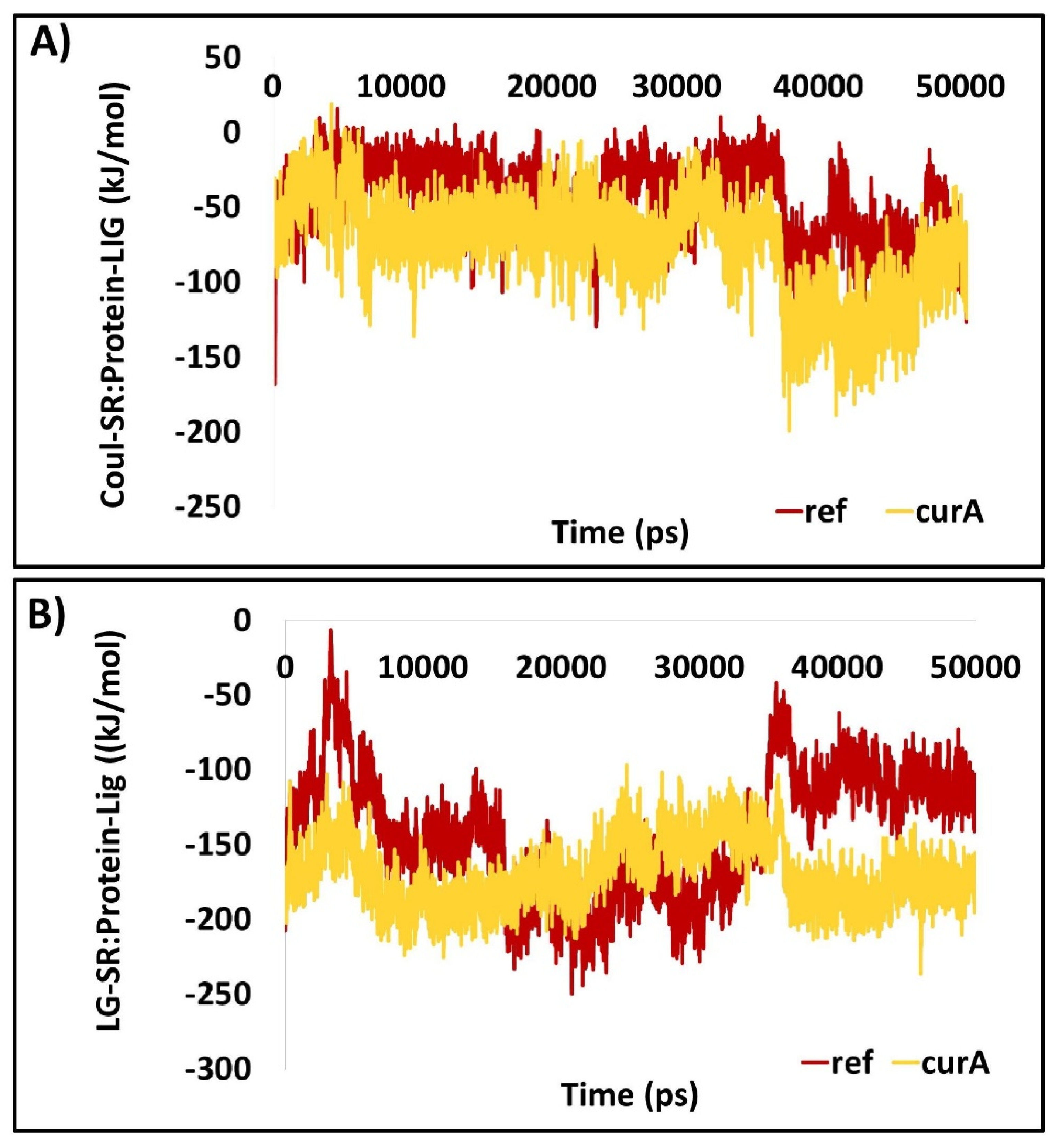
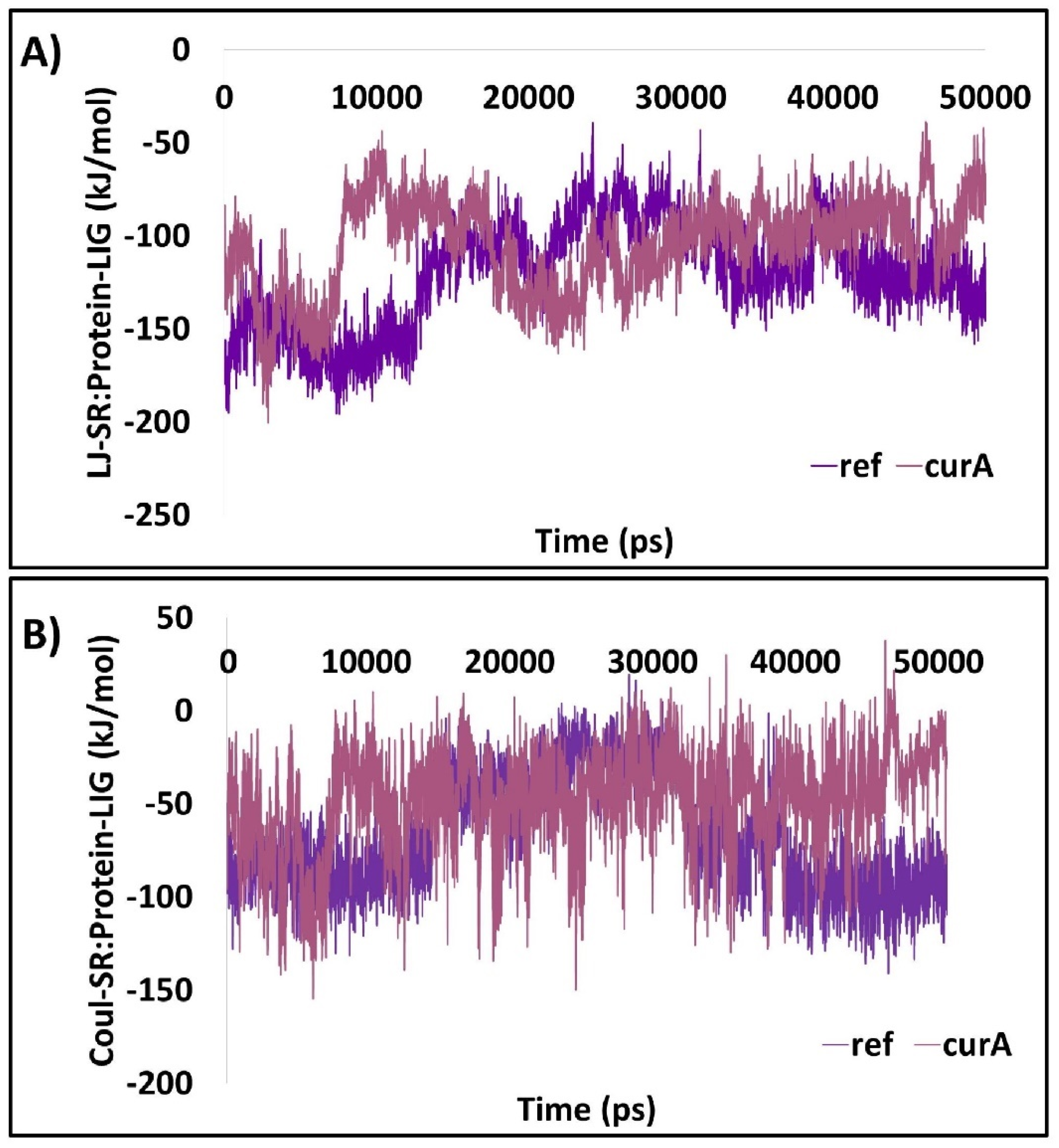
2.1.8. The Protein–Ligand Complex Projects the Better Interaction Energy
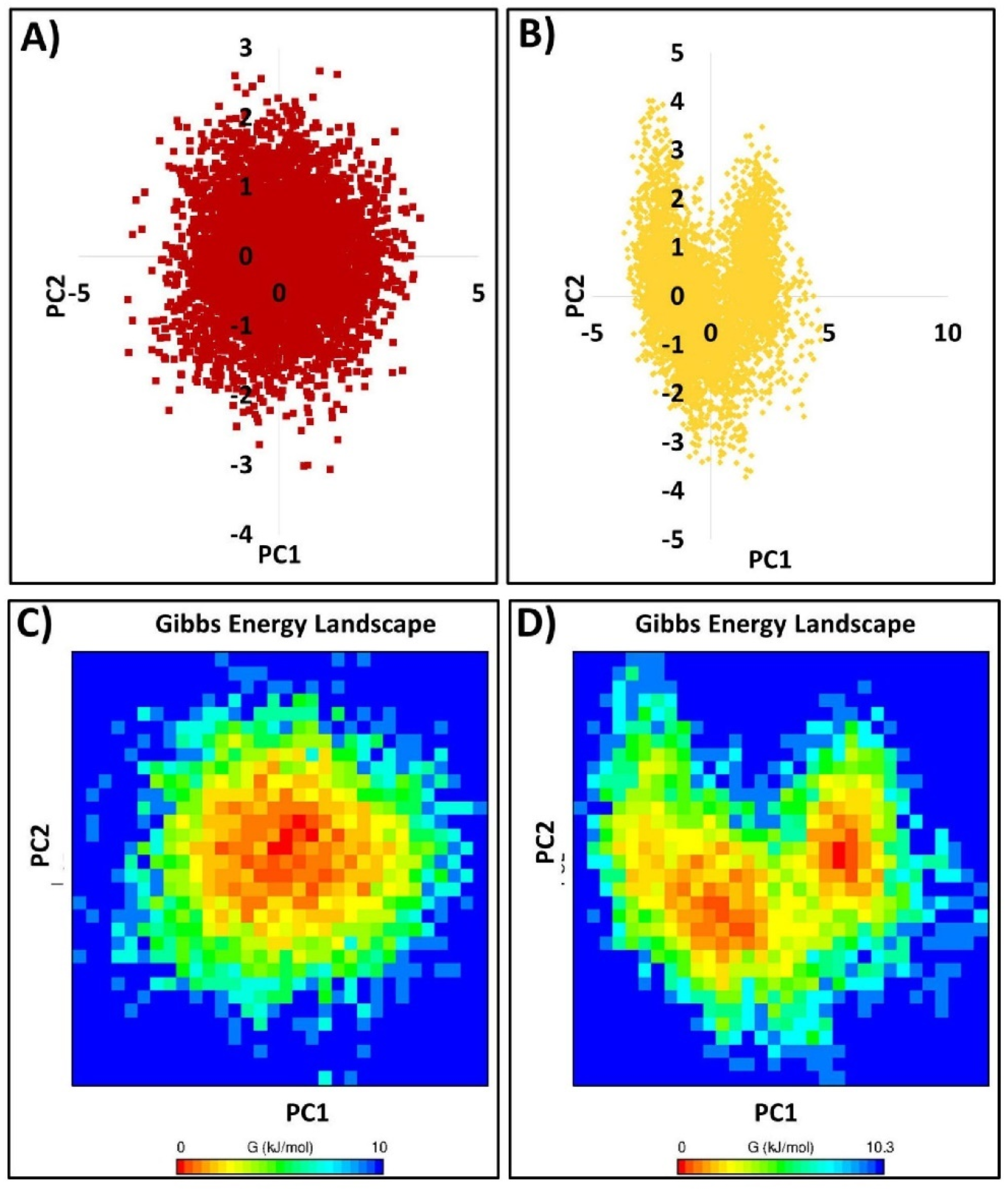
2.1.9. Essential Dynamics and Free Energy Landscape
2.2. Effect of Natural Compound Analogues on DDX3
2.2.1. Binding Affinity Studies
2.2.2. Molecular Dynamics Simulation Studies
2.2.3. Stability Analysis by Root Mean Square Deviation
2.2.4. Radius of Gyration
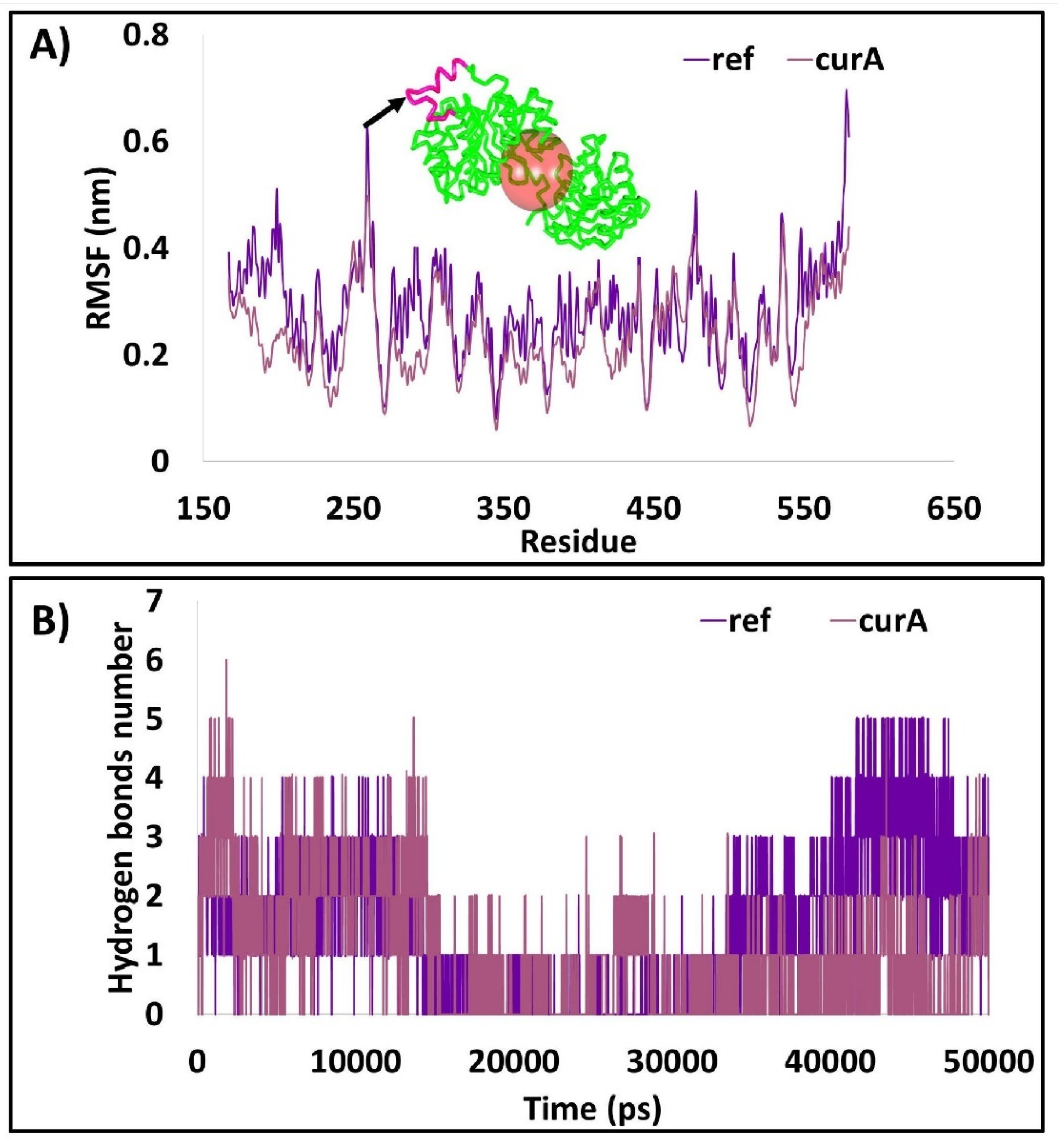
2.2.5. Root Mean Square Fluctuations
2.2.6. The Compound Accommodates at the Same Binding Pocket as the Cocrystallised Ligand
2.2.7. Distance Measure between the Interacting Residues and the Ligand
2.2.8. The Protein–Ligand Complex Projects the Better Interaction Energy
2.2.9. Essential Dynamics
3. Discussion
4. Materials and Methods
4.1. Target Structure
4.2. Selection of the Small Molecules
4.3. Molecular Docking to Predict the Binding Mode of the Ligands
4.4. Molecular Dynamics Simulation (MDS) Studies
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lin, L.-T.; Hsu, W.-C.; Lin, C.-C. Antiviral natural products and herbal medicines. J. Tradit. Complement. Med. 2014, 4, 24–35. [Google Scholar] [CrossRef]
- Noureddin, S.A.; El-Shishtawy, R.M.; Al-Footy, K.O. Curcumin analogues and their hybrid molecules as multifunctional drugs. Eur. J. Med. Chem. 2019, 182, 111631. [Google Scholar] [CrossRef]
- Mathew, D.; Hsu, W.-L. Antiviral potential of curcumin. J. Funct. Foods 2018, 40, 692–699. [Google Scholar] [CrossRef]
- Yang, Y.; Xiao, Z.; Ye, K.; He, X.; Sun, B.; Qin, Z.; Yu, J.; Yao, J.; Wu, Q.; Bao, Z.; et al. SARS-CoV-2: Characteristics and current advances in research. Virol. J. 2020, 17, 117. [Google Scholar] [CrossRef]
- Naqvi, A.A.T.; Fatima, K.; Mohammad, T.; Fatima, U.; Singh, I.K.; Singh, A.; Atif, S.M.; Hariprasad, G.; Hasan, G.M.; Hassan, M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165878. [Google Scholar] [CrossRef]
- Wu, D.; Wu, T.; Liu, Q.; Yang, Z. The SARS-CoV-2 outbreak: What we know. Int. J. Infect. Dis. 2020, 94, 44–48. [Google Scholar] [CrossRef]
- Kumar, S.; Nyodu, R.; Maurya, V.K.; Saxena, S.K. Morphology, Genome Organization, Replication, and Pathogenesis of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Coronavirus Dis. 2019 Epidemiol. Pathog. Diagn. Ther. 2020, 23–31. [Google Scholar] [CrossRef]
- Gahlawat, A.; Kumar, N.; Kumar, R.; Sandhu, H.; Singh, I.P.; Singh, S.; Sjöstedt, A.; Garg, P. Structure-Based Virtual Screening to Discover Potential Lead Molecules for the SARS-CoV-2 Main Protease. J. Chem. Inf. Model. 2020, 60, 5781–5793. [Google Scholar] [CrossRef]
- Alexpandi, R.; De Mesquita, J.F.; Pandian, S.K.; Ravi, A.V. Quinolines-Based SARS-CoV-2 3CLpro and RdRp Inhibitors and Spike-RBD-ACE2 Inhibitor for Drug-Repurposing Against COVID-19: An in silico Analysis. Front. Microbiol. 2020, 11, 1796. [Google Scholar] [CrossRef]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef]
- Sacco, M.D.; Ma, C.; Lagarias, P.; Gao, A.; Townsend, J.A.; Meng, X.; Dube, P.; Zhang, X.; Hu, Y.; Kitamura, N.; et al. Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against Mpro and Cathepsin L. Sci. Adv. 2020, 6, eabe0751. [Google Scholar] [CrossRef]
- Ghahremanpour, M.M.; Tirado-Rives, J.; Deshmukh, M.; Ippolito, J.A.; Zhang, C.-H.; de Vaca, I.C.; Liosi, M.-E.; Anderson, K.S.; Jorgensen, W.L. Identification of 14 Known Drugs as Inhibitors of the Main Protease of SARS-CoV-2. ACS Med. Chem. Lett. 2020, 11, 2526–2533. [Google Scholar] [CrossRef]
- Li, Z.; Li, X.; Huang, Y.; Wu, Y.; Liu, R.; Zhou, L.; Lin, Y. Identify potent SARS-CoV-2 main protease inhibitors via accelerated free energy perturbation-based virtual screening of existing drugs. BioRxiv 2020, 117, 27381–27387. [Google Scholar] [CrossRef]
- Guo, S.; Xie, H.; Lei, Y.; Liu, B.; Zhang, L.; Xu, Y.; Zuo, Z. Discovery of novel inhibitors against main protease (Mpro) of SARS-CoV-2 via virtual screening and biochemical evaluation. Bioorg. Chem. 2021, 110, 104767. [Google Scholar] [CrossRef]
- He, Y.; Zhang, D.; Yang, Y.; Wang, X.; Zhao, X.; Zhang, P.; Zhu, H.; Xu, N.; Liang, S. A double-edged function of DDX3, as an oncogene or tumor suppressor, in cancer progression (Review). Oncol. Rep. 2018, 39, 883–892. [Google Scholar] [CrossRef]
- Mo, J.; Liang, H.; Su, C.; Li, P.; Chen, J.; Zhang, B. DDX3X: Structure, physiologic functions and cancer. Mol. Cancer 2021, 20, 38. [Google Scholar] [CrossRef]
- Quaranta, P.; Lottini, G.; Chesi, G.; Contrafatto, F.; Russotto, R.; Macera, L.; Lai, M.; Spezia, P.G.; Brai, A.; Botta, M.; et al. DDX3 inhibitors show antiviral activity against positive-sense single-stranded RNA viruses but not against negative-sense single-stranded RNA viruses: The coxsackie B model. Antivir. Res. 2020, 178, 104750. [Google Scholar] [CrossRef]
- Valiente-Echeverría, F.; Hermoso, M.A.; Soto-Rifo, R. RNA helicase DDX3: At the crossroad of viral replication and antiviral immunity. Rev. Med. Virol. 2015, 25, 286–299. [Google Scholar] [CrossRef]
- Thulasi Raman, S.N.; Liu, G.; Pyo, H.M.; Cui, Y.C.; Xu, F.; Ayalew, L.E.; Tikoo, S.K.; Zhou, Y. DDX3 Interacts with Influenza A Virus NS1 and NP Proteins and Exerts Antiviral Function through Regulation of Stress Granule Formation. J. Virol. 2016, 90, 3661–3675. [Google Scholar] [CrossRef]
- Ciccosanti, F.; Di Rienzo, M.; Romagnoli, A.; Colavita, F.; Refolo, G.; Castilletti, C.; Agrati, C.; Brai, A.; Manetti, F.; Botta, L.; et al. Proteomic analysis identifies the RNA helicase DDX3X as a host target against SARS-CoV-2 infection. Antivir. Res. 2021, 190, 105064. [Google Scholar] [CrossRef]
- Van Voss, M.R.H.; Vesuna, F.; Trumpi, K.; Brilliant, J.; Berlinicke, C.; de Leng, W.; Kranenburg, O.; Offerhaus, G.J.; Bürger, H.; van der Wall, E.; et al. Identification of the DEAD box RNA helicase DDX3 as a therapeutic target in colorectal cancer. Oncotarget 2015, 6, 28312–28326. [Google Scholar] [CrossRef]
- Högbom, M.; Collins, R.; van den Berg, S.; Jenvert, R.M.; Karlberg, T.; Kotenyova, T.; Flores, A.; Hedestam, G.B.K.; Schiavone, L.H. Crystal Structure of Conserved Domains 1 and 2 of the Human DEAD-box Helicase DDX3X in Complex with the Mononucleotide AMP. J. Mol. Biol. 2007, 372, 150–159. [Google Scholar] [CrossRef]
- Jena, A.B.; Kanungo, N.; Nayak, V.; Chainy, G.B.N.; Dandapat, J. Catechin and curcumin interact with S protein of SARS-CoV2 and ACE2 of human cell membrane: Insights from computational studies. Sci. Rep. 2021, 11, 2043. [Google Scholar] [CrossRef]
- Shanmugarajan, D.; Prabitha, P.; Kumar, B.R.P.; Suresh, B. Curcumin to inhibit binding of spike glycoprotein to ACE2 receptors: Computational modelling, simulations, and ADMET studies to explore curcuminoids against novel SARS-CoV-2 targets. RSC Adv. 2020, 10, 31385–31399. [Google Scholar] [CrossRef]
- Patel, A.; Rajendran, M.; Shah, A.; Patel, H.; Pakala, S.B.; Karyala, P. Virtual screening of curcumin and its analogs against the spike surface glycoprotein of SARS-CoV-2 and SARS-CoV. J. Biomol. Struct. Dyn. 2021, 1–9. [Google Scholar] [CrossRef]
- Marín-Palma, D.; Tabares-Guevara, J.H.; Zapata-Cardona, M.I.; Flórez-Álvarez, L.; Yepes, L.M.; Rugeles, M.T.; Zapata-Builes, W.; Hernandez, J.C.; Taborda, N.A. Curcumin Inhibits in Vitro SARS-CoV-2 Infection in Vero E6 Cells through Multiple Antiviral Mechanisms. Molecules 2021, 26, 6900. [Google Scholar] [CrossRef]
- Kumar, M.; Sodhi, K.K.; Singh, D.K. Addressing the potential role of curcumin in the prevention of COVID-19 by targeting the Nsp9 replicase protein through molecular docking. Arch. Microbiol. 2021, 203, 1691–1696. [Google Scholar] [CrossRef]
- Liu, Z.; Ying, Y. The Inhibitory Effect of Curcumin on Virus-Induced Cytokine Storm and Its Potential Use in the Associated Severe Pneumonia. Front. Cell Dev. Biol. 2020, 8, 479. [Google Scholar] [CrossRef]
- Thimmulappa, R.K.; Mudnakudu-Nagaraju, K.K.; Shivamallu, C.; Subramaniam, K.J.T.; Radhakrishnan, A.; Bhojraj, S.; Kuppusamy, G. Antiviral and immunomodulatory activity of curcumin: A case for prophylactic therapy for COVID-19. Heliyon 2021, 7, e06350. [Google Scholar] [CrossRef]
- Manoharan, Y.; Haridas, V.; Vasanthakumar, K.C.; Muthu, S.; Thavoorullah, F.F.; Shetty, P. Curcumin: A Wonder Drug as a Preventive Measure for COVID19 Management. Indian J. Clin. Biochem. 2020, 35, 373–375. [Google Scholar] [CrossRef]
- Giordano, A.; Tommonaro, G. Curcumin and Cancer. Nutrients 2019, 11, 2376. [Google Scholar] [CrossRef] [PubMed]
- Mansouri, K.; Rasoulpoor, S.; Daneshkhah, A.; Abolfathi, S.; Salari, N.; Mohammadi, M.; Rasoulpoor, S.; Shabani, S. Clinical effects of curcumin in enhancing cancer therapy: A systematic review. BMC Cancer 2020, 20, 791. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Xu, Y.; Meng, L.; Huang, L.; Sun, H. Curcumin inhibits proliferation and promotes apoptosis of breast cancer cells. Exp. Ther. Med. 2018, 16, 1266–1272. [Google Scholar] [CrossRef]
- Mohammed, F.; Rashid-Doubell, F.; Taha, S.; Cassidy, S.; Fredericks, S. Effects of curcumin complexes on MDA-MB-231 breast cancer cell proliferation. Int. J. Oncol. 2020, 57, 445–455. [Google Scholar] [CrossRef]
- Rampogu, S.; Kim, S.M.; Son, M.; Baek, A.; Park, C.; Lee, G.; Kim, Y.; Kim, G.S.; Kim, J.H.; Lee, K.W. A computational approach with biological evaluation: Combinatorial treatment of curcumin and exemestane synergistically regulates ddx3 expression in cancer cell lines. Biomolecules 2020, 10, 857. [Google Scholar] [CrossRef]
- Mishra, C.B.; Pandey, P.; Sharma, R.D.; Malik, M.Z.; Mongre, R.K.; Lynn, A.M.; Prasad, R.; Jeon, R.; Prakash, A. Identifying the natural polyphenol catechin as a multi-targeted agent against SARS-CoV-2 for the plausible therapy of COVID-19: An integrated computational approach. Brief. Bioinform. 2020, 22, 1346–1360. [Google Scholar] [CrossRef]
- Hospital, A.; Goñi, J.R.; Orozco, M.; Gelpí, J.L. Molecular dynamics simulations: Advances and applications. Adv. Appl. Bioinform. Chem. 2015, 8, 37–47. [Google Scholar] [CrossRef]
- Kumar, C.V.; Swetha, R.G.; Anbarasu, A.; Ramaiah, S. Computational analysis reveals the association of threonine 118 methionine mutation in PMP22 resulting in CMT-1A. Adv. Bioinform. 2014, 2014, 502618. [Google Scholar] [CrossRef]
- Aier, I.; Varadwaj, P.K.; Raj, U. Structural insights into conformational stability of both wild-type and mutant EZH2 receptor. Sci. Rep. 2016, 6, 34984. [Google Scholar] [CrossRef]
- Rampogu, S.; Lemuel, M.R.; Lee, K.W. Virtual screening, molecular docking, molecular dynamics simulations and free energy calculations to discover potential DDX3 inhibitors. Adv. Cancer Biol.-Metastasis 2021, 4, 100022. [Google Scholar] [CrossRef]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Orhan, I.E.; Banach, M.; Rollinger, J.M.; Barreca, D.; Weckwerth, W.; Bauer, R.; Bayer, E.A.; et al. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef]
- Soni, V.K.; Mehta, A.; Ratre, Y.K.; Tiwari, A.K.; Amit, A.; Singh, R.P.; Sonkar, S.C.; Chaturvedi, N.; Shukla, D.; Vishvakarma, N.K. Curcumin, a traditional spice component, can hold the promise against COVID-19? Eur. J. Pharmacol. 2020, 886, 173551. [Google Scholar] [CrossRef] [PubMed]
- Bimonte, S.; Barbieri, A.; Palma, G.; Rea, D.; Luciano, A.; D’Aiuto, M.; Arra, C.; Izzo, F. Dissecting the Role of Curcumin in Tumour Growth and Angiogenesis in Mouse Model of Human Breast Cancer. Biomed Res. Int. 2015, 2015, 878134. [Google Scholar] [CrossRef] [PubMed]
- Frecer, V.; Miertus, S. Antiviral agents against COVID-19: Structure-based design of specific peptidomimetic inhibitors of SARS-CoV-2 main protease. RSC Adv. 2020, 10, 40244–40263. [Google Scholar] [CrossRef]
- Fakhar, Z.; Khan, S.; AlOmar, S.Y.; Alkhuriji, A.; Ahmad, A. ABBV-744 as a potential inhibitor of SARS-CoV-2 main protease enzyme against COVID-19. Sci. Rep. 2021, 11, 234. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Bhardwaj, T.; Kumar, A.; Gehi, B.R.; Kapuganti, S.K.; Garg, N.; Nath, G.; Giri, R. Reprofiling of approved drugs against SARS-CoV-2 main protease: An in-silico study. J. Biomol. Struct. Dyn. 2020, 1–15. [Google Scholar] [CrossRef]
- Nguyen, D.D.; Gao, K.; Chen, J.; Wang, R.; Wei, G.-W. Unveiling the molecular mechanism of SARS-CoV-2 main protease inhibition from 137 crystal structures using algebraic topology and deep learning. Chem. Sci. 2020, 11, 12036–12046. [Google Scholar] [CrossRef]
- Bhardwaj, V.K.; Singh, R.; Das, P.; Purohit, R. Evaluation of acridinedione analogs as potential SARS-CoV-2 main protease inhibitors and their comparison with repurposed anti-viral drugs. Comput. Biol. Med. 2021, 128, 104117. [Google Scholar] [CrossRef]
- Bol, G.M.; Xie, M.; Raman, V. DDX3, a potential target for cancer treatment. Mol. Cancer 2015, 14, 188. [Google Scholar] [CrossRef]
- Samal, S.K.; Routray, S.; Veeramachaneni, G.K.; Dash, R.; Botlagunta, M. Ketorolac salt is a newly discovered DDX3 inhibitor to treat oral cancer. Sci. Rep. 2015, 5, 9982. [Google Scholar] [CrossRef]
- Botlagunta, M.; Kollapalli, B.; Kakarla, L.; Gajarla, S.P.; Gade, S.P.; Dadi, C.L.; Penumadu, A.; Javeed, S. In vitro anti-cancer activity of doxorubicin against human RNA helicase, DDX3. Bioinformation 2016, 12, 347–353. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vyas, A.; Dandawate, P.; Padhye, S.; Ahmad, A.; Sarkar, F. Perspectives on new synthetic curcumin analogs and their potential anticancer properties. Curr. Pharm. Des. 2013, 19, 2047–2069. [Google Scholar] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Robertson, D.H.; Brooks, C.L.; Vieth, M. Detailed analysis of grid-based molecular docking: A case study of CDOCKER—A CHARMm-based MD docking algorithm. J. Comput. Chem. 2003, 24, 1549–1562. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Zoete, V.; Cuendet, M.A.; Grosdidier, A.; Michielin, O. SwissParam: A fast force field generation tool for smallorganic molecules. J. Comput. Chem. 2011, 32, 2359–2368. [Google Scholar] [CrossRef]
- Rampogu, S.; Lee, K.W. Old Drugs for New Purpose—Fast Pace Therapeutic Identification for SARS-CoV-2 Infections by Pharmacophore Guided Drug Repositioning Approach. Bull. Korean Chem. Soc. 2021, 42, 212–226. [Google Scholar] [CrossRef]
- Rampogu, S.; Parameswaran, S.; Lemuel, M.R.; Lee, K.W. Exploring the Therapeutic Ability of Fenugreek against Type 2 Diabetes and Breast Cancer Employing Molecular Docking and Molecular Dynamics Simulations. Evid.-Based Complement. Altern. Med. 2018, 2018, 1943203. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rampogu, S.; Lee, G.; Park, J.S.; Lee, K.W.; Kim, M.O. Molecular Docking and Molecular Dynamics Simulations Discover Curcumin Analogue as a Plausible Dual Inhibitor for SARS-CoV-2. Int. J. Mol. Sci. 2022, 23, 1771. https://doi.org/10.3390/ijms23031771
Rampogu S, Lee G, Park JS, Lee KW, Kim MO. Molecular Docking and Molecular Dynamics Simulations Discover Curcumin Analogue as a Plausible Dual Inhibitor for SARS-CoV-2. International Journal of Molecular Sciences. 2022; 23(3):1771. https://doi.org/10.3390/ijms23031771
Chicago/Turabian StyleRampogu, Shailima, Gihwan Lee, Jun Sung Park, Keun Woo Lee, and Myeong Ok Kim. 2022. "Molecular Docking and Molecular Dynamics Simulations Discover Curcumin Analogue as a Plausible Dual Inhibitor for SARS-CoV-2" International Journal of Molecular Sciences 23, no. 3: 1771. https://doi.org/10.3390/ijms23031771
APA StyleRampogu, S., Lee, G., Park, J. S., Lee, K. W., & Kim, M. O. (2022). Molecular Docking and Molecular Dynamics Simulations Discover Curcumin Analogue as a Plausible Dual Inhibitor for SARS-CoV-2. International Journal of Molecular Sciences, 23(3), 1771. https://doi.org/10.3390/ijms23031771




