Multi Platforms Strategies and Metabolomics Approaches for the Investigation of Comprehensive Metabolite Profile in Dogs with Babesia canis Infection
Abstract
:1. Introduction
2. Results
2.1. The Metabolomics Dataset in Dogs Infected with B. canis
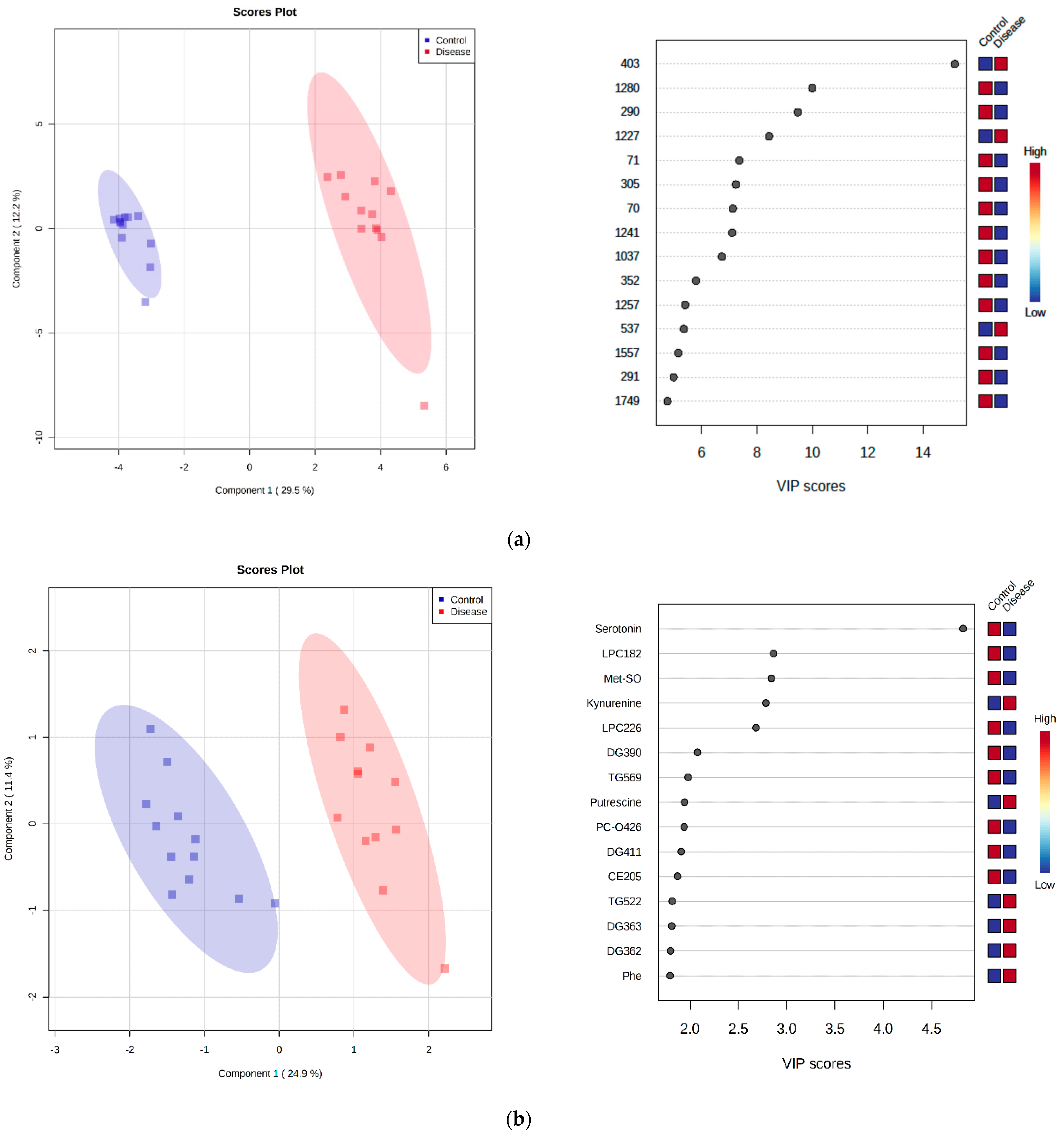
2.2. Untargeted LC-MS Metabolomics Analysis
2.3. Targeted LC-MS Metabolomics Analysis (Biocrates Analysis)
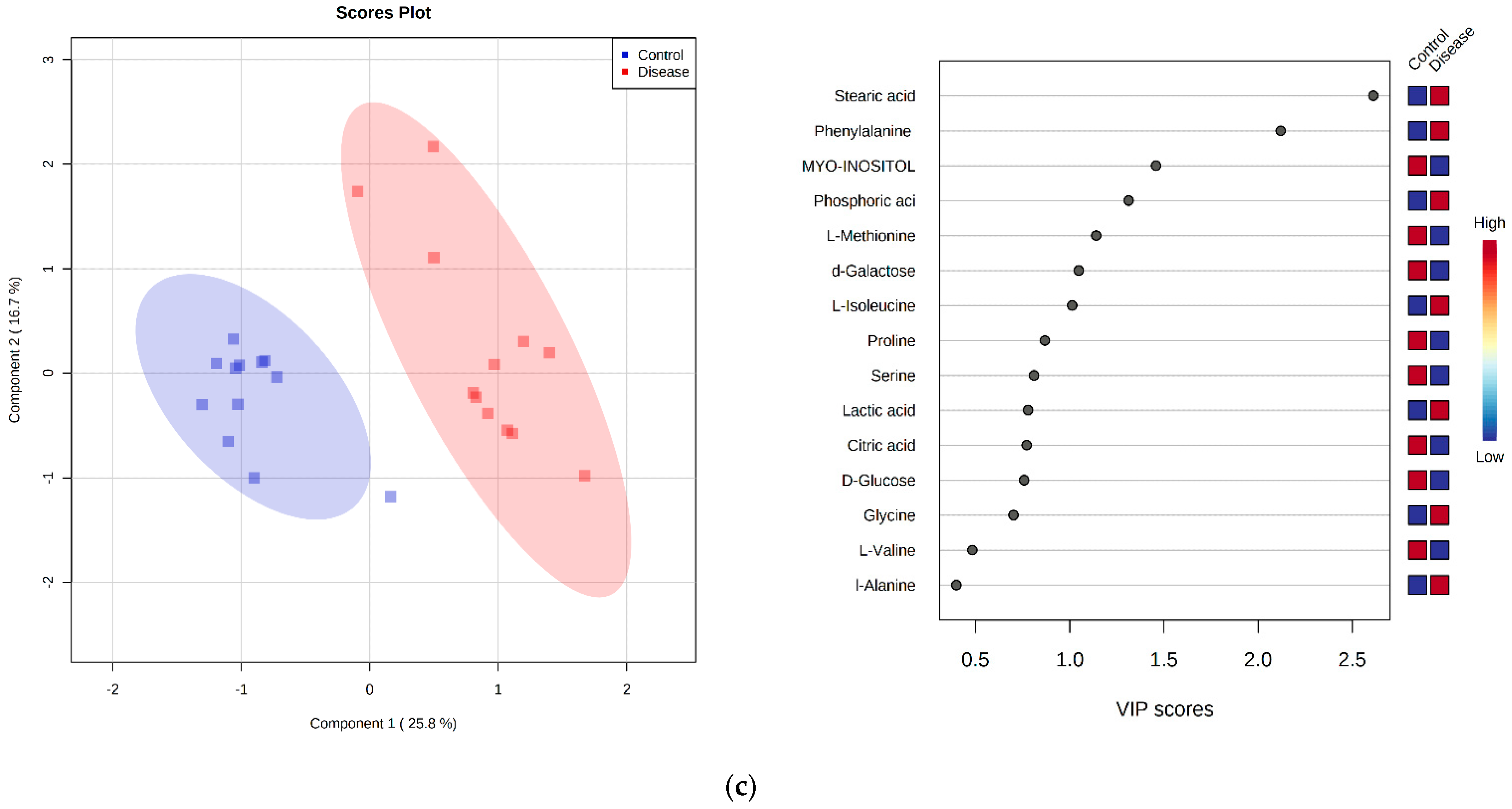
2.4. GC-MS Metabolomics Analysis
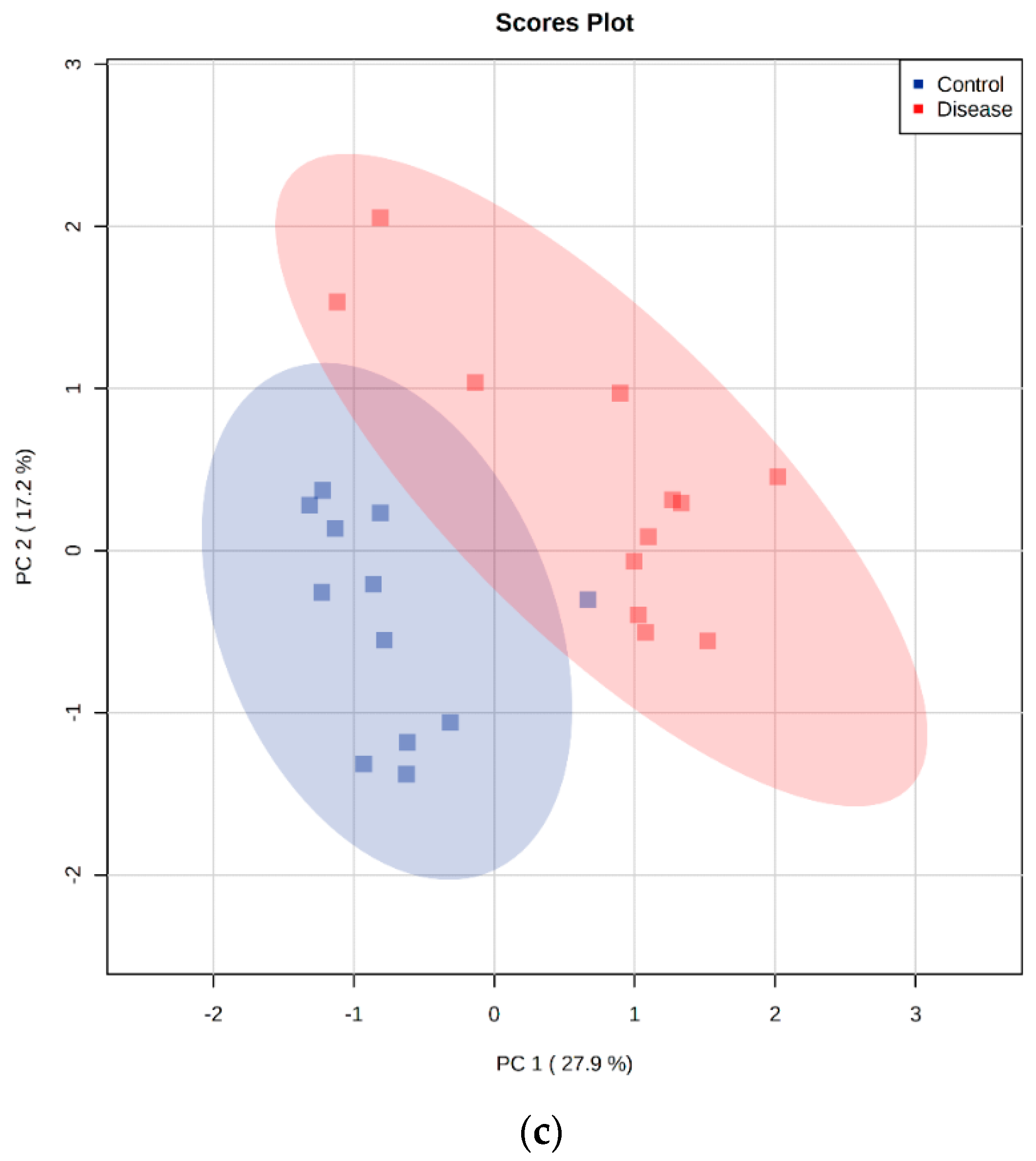
2.5. Principal Component Analysis
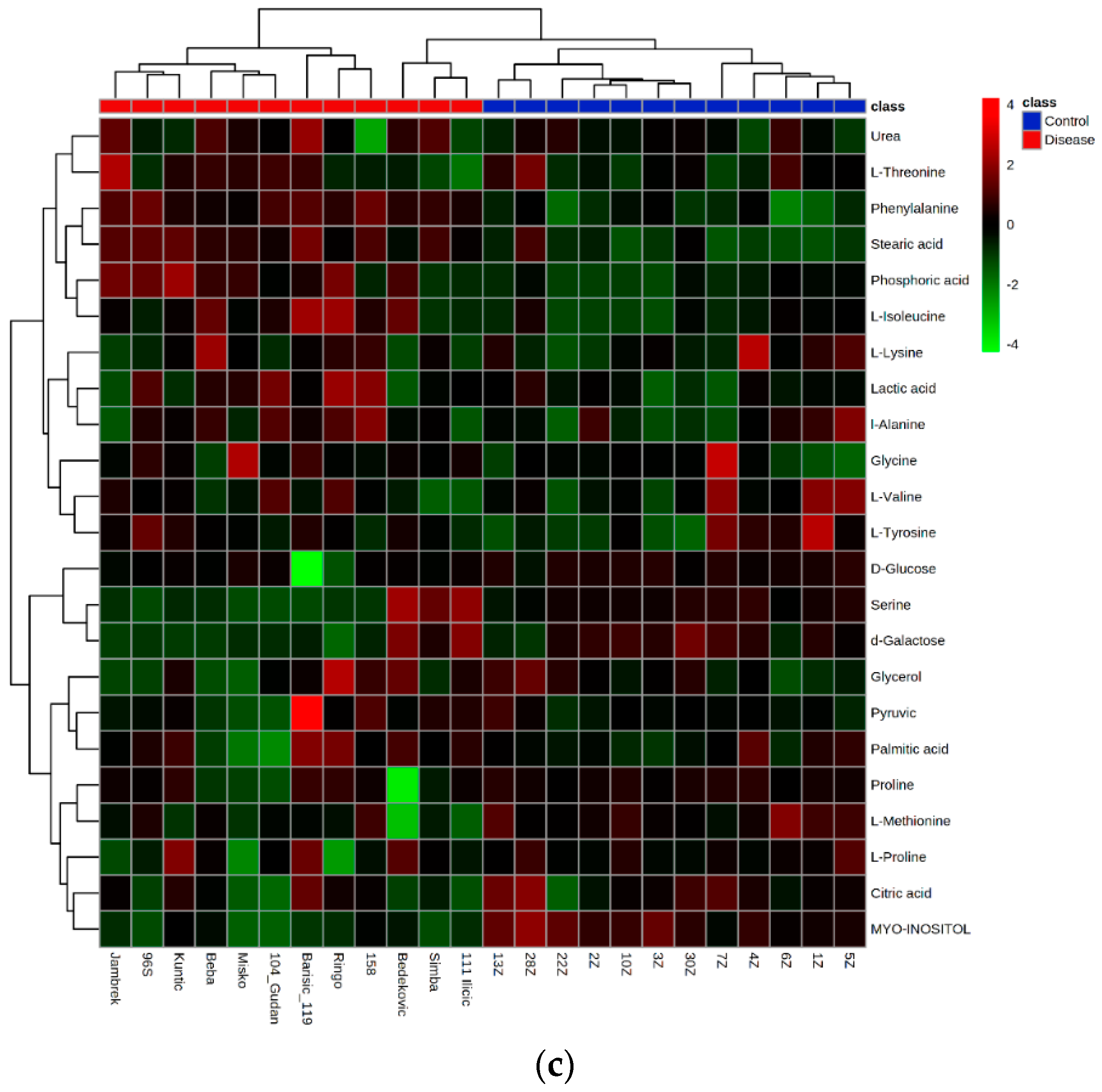
2.6. Hierarchical Clustering Analysis
2.7. Discriminant Metabolite Identification
2.8. Pathway Analysis and Enrichment Analysis
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Experimental Design
4.3. Blood Sample Preparation and Analysis
4.4. Untargeted LC-MS Metabolomics Analysis
4.5. Targeted LC-MS Metabolomics Analysis
4.6. GC-MS Metabolomics Analysis
4.7. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bilić, P.; Kuleš, J.; Barić Rafaj, R.; Mrljak, V. Canine babesiosis: Where do we stand? Acta Vet. Beogr. 2018, 68, 127–160. [Google Scholar] [CrossRef] [Green Version]
- Solano-Gallego, L.; Sainz, Á.; Roura, X.; Estrada-Peña, A.; Miró, G. A review of canine babesiosis: The European perspective. Parasites Vectors 2016, 9, 336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beck, R.; Vojta, L.; Mrljak, V.; Marinculić, A.; Beck, A.; Živičnjak, T.; Cacciò, S.M. Diversity of Babesia and Theileria species in symptomatic and asymptomatic dogs in Croatia. Int. J. Parasitol. 2009, 39, 843–848. [Google Scholar] [CrossRef] [PubMed]
- Matijatko, V.; Torti, M.; Schetters, T.P. Canine babesiosis in Europe: How many diseases? Trends Parasitol. 2012, 28, 99–105. [Google Scholar] [CrossRef]
- Jacobson, L.S.; A Clark, I. The pathophysiology of canine babesiosis: New approaches to an old puzzle. J. S. Afr. Vet. Assoc. 1994, 65, 134–145. [Google Scholar]
- Taboada, J.; Merchant, S.R. Babesiosis of Companion Animals and Man. Vet. Clin. N. Am. Small Anim. Pract. 1991, 21, 103–123. [Google Scholar] [CrossRef]
- Bone, R.C.; Balk, R.A.; Cerra, F.B.; Dellinger, R.P.; Fein, A.M.; Knaus, W.A.; Schein, R.M.; Sibbald, W.J. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Chest 1992, 101, 1644–1655. [Google Scholar] [CrossRef] [Green Version]
- Greene, C.E. Babesiosis. In Infectious Diseases of the Dog and Cat; Taboada, J., Ed.; W.B. Saunders Company: St. Louis, MO, USA, 1998; pp. 473–481. ISBN 9780323266215. [Google Scholar]
- Schetters, T.; Kleuskens, J.; Van De Crommert, J.; De Leeuw, P.; Finizio, A.-L.; Gorenflot, A. Systemic inflammatory responses in dogs experimentally infected with Babesia canis; a haematological study. Vet. Parasitol. 2009, 162, 7–15. [Google Scholar] [CrossRef]
- Matijatko, V.; Mrljak, V.; Kiš, I.; Kučer, N.; Foršek, J.; Živičnjak, T.; Romić, Ž.; Šimec, Z.; Ceron, J.J. Evidence of an acute phase response in dogs naturally infected with Babesia canis. Vet. Parasitol. 2007, 144, 242–250. [Google Scholar] [CrossRef]
- Boozer, A.L.; Macintire, D.K. Canine babesiosis. Vet. Clin. N. Am. Small Anim. Pract. 2003, 33, 885–904. [Google Scholar] [CrossRef]
- Muller-Werdan, U.; Schuster, H.P. Abriss der Pathophysiologie als Grundlage der Therapie; Sepsis und MODS; Werdan, K., Schuster, H.P., Muller-Werdan, U., Eds.; Springer: Berlin/Heidelberg, Germany, 2005; pp. 23–61. ISBN 3540265872. [Google Scholar]
- Clark, I.A.; Jacobson, L.S. Do babesiosis and malaria share a common disease process? Ann. Trop. Med. Parasitol. 1998, 92, 483–488. [Google Scholar] [CrossRef] [PubMed]
- Moore, R.E.; Kirwan, J.; Doherty, M.K.; Whitfield, P.D. Biomarker discovery in animal health and disease: The application of post-genomic technologies. Biomark. Insights 2007, 2, 185–196. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2717813/ (accessed on 25 November 2021). [CrossRef] [PubMed] [Green Version]
- Karakitsou, E.; Foguet, C.; de Atauri, P.; Kultima, K.; Khoonsari, P.E.; Martins dos Santos, V.A.P.; Rosato, A.; Cascante, M. Metabolomics in systems medicine: An overview of methods and applications. Curr. Opin. Syst. Biol. 2019, 15, 91–99. [Google Scholar] [CrossRef]
- Roberts, L.D.; Souza, A.L.; Gerszten, R.E.; Clish, C.B. Targeted Metabolomics. Curr. Protoc. Mol. Biol. 2012, 30, 30.2.1–30.2.24. [Google Scholar] [CrossRef] [PubMed]
- Klupczynska, A.; Derezinski, P.; Kokot, Z.J. Metabolomics in medical sciences—trends, challenges and perspectives. Acta Pol. Pharm. 2015, 72, 629–641. Available online: https://pubmed.ncbi.nlm.nih.gov/26647618/ (accessed on 25 November 2021).
- Issaq, H.J.; Van, Q.N.; Waybright, T.J.; Muschik, G.M.; Veenstra, T.D. Analytical and statistical approaches to metabolomics research. J. Sep. Sci. 2009, 32, 2183–2199. [Google Scholar] [CrossRef]
- Gowda, G.A.N.; Djukovic, D. Overview of mass spectrometry-based metabolomics: Opportunities and challenges. Methods Mol. Biol. 2014, 1198, 3–12. [Google Scholar] [CrossRef] [Green Version]
- Tamai, R.; Furuya, M.; Hatoya, S.; Akiyoshi, H.; Yamamoto, R.; Komori, Y.; Yokoi, S.I.; Tani, K.; Hirano, Y.; Komori, M.; et al. Profiling of serum metabolites in canine lymphoma using gas chromatography mass spectrometry. J. Vet. Med. Sci. 2014, 76, 1513–1518. [Google Scholar] [CrossRef] [Green Version]
- Minamoto, Y.; Otoni, C.C.; Steelman, S.M.; Büyükleblebici, O.; Steiner, J.M.; Jergens, A.E.; Suchodolski, J.S. Alteration of the fecal microbiota and serum metabolite profiles in dogs with idiopathic inflammatory bowel disease. Gut Microbes 2015, 6, 33–47. [Google Scholar] [CrossRef] [Green Version]
- O’Kell, A.L.; Garrett, T.J.; Wasserfall, C.; Atkinson, M.A. Untargeted metabolomic analysis in naturally occurring canine diabetes mellitus identifies similarities to human Type 1 Diabetes. Sci. Rep. 2017, 7, 9467. [Google Scholar] [CrossRef] [Green Version]
- Abdullah, M.; Kornegay, J.N.; Honcoop, A.; Parry, T.L.; Balog-Alvarez, C.J.; O’Neal, S.K.; Bain, J.R.; Muehlbauer, M.J.; Newgard, C.B.; Patterson, C.; et al. Non-targeted metabolomics analysis of Golden Retriever Muscular Dystrophy-affected muscles reveals alterations in arginine and proline metabolism, and elevations in glutamic and oleic acid in vivo. Metabolites 2017, 7, 38. [Google Scholar] [CrossRef] [PubMed]
- De Souza, A.I.; Cardin, S.; Wait, R.; Chung, Y.L.; Vijayakumar, M.; Maguy, A.; Camm, A.J.; Nattel, S. Proteomic and metabolomic analysis of atrial profibrillatory remodelling in congestive heart failure. J. Mol. Cell. Cardiol. 2010, 49, 851–863. [Google Scholar] [CrossRef] [PubMed]
- Shibayama, J.; Yuzyuk, T.N.; Cox, J.; Makaju, A.; Miller, M.; Lichter, J.; Li, H.; Leavy, J.D.; Franklin, S.; Zaitsev, A.V. Metabolic remodeling in moderate synchronous versus dyssynchronous pacing-induced heart failure: Integrated metabolomics and proteomics study. PLoS ONE 2015, 10, e0118974. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wei, S.; Liu, L.; Gowda, G.N.; Bonney, P.; Stewart, J.; Knapp, D.W.; Raftery, D. NMR-based metabolomics study of canine bladder cancer. Biochim. Biophys. Acta Mol. Basis Dis. 2012, 1822, 1807–1814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlos, G.; Dos Santos, F.P.; Fröehlich, P.E. Canine metabolomics advances. Metabolomics 2020, 16, 16. [Google Scholar] [CrossRef]
- Kuleš, J.; Rubić, I.; Beer Ljubić, B.; Bilić, P.; Barić Rafaj, R.; Brkljačić, M.; Burchmore, R.; Eckersall, P.D.; Mrljak, V. Combined Untargeted and Targeted Metabolomics Approaches Reveal Urinary Changes of Amino Acids and Energy Metabolism in Canine Babesiosis With Different Levels of Kidney Function. Front. Microbiol. 2021, 12, 715701. [Google Scholar] [CrossRef] [PubMed]
- Paget, T.; Haroune, N.; Bagchi, S.; Jarroll, E. Metabolomics and protozoan parasites. Acta Parasitol. 2013, 58, 127–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akpunarlieva, S.; Weidt, S.; Lamasudin, D.; Naula, C.; Henderson, D.; Barrett, M.; Burchmore, R. Integration of proteomics and metabolomics to elucidate metabolic adaptation in Leishmania. J. Proteom. 2017, 155, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Atan, N.A.D.; Koushki, M.; Ahmadi, N.A.; Rezaei-Tavirani, M. Metabolomics-based studies in the field of Leishmania/leishmaniasis. Alex. J. Med. 2018, 54, 383–390. [Google Scholar] [CrossRef] [Green Version]
- Vincent, I.M.; Creek, D.J.; Burgess, K.; Woods, D.J.; Burchmore, R.J.S.; Barrett, M.P. Untargeted Metabolomics Reveals a Lack of Synergy between Nifurtimox and Eflornithine against Trypanosoma brucei. PLoS Negl. Trop. Dis. 2012, 6, e1618. [Google Scholar] [CrossRef] [Green Version]
- Barrett, M.P.; Bakker, B.M.; Breitling, R. Metabolomic systems biology of trypanosomes. Parasitology 2010, 137, 1285–1290. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Feng, G.; Zhang, Q.; Cao, J. From Metabolite to Metabolome: Metabolomics Applications in Plasmodium Research. Front. Microbiol. 2021, 11, 626183. [Google Scholar] [CrossRef] [PubMed]
- Na, J.; Khan, A.; Kim, J.K.; Wadood, A.; Choe, Y.L.; Walker, D.I.; Jones, D.P.; Lim, C.S.; Park, Y.H. Discovery of metabolic alterations in the serum of patients infected with Plasmodium spp. by high-resolution metabolomics. Metabolomics 2020, 16, 9. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef] [PubMed]
- Pederzolli, C.D.; Sgaravatti, Â.M.; Braum, C.A.; Prestes, C.C.; Zorzi, G.K.; Sgarbi, M.B.; Wyse, A.T. 5-Oxoproline Reduces Non-Enzymatic Antioxidant Defenses in vitro in Rat Brain. Metab. Brain Dis. 2007, 22, 51–65. [Google Scholar] [CrossRef]
- Crnogaj, M.; Cerón, J.J.; Šmit, I.; Kiš, I.; Gotić, J.; Brkljačić, M.; Matijatko, V.; Rubio, C.P.; Kučer, N.; Mrljak, V. Relation of antioxidant status at admission and disease severity and outcome in dogs naturally infected with Babesia canis canis. BMC Vet. Res. 2017, 13, 114. [Google Scholar] [CrossRef] [Green Version]
- Petrat, F.; Boengler, K.; Schulz, R.; de Groot, H. Glycine, a simple physiological compound protecting by yet puzzling mechanism(s) against ischaemia-reperfusion injury: Current knowledge. Br. J. Pharmacol. 2012, 165, 2059–2072. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Wu, Z.; Lin, G.; Hu, S.; Wang, B.; Dai, Z.; Wu, G. Glycine stimulates protein synthesis and inhibits oxidative stress in pig small intestinal epithelial cells. J. Nutr. 2014, 144, 1540–1548. [Google Scholar] [CrossRef] [Green Version]
- Effenberger-Neidnicht, K.; Jägers, J.; Verhaegh, R.; de Groot, H. Glycine selectively reduces intestinal injury during endotoxemia. J. Surg. Res. 2014, 192, 592–598. [Google Scholar] [CrossRef]
- Meyer, K.F.; Martins, J.L.; de Freitas Filho, L.G.; Oliva, M.L.; Patrício, F.R.; Macedo, M.; Wang, L. Glycine reduces tissue lipid peroxidation in hypoxia-reoxygenation-induced necrotizing enterocolitis in rats. Acta Cir. Bras. 2006, 21, 161–167. [Google Scholar] [CrossRef] [Green Version]
- Zhong, Z.; Wheeler, M.D.; Li, X.; Froh, M.; Schemmer, P.; Yin, M.; Bunzendaul, H.; Bradford, B.; Lemasters, J.J. L-Glycine: A novel antiinflammatory, immunomodulatory, and cytoprotective agent. Curr. Opin. Clin. Nutr. Metab. Care 2003, 6, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, X.; Wu, H.; Chen, S.; Zhu, H.; Zhang, J.; Hou, Y.; Hu, C.-A.A.; Zhang, G. Glycine enhances muscle protein mass associated with maintaining Akt-mTOR-FOXO1 signaling and suppressing TLR4 and NOD2 signaling in piglets challenged with LPS. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2016, 311, R365–R373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.T. Regulative Effect of Glycine on Intestinal Injury and Muscle Protein Sythesis and Degradation of Piglets after Lipopolysaccharide Challenge. Master’s Thesis, Wuhan Polytechnic University, Wuhan, China, 2015. Available online: https://pubmed.ncbi.nlm.nih.gov/28832517/ (accessed on 25 November 2021).
- Liu, Y.; Wang, X.; Hu, C.A. Therapeutic Potential of Amino Acids in Inflammatory Bowel Disease. Nutrients 2017, 9, 920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsune, I.; Ikejima, K.; Hirose, M.; Yoshikawa, M.; Enomoto, N.; Takei, Y.; Sato, N. Dietary glycine prevents chemical-induced experimental colitis in the rat. Gastroenterology 2003, 125, 775–785. [Google Scholar] [CrossRef]
- Cordy, R.J.; Patrapuvich, R.; Lili, L.N.; Cabrera-Mora, M.; Chien, J.T.; Tharp, G.K.; Khadka, M.; Meyer, E.V.; Lapp, S.A. Distinct amino acid and lipid perturbations characterize acute versus chronic malaria. JCI Insight 2019, 4, e125156. [Google Scholar] [CrossRef]
- Sherman, I.W. Amino acid metabolism and protein synthesis in malarial parasites. Bull World Health Organ. 1977, 55, 265–276. Available online: https://pubmed.ncbi.nlm.nih.gov/338183/ (accessed on 25 November 2021).
- Algranati, I.D. Polyamine metabolism in Trypanosoma cruzi: Studies on the expression and regulation of heterologous genes involved in polyamine biosynthesis. Amino Acids 2010, 38, 645–651. [Google Scholar] [CrossRef]
- Rojas-Martínez, C.; Rodríguez-Vivas, R.I.; Figueroa Millán, J.V.; Acosta Viana, K.Y.; Gutiérrez Ruiz, E.J.; Álvarez Martínez, J.A. Putrescine: Essential factor for in vitro proliferation of Babesia bovis. Exp. Parasitol. 2017, 175, 79–84. [Google Scholar] [CrossRef]
- Cook, T.; Roos, D.; Morada, M.; Zhu, G.; Keithly, J.S.; Feagin, J.E.; Wu, G.; Yarlett, N. Divergent polyamine metabolism in the Apicomplexa. Microbiology 2007, 153, 1123–1130. [Google Scholar] [CrossRef] [Green Version]
- Clark, K.; Niemand, J.; Reeksting, S.; Smit, S.; van Brummelen, A.C.; Williams, M.; Louw, A.I.; Birkholtz, L. Functional consequences of perturbing polyamine metabolism in the malaria parasite. Plasmodium Falciparum Amino Acids 2010, 38, 633–644. [Google Scholar] [CrossRef]
- Lindell, S.E.; Schayer, R.W. Formation of histamine in the kidney of the dogs. Brit. J. Pharmacol. 1958, 13, 89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Branco, A.C.C.C.; Yoshikawa, F.S.Y.; Pietrobon, A.J.; Sato, M.N. Role of Histamine in Modulating the Immune Response and Inflammation. Mediat. Inflamm. 2018, 2018, 1–10. [Google Scholar] [CrossRef]
- MacGlashan, D. Histamine: A mediator of inflammation. J. Allergy Clin. Immunol. 2003, 112 (Suppl. 4), S53–S59. [Google Scholar] [CrossRef]
- Atkinson, D.E. Citrate and the citrate cycle in the regulation of energy metabolism. Biochem. Soc. Symp. 1968, 27, 23–40. Available online: https://pubmed.ncbi.nlm.nih.gov/5759705/ (accessed on 25 November 2021). [PubMed]
- Akram, M. Citric Acid Cycle and Role of its Intermediates in Metabolism. Cell Biochem. Biophys. 2013, 68, 475–478. [Google Scholar] [CrossRef] [PubMed]
- Williams, N.C.; O’Neill, L.A.J. A Role for the Krebs Cycle Intermediate Citrate in Metabolic Reprogramming in Innate Immunity and Inflammation. Front. Immunol. 2018, 9, 141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Infantino, V.; Iacobazzi, V.; Palmieri, F.; Menga, A. ATP-citrate lyase is essential for macrophage inflammatory response. Biochem. Biophys. Res. Commun. 2013, A440, 105–111. [Google Scholar] [CrossRef]
- Ashbrook, M.J.; McDonough, K.L.; Pituch, J.J.; Christopherson, P.L.; Cornell, T.T.; Selewski, D.T.; Shanley, T.P.; Blatt, N.B. Citrate modulates lipopolysaccharide-induced monocyte inflammatory responses. Clin. Exp. Immunol. 2015, 180, 520–530. [Google Scholar] [CrossRef] [Green Version]
- Zotta, A.; Zaslona, Z.; O’Neill, L.A. Is Citrate A Critical Signal in Immunity and Inflammation? J. Cell. Signal. 2020, 1, 87–96. [Google Scholar] [CrossRef]
- Zhong, P.; Zhang, J.; Cui, X. Abnormal metabolites related to bone marrow failure in aplastic anemia patients. Genet. Mol. Res. 2015, 14, 13709–13718. [Google Scholar] [CrossRef]
- Lent-Schochet, D.; McLaughlin, M.; Ramakrishnan, N.; Jialal, I. Exploratory metabolomics of metabolic syndrome: A status report. World J. Diabetes 2019, 10, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Tavares, G.; Venturini, G.; Padilha, K.; Zatz, R.; Pereira, A.C.; Thadhani, R.I.; Rhee, E.P.; Titan, S.M.O. 1,5-Anhydroglucitol predicts CKD progression in macroalbuminuric diabetic kidney disease: Results from non-targeted metabolomics. Metabolomics 2018, 14, 39. [Google Scholar] [CrossRef] [PubMed]
- Newgard, C.B.; An, J.; Bain, J.R.; Muehlbauer, M.J.; Stevens, R.D.; Lien, L.F. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009, 9, 311–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Y.; Yang, X.; Deng, X.; Zhang, X.; Li, P.; Tao, J.; Qin, C.; Wei, J.; Lu, Q. Metabolomics in bladder cancer: A systematic review. Int. J. Clin. Exp. Med. 2015, 8, 11052–11063. Available online: https://pubmed.ncbi.nlm.nih.gov/26379905/ (accessed on 25 November 2021). [PubMed]
- Akimoto, H.; Oshima, S.; Sugiyama, T.; Negishi, A.; Nemoto, T.; Kobayashi, D. Changes in brain metabolites related to stress resilience: Metabolomic analysis of the hippocampus in a rat model of depression. Behav. Brain Res. 2019, 359, 342–352. [Google Scholar] [CrossRef]
- Lee, Y.J.; Kang, I.J.; Bünger, R.; Kang, Y.H. Mechanisms of pyruvate inhibition of oxidant-induced apoptosis in human endothelial cells. Microvasc. Res. 2003, 66, 91–101. [Google Scholar] [CrossRef]
- Knott, E.M.; Sun, J.; Lei, Y.; Ryou, M.G.; Olivencia-Yurvati, A.H.; Mallet, R.T. Pyruvate Mitigates Oxidative Stress During Reperfusion of Cardioplegia-Arrested Myocardium. Ann. Thorac. Surg. 2006, 81, 928–934. [Google Scholar] [CrossRef]
- Jacobson, L.S.; Lobetti, R.G. Glucose, lactate, and pyruvate concentrations in dogs with babesiosis. AJVR 2005, 66, 244–250. [Google Scholar] [CrossRef]
- Wright, I.G.; Waltisbuhl, D.J.; Mahoney, D.F.; Goodger, B.V. Acute Babesia bovis infections: Metabolic and blood gas changes during infection. Br. Vet. J. 1982, 138, 61–69. [Google Scholar] [CrossRef]
- Krishnan, N.; Dickman, M.B.; Becker, D.F. Proline modulates the intracellular redox environment and protects mammalian cells against oxidative stress. Free Radic. Biol. Med. 2008, 44, 671–681. [Google Scholar] [CrossRef] [Green Version]
- Abuduli, M.; Ohminami, H.; Otani, T.; Kubo, H.; Ueda, H.; Kawai, Y.; Masuda, M.; Okumura, H.Y.; Sakaue, H.; Yamamoto, H.; et al. Effects of dietary phosphate on glucose and lipid metabolism. Am. J. Physiol. Endocrinol. Metab. 2016, 310, E526–E538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serna, J.; Bergwitz, C. Importance of Dietary Phosphorus for Bone Metabolism and Healthy Aging. Nutrients 2020, 12, 3001. [Google Scholar] [CrossRef] [PubMed]
- Hoak, J.C. Stearic acid, clotting, and thrombosis. Am. J. Clin. Nutr. 1994, 60, 1050S–1053S. [Google Scholar] [CrossRef] [PubMed]
- Barić Rafaj, R.; Kuleš, J.; Selanec, J.; Vrkić, N.; Zovko, V.; Zupančič, M.; Trampuš Bakija, A.; Matijatko, V.; Crnogaj, M.; Mrljak, V. Markers of coagulation activation, endothelial stimulation, and inflammation in dogs with babesiosis. J. Vet. Intern. Med. 2013, 27, 1172–1178. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Huang, C.; Gao, L.; Xu, Y.J.; Chia, S.E.; Chen, S.; Li, N.; Yu, K.; Ling, Q.; Cheng, Q.; et al. Identification of serum biomarkers associated with hepatitis B virus-related hepatocellular carcinoma and liver cirrhosis using mass-spectrometry-based metabolomics. Metabolomics 2015, 11, 1526–1538. [Google Scholar] [CrossRef]
- Lawrence, Y.A.; Guard, B.C.; Steiner, J.M.; Suchodolski, J.S.; Lidbury, J.A. Untargeted metabolomic profiling of urine from healthy dogs and dogs with chronic hepatic disease. PLoS ONE 2019, 14, e0217797. [Google Scholar] [CrossRef]
- Page, I.H.; McCubbin, J.W. The variable arterial pressure response to serotonin in laboratory animals and man. Circ. Res. 1953, 1, 354–362. [Google Scholar] [CrossRef] [Green Version]
- Deza-Araujo, Y.I.; Neukam, P.T.; Marxen, M.; Muller, D.K.; Henle, T.; Smolka, M.N. Acute tryptophan loading decreases functional connectivity between the default mode network and emotion-related brain regions. Hum. Brain Mapp. 2019, 40, 1844–1855. [Google Scholar] [CrossRef]
- Ryding, E.; Lindstrom, M.; Traskman-Bendz, L. The role of dopamine and serotonin in suicidal behaviour and aggression. Prog. Brain Res. 2008, 172, 307–315. [Google Scholar] [CrossRef]
- Gonzalez-Burgos, I.; Feria-Velasco, A. Serotonin/dopamine interaction in memory formation. Prog. Brain Res. 2008, 172, 603–623. [Google Scholar] [CrossRef]
- Monti, J.M.; Jantos, H. The roles of dopamine and serotonin, and of their receptors, in regulating sleep and waking. Prog. Brain Res. 2008, 172, 625–646. [Google Scholar] [CrossRef] [PubMed]
- Kirtz, G.; Leschnik, M.; Hooijberg, E.; Tichy, A.; Leidinger, E. In-clinic laboratory diagnosis of canine babesiosis (Babesia canis canis) for veterinary practitioners in Central Europe. Tierärztliche Prax. Ausg. K Kleintiere Heimtiere 2012, 40, 87–94. [Google Scholar] [CrossRef] [Green Version]
- Cleare, A.J. Reduced whole blood serotonin in major depression. Depress. Anxiety 1997, 5, 108–111. [Google Scholar] [CrossRef]
- Pawlak, K.; Mysliwiec, M.; Pawlak, D. Haemostatic system, biochemical profiles, kynurenines and the prevalence of cardiovascular disease in peritoneally dialyzed patients. Thromb. Res. 2010, 125, e40–e45. [Google Scholar] [CrossRef] [PubMed]
- Pawlak, K.; Domaniewski, T.; Myśliwiec, M.; Pawlak, D. Kynurenines and oxidative status are independently associated with thrombomodulin and von Willebrand factor levels in patient with endstage renal disease. Thromb. Res. 2009, 124, 452–457. [Google Scholar] [CrossRef] [PubMed]
- Kuleš, J.; Gotić, J.; Mrljak, V.; Rafaj, R.B. Blood markers of fibrinolysis and endothelial activation in canine babesiosis. BMC Vet. Res. 2017, 13, 82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michiels, C. Physiological and pathological responses to hypoxia. Am. J. Pathol 2004, 164, 1875–1882. [Google Scholar] [CrossRef] [Green Version]
- Furuhashi, M.; Koyama, M.; Higashiura, Y.; Murase, T.; Nakamura, T.; Matsumoto, M.; Sakai, A.; Ohnishi, H.; Tanaka, M.; Saitoh, S.; et al. Differential regulation of hypoxanthine and xanthine by obesity in a general population. J. Diabetes Investig. 2020, 11, 878–887. [Google Scholar] [CrossRef] [Green Version]
- Wung, W.E.; Howell, S.B. Simultaneous liquid chromatography of 5-flurouracil, uridine, hypoxanthine, xanthine, allopurinol, and oxypurinol in plasma. Clin. Chem. 1980, 26, 1704–1708. Available online: https://pubmed.ncbi.nlm.nih.gov/7428153/ (accessed on 25 November 2021). [CrossRef]
- Neumman, S.; Welling, H.; Thuere, S. Evaluation of Serum L-phenylalanine Concentration as Indicator of Liver Disease in Dogs: A Pilot Study. J. Am. Anim. Hosp. Assoc. 2007, 43, 193–200. [Google Scholar] [CrossRef]
- Bürgi, S.S.; Schroecksnadel, S.; Jenny, M.; Karall, D.; Fuchs, D. Chronic Immune Stimulation May Cause Moderate Impairment of Phenylalanine 4-hydroxylase. Pteridines 2011, 22, 120–125. [Google Scholar] [CrossRef] [Green Version]
- Dawiskiba, T. Serum and urine metabolomic fingerprinting in diagnostics of inflammatory bowel diseases. World J. Gastroenterol. 2014, 20, 163. [Google Scholar] [CrossRef] [PubMed]
- Florin-Christensen, J.; Suarez, C.E.; Florin-Christensen, M.; Hines, S.A.; McElwain, T.F.; Palmer, G.H. Phosphatidylcholine formation is the predominant lipid biosynthetic event in the hemoparasite Babesia Bovis. Mol. Biochem. Parasitol. 2000, 106, 147–156. [Google Scholar] [CrossRef]
- Khovidhunkit, W.; Kim, M.S.; Memon, R.A.; Shigenaga, J.K.; Moser, A.H.; Feingold, K.R.; Grunfeld, C. Thematic review series: The Pathogenesis of Atherosclerosis.Effects of infection and inflammation on lipid and lipoprotein metabolism mechanisms and consequences to the host. J. Lipid Res. 2004, 45, 1169–1196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sargis, R.M.; Subbaiah, P.V. Protection of membrane cholesterol by sphingomyelin against free radical-mediated oxidation. Free Radic. Biol. Med. 2006, 40, 2092–2102. [Google Scholar] [CrossRef] [Green Version]
- Oborina, E.M.; Yappert, M.C. E_ect of sphingomyelin versus dipalmitoylphosphatidylcholine on the extent of lipid oxidation. Chem. Phys. Lipids 2003, 123, 223–232. [Google Scholar] [CrossRef]
- Lupanova, T.; Stefanova, N.; Petkova, D.; Staneva, G.; Jordanova, A.; Koumanov, K.; Pankov, R.; Momchilova, A. Alterations in the content and physiological role of sphingomyelin in plasma membranes of cells cultured in three-dimensional matrix. Mol. Cell Biochem. 2010, 340, 215–222. [Google Scholar] [CrossRef]
- Coliva, G.; Lange, M.; Colombo, S.; Chervet, J.P.; Domingues, M.R.; Fedorova, M. Sphingomyelins Prevent Propagation of Lipid Peroxidation—LC-MS/MS Evaluation of Inhibition Mechanisms. Molecules 2020, 25, 1925. [Google Scholar] [CrossRef] [Green Version]
- Crnogaj, M.; Kiš, I.; Kučer, N.; Šmit, I.; Mayer, I.; Brkljačić, M.; Selanec, J.; Mrljak, V. Lipid peroxidation in dogs naturally infected with Babesia canis canis. Vet. Arhiv 2015, 85, 37–48. Available online: https://hrcak.srce.hr/135265 (accessed on 25 November 2021).
- Gloaguen, Y.; Morton, F.; Daly, R.; Gurden, R.; Rogers, S.; Wandy, J.; Wilson, D.; Barrett, M.; Burgess, K. PiMP my metabolome: An integrated, web-based tool for LC-MS metabolomics data. Bioinformatics 2017, 33, 4007–4009. [Google Scholar] [CrossRef] [Green Version]
- Holman, J.D.; Tabb, D.L.; Mallick, P. Employing ProteoWizard to Convert Raw Mass Spectrometry Data. Curr. Protoc. Bioinform. 2014, 46, 13–24. [Google Scholar] [CrossRef] [PubMed]

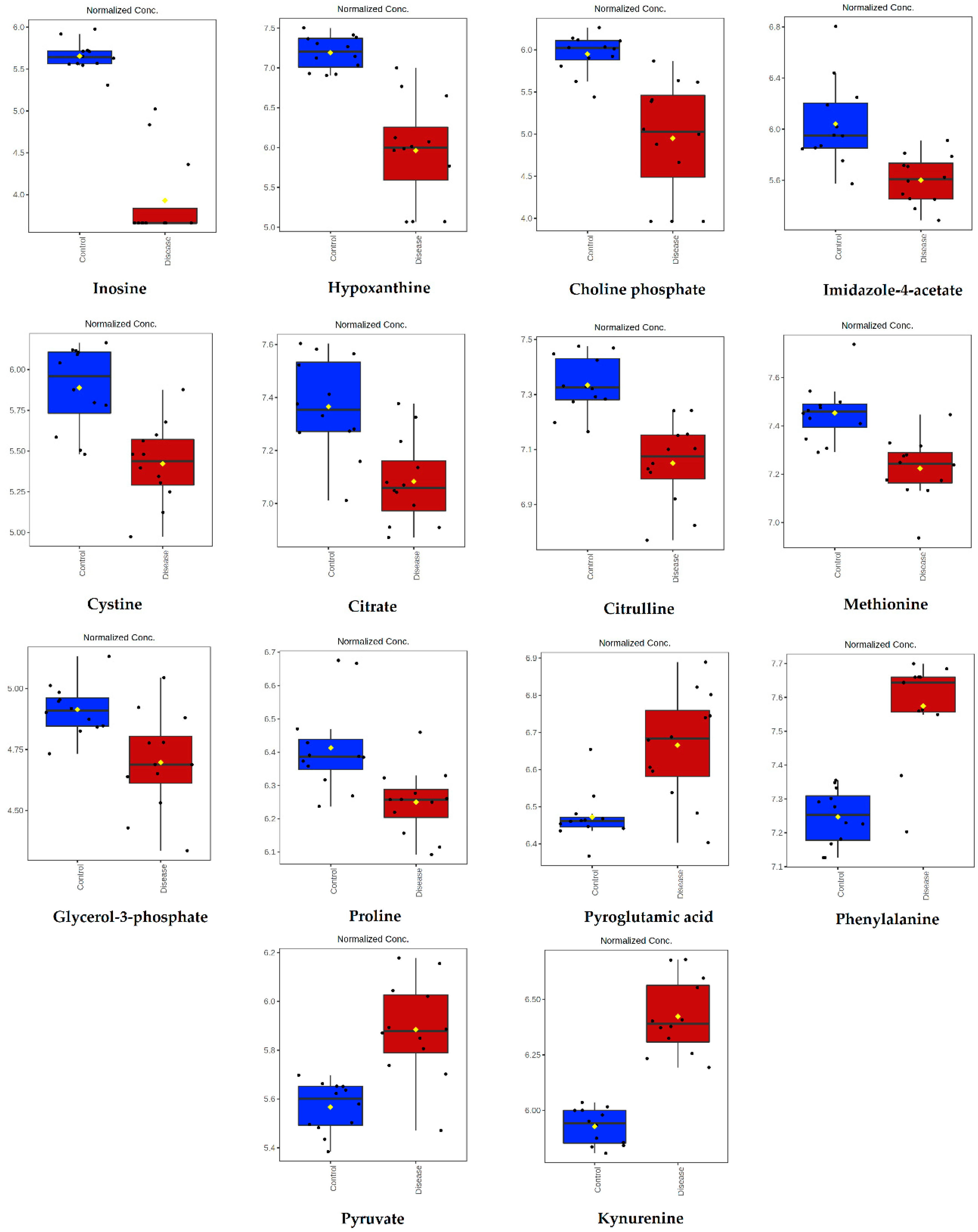
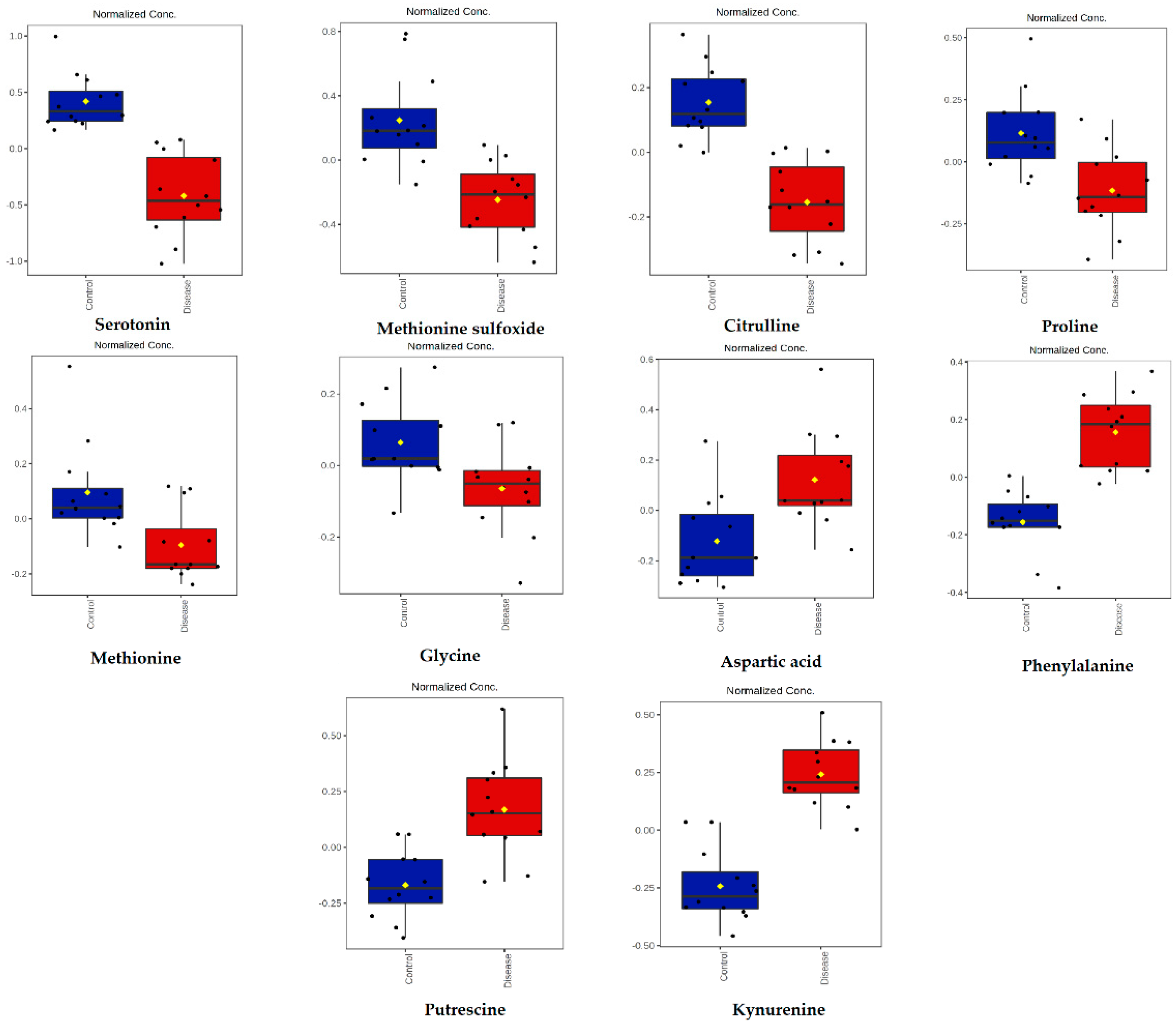
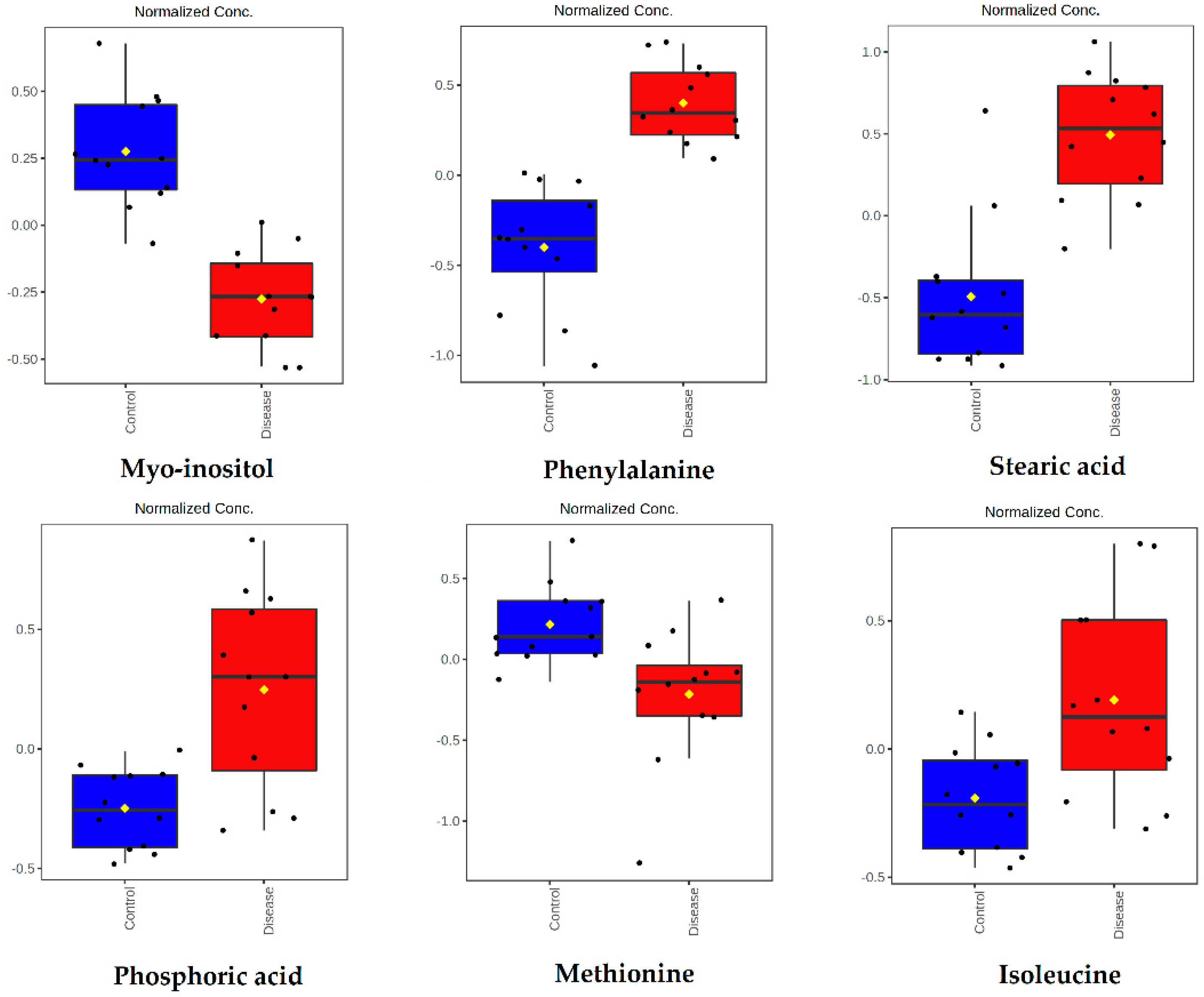
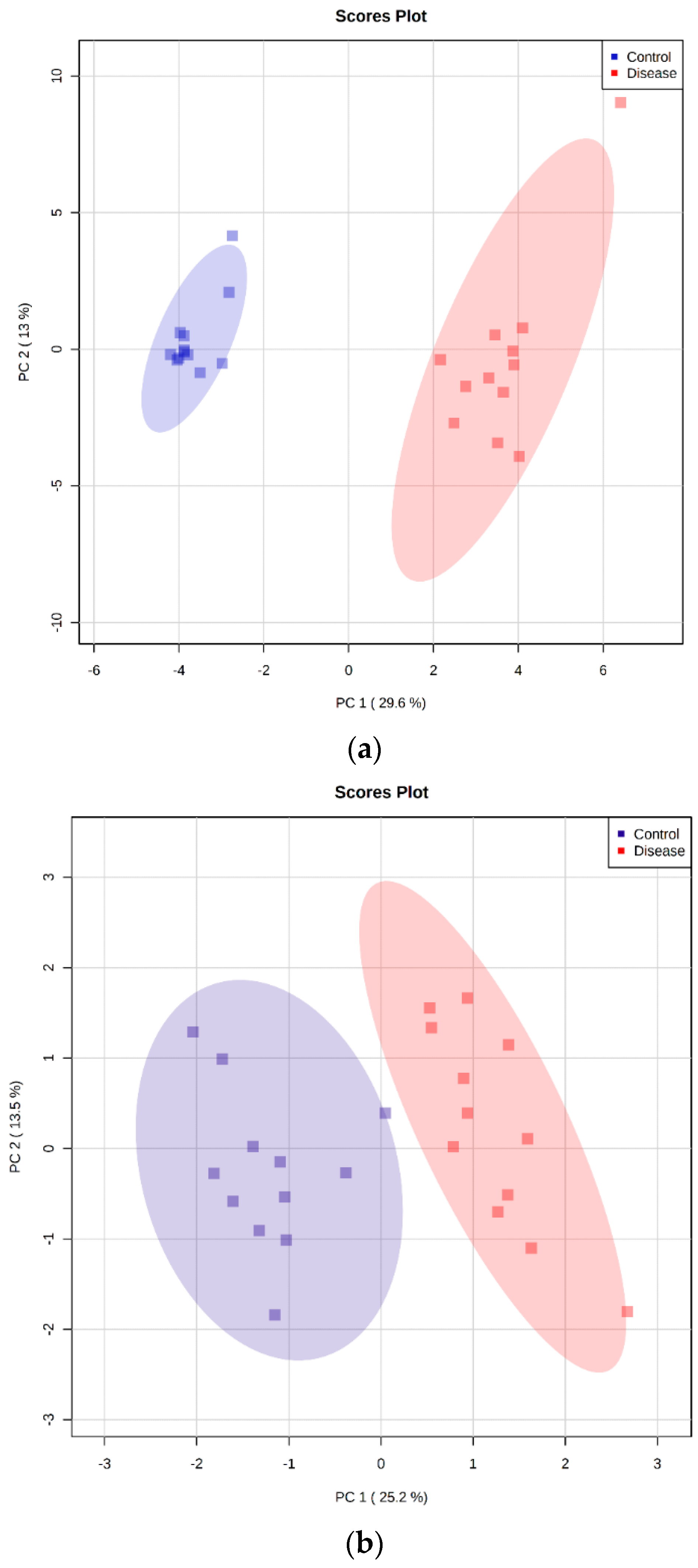

| Metabolite | Peak ID | Mass | RT(s) | p-Value | Log2 (FC) |
|---|---|---|---|---|---|
| Inosine | 1280 | 267.0736 | 591.68 | 1.74 × 10−10 | 4.62 |
| Hypoxanthine | 70 | 137.0458 | 567.72 | 3.00 × 10−6 | 2.95 |
| Choline phosphate | 352 | 184.0734 | 714.56 | 8.90 × 10−5 | 2.29 |
| Imidazole-4-acetate | 169 | 127.0502 | 624.14 | 6.69 × 10−4 | 1.81 |
| Cystine | 407 | 241.031 | 771.48 | 1.59 × 10−4 | 1.54 |
| Citrate | 848 | 191.02 | 828.92 | 5.74 × 10−4 | 0.95 |
| Citrulline | 73 | 176.103 | 764.51 | 1.94 × 10−5 | 0.91 |
| Methionine | 51 | 150.0584 | 610.06 | 1.50 × 10-4 | 0.76 |
| Glycerol-3-phosphate | 1657 | 171.0066 | 713.52 | 3.26 × 10−3 | 0.62 |
| Proline | 1004 | 114.0561 | 659.98 | 2.94 × 10−3 | 0.58 |
| Pyroglutamic acid | 951 | 128.0354 | 575.74 | 3.86 × 10−4 | −0.70 |
| Phenylalanine | 54 | 166.0863 | 551.14 | 9.88 × 10−7 | −1.13 |
| Pyruvate | 1209 | 87.0088 | 501.48 | 3.05 × 10−5 | −1.15 |
| Kynurenine | 199 | 209.0921 | 580.41 | 5.04 × 10−9 | −1.72 |
| Metabolites | Short Name | p-Value | Log2 (FC) | Classification |
|---|---|---|---|---|
| Serotonin | Serotonin | 1.14 × 10−6 | 2.59 | Biogenic amines |
| Methionine sulfoxide | Met-SO | 1.39 × 10−4 | 1.79 | Biogenic amines |
| Citrulline | Cit | 2.50 × 10−6 | 1.01 | Amino acids |
| Proline | Pro | 2.34 × 10−3 | 0.78 | Amino acids |
| Methionine | Met | 5.87 × 10−3 | 0.70 | Amino acids |
| Glycine | Gly | 1.45 × 10−2 | 0.42 | Amino acids |
| Aspartic acid | Asp | 4.02 × 10−3 | −0.83 | Amino acids |
| Phenylalanine | Phe | 2.32 × 10−6 | −1.06 | Amino acids |
| Putrescine | Putrescine | 2.09 × 10−4 | −1.22 | Biogenic amines |
| Kynurenine | Kynurenine | 7.76 × 10−8 | −1.59 | Biogenic amines |
| Metabolite | Short Name | HMDB ID | Chemical Class |
|---|---|---|---|
| Lactic acid | Lac | 00190 | Organic acid |
| Alanine | Ala | 00161 | Amino acid |
| Glycine | Gly | 00123 | Amino acid |
| Pyruvic acid | Pyruvic acid | 00243 | Organic acid |
| 3-Hydroxybutyric acid | 3-Hydroxybutyric acid | 00011 | Organic acid |
| Valine | Val | 00883 | Amino acid |
| Urea | Ur | 00294 | Organic acid |
| Glycerol | Glycerol | 00131 | Carbohydrate |
| Phosphoric acid | Phosphoric acid | NA | Organic acid |
| Isoleucine | Ile | 00172 | Amino acid |
| Proline | Pro | 00162 | Amino acid |
| Serine | Ser | 00187 | Amino acid |
| Threonine | Thr | 00167 | Amino acid |
| Methionine | Met | 00696 | Amino acid |
| L-Proline | L-Pro | 00162 | Amino acid |
| Glutamic acid | Glu | 00148 | Organic acid |
| Phenylalanine | Phe | 00159 | Amino acid |
| Isoleucyl-Glutamine | Isoleucyl-Glutamine | 28905 | Amino acid |
| Citric acid | Citric acid | 00094 | Organic acid |
| Glucose | Glc | 00122 | Carbohydrate |
| Lysine | Lys | 00182 | Amino acid |
| Tyrosine | Tyr | 00158 | Amino acid |
| Galactose | Gal | 00143 | Carbohydrate |
| Palmitic acid | Palmitic acid | 00220 | Fatty acid |
| Myo-inositol | Myo-inositol | 00211 | Sugar alcohol |
| Stearic acid | Stearic acid | 00827 | Fatty acid |
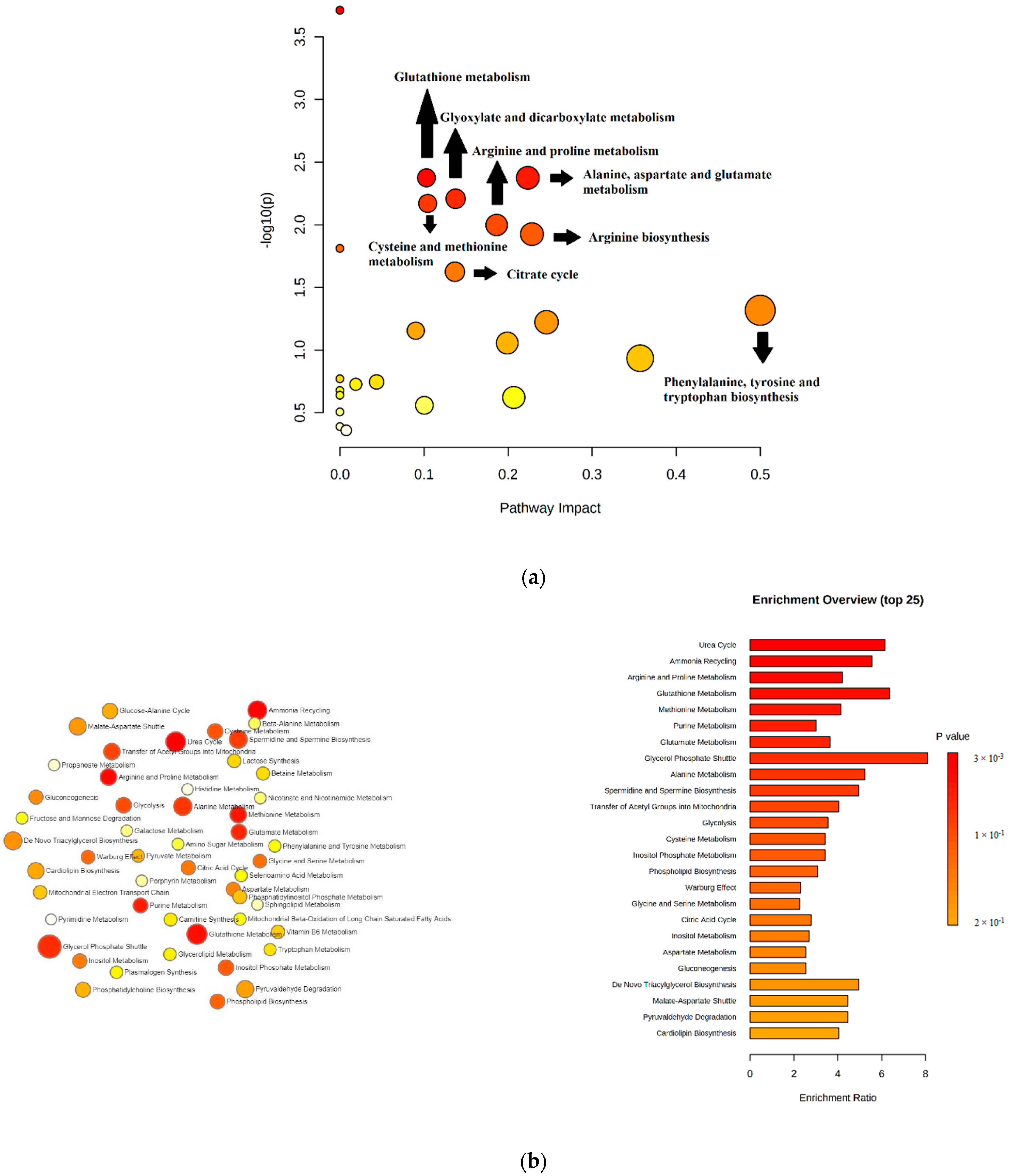
| Metabolic Pathways | Significant Metabolites (Fold Change) | p-Value | Pathway Impact |
|---|---|---|---|
| Glutathione metabolism | 5-oxoproline (up) Glycine (down) Putrescine (up) | 0.0073447 | 0.10 |
| Alanine, aspartate, and glutamate metabolism | Citrate (down) Aspartate (up) Pyruvate (up) | 0.0073447 | 0.22 |
| Glyoxylate and dicarboxylate metabolism | Pyruvate (up) Glycine (down) Citrate (down) | 0.010697 | 0.14 |
| Cysteine and methionine metabolism | Cystine (down) Methionine (down) pyruvate (up) | 0.011653 | 0.10 |
| Arginine and proline metabolism | Proline (down) Putrescine (up) Pyruvate (up) | 0.017166 | 0.19 |
| Arginine biosynthesis | Citrulline (down) Aspartate (up) | 0.017204 | 0.23 |
| Citrate cycle | Pyruvate (up) Citrate (down) | 0.034034 | 0.14 |
| Phenylalanine, tyrosine, and tryptophan biosynthesis | Phenylalanine (up) | 0.048184 | 0.50 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rubić, I.; Burchmore, R.; Weidt, S.; Regnault, C.; Kuleš, J.; Barić Rafaj, R.; Mašek, T.; Horvatić, A.; Crnogaj, M.; Eckersall, P.D.; et al. Multi Platforms Strategies and Metabolomics Approaches for the Investigation of Comprehensive Metabolite Profile in Dogs with Babesia canis Infection. Int. J. Mol. Sci. 2022, 23, 1575. https://doi.org/10.3390/ijms23031575
Rubić I, Burchmore R, Weidt S, Regnault C, Kuleš J, Barić Rafaj R, Mašek T, Horvatić A, Crnogaj M, Eckersall PD, et al. Multi Platforms Strategies and Metabolomics Approaches for the Investigation of Comprehensive Metabolite Profile in Dogs with Babesia canis Infection. International Journal of Molecular Sciences. 2022; 23(3):1575. https://doi.org/10.3390/ijms23031575
Chicago/Turabian StyleRubić, Ivana, Richard Burchmore, Stefan Weidt, Clement Regnault, Josipa Kuleš, Renata Barić Rafaj, Tomislav Mašek, Anita Horvatić, Martina Crnogaj, Peter David Eckersall, and et al. 2022. "Multi Platforms Strategies and Metabolomics Approaches for the Investigation of Comprehensive Metabolite Profile in Dogs with Babesia canis Infection" International Journal of Molecular Sciences 23, no. 3: 1575. https://doi.org/10.3390/ijms23031575
APA StyleRubić, I., Burchmore, R., Weidt, S., Regnault, C., Kuleš, J., Barić Rafaj, R., Mašek, T., Horvatić, A., Crnogaj, M., Eckersall, P. D., Novak, P., & Mrljak, V. (2022). Multi Platforms Strategies and Metabolomics Approaches for the Investigation of Comprehensive Metabolite Profile in Dogs with Babesia canis Infection. International Journal of Molecular Sciences, 23(3), 1575. https://doi.org/10.3390/ijms23031575










