Abstract
Aging is a complex process indicated by low energy levels, declined physiological activity, stress induced loss of homeostasis leading to the risk of diseases and mortality. Recent developments in medical sciences and an increased availability of nutritional requirements has significantly increased the average human lifespan worldwide. Several environmental and physiological factors contribute to the aging process. However, about 40% human life expectancy is inherited among generations, many lifespan associated genes, genetic mechanisms and pathways have been demonstrated during last decades. In the present review, we have evaluated many human genes and their non-human orthologs established for their role in the regulation of lifespan. The study has included more than fifty genes reported in the literature for their contributions to the longevity of life. Intact genomic DNA is essential for the life activities at the level of cell, tissue, and organ. Nucleic acids are vulnerable to oxidative stress, chemotherapies, and exposure to radiations. Efficient DNA repair mechanisms are essential for the maintenance of genomic integrity, damaged DNA is not replicated and transferred to next generations rather the presence of deleterious DNA initiates signaling cascades leading to the cell cycle arrest or apoptosis. DNA modifications, DNA methylation, histone methylation, histone acetylation and DNA damage can eventually lead towards apoptosis. The importance of calorie restriction therapy in the extension of lifespan has also been discussed. The role of pathways involved in the regulation of lifespan such as DAF-16/FOXO (forkhead box protein O1), TOR and JNK pathways has also been particularized. The study provides an updated account of genetic factors associated with the extended lifespan and their interactive contributory role with cellular pathways.
1. Introduction
Aging is a multifaceted, complex process represented by the functional decline of tissues and organs due to changes in molecular composition and physiology of cells. An increase in the human lifespan has been reported for last many decades [1]. According to the estimates of United Nations, there were more than 63 million people above 90 years of age in 2020 [2]. Longevity of human life is heritable from 20% to 40% in the modern human populations [3]. Survival into extremely long ages is a characteristic clustered in families [4]. Better immune systems, metabolic health such as enhanced insulin sensitivity [5], lipid metabolism leading to healthy levels of plasma lipids [6], and delay or ability to avoid age related diseases has also been observed in the longevous families [7,8]. As longevity of life exhibits high heritability, insights into the genetic factors may improve our present understanding of mechanisms responsible to promote health and reduce the risk of diseases [3,9]. However, a few genes and genetic loci have been identified for this trait in the recent studies [10,11,12]. Gene coding for apolipoprotein (APOE) has been reported by multiple genome-wide association studies (GWAS) [10,11,12]. Telomeres are the protective caps at the termini of human and other eukaryotic chromosomes. Their length decreases with age, though it is dependent on many internal and environmental factors. According to reports based on experimental models, the overall length or rate of telomere shortening has been reported to have a link with age expectancy [13]. The shortening of telomere length is rapid in men as compared to women, leading to a comparatively high longevity in women [14]. The application of the well-known antioxidant red wine has been reported to promote the age longevity related genes. Overall, 115 aging associated genes have been studied among 25 mammalian species [15]. Studies on the controlled human populations have shown an increase in the expression levels of Sirt1, catalase, p53 and manganese-superoxide dismutase, after 14 days of red wine use, with no serious side effects. Further studies on Drosophila melanogaster have shown up to 7% increase in lifespan after moderate red wine applications [16]. There are several other genes, such as the kinase signaling gene MAP3K5 [17], and variants of genes in the insulin/IGF1 pathway [18]. Studies on bats have shown that DNA methylation is negatively linked with longevity of lifespan, and that DNA damage is positively linked with it [19,20]. Studies on monkeys have shown that the genome flexibility and environmental adaptations also contribute to the lifespan [21]. In context with the above information, we aimed to conduct a systematic review of human genes and their non-human orthologs associated with the life expectancy.
2. Genes and Longevity of Lifespan
During past three- or four-decades, genetic researchers have identified many genes that promote lifespan in different species [22,23,24]. Some examples of candidate genes and their orthologs reported for their association with longevity of lifespan have been tabulated in Table 1.

Table 1.
Human genes and their orthologues associated with the longevity of lifespan. The main physiological roles of and reported association with life threatening diseases have been discussed for each gene.
2.1. Aging and Genomic Modulation
2.1.1. Apolipoprotein E (APOE)
APOE gene codes for a protein known as apolipoprotein E that binds with the lipid molecules to make lipoproteins. APOE is a pleotropic gene contributing to packing and transport of fats and cholesterol along with human blood, maintenance of normal blood glucose levels in the human body, neuronal cell homeostasis, adrenal and brain physiology and cardiovascular health [229]. There are three extensively studied alleles of the APOE gene, i.e., e2, e3 and e4 coding for Apo E2, E3 and E4 proteins [230,231]. Minor variations of specific amino acids give rise to binding variability in ApoE proteins for multiple corresponding molecules including LDLRs (LDL receptors), HSPGs (cell-surface heparin sulfate proteoglycans), and ABCA1 (ATP-binding cassette protein 1). The amino acid variations also affect protein stability and protein folding [232,233]. About 50% of the world’s population has e3 genotype [234]. APOE is one of the well demonstrated genes that have been clearly linked with human mortality. Studies on European populations have shown up to 3.5% contribution of APOE gene in human lifespan [235]. There are consistent studies demonstrating the role of allele e4 in the onset of Alzheimer disease and cardiovascular health [236,237]. Approximately a 4.2 year shorter lifespan has been found among the populations having high frequency of e4 allele [238]. In a series of experiments Finch, Sapolsky, and Stanford have shown that an interaction between diet and genotype play an important role in the longevity of human lifespan [239,240,241]. They also emphasized that the carriers of e4 alleles have higher cholesterol levels with greater chances of plaque formation in the arteries and enhanced risk of cardiovascular diseases, dementia, stroke and Alzheimer’s disease [230]. There are many factors that contribute to the longevity of lifespan in association with genotypes. Aerobic exercise improves the longevity of human lifespan among e4 allele carriers [242]. The impact of diabetes on the human lifespan is also genotype dependent [243]. There is evidence that the increase in human lifespan started about 1.8 million years ago, at those times the human populations were likely to be homozygous for the e4 allele [244]. Based on several studies, a relationship between cognitive decline and e4 genotype [245], high mortality rate due to pneumonia and severe dementia have been reported in old men with e4 genotype [246]. The e4 type have been reported to have short telomeres that have a role in life expectancy [247]. In Japanese and Italian populations, e2 allele is found to favor and e4 allele decrease the chances of exceptional longevity of lifespan [248]. The main hurdle in the development of ApoE based therapeutics is due to limited understanding of each ApoE (subtype) mediated pathway and its impacts on overall human physiology [25].
2.1.2. p53
p53 is a gene that codes for cell cycle regulatory protein responsible to maintain the genome stability, reduce the mutation rate and suppress cancer. Human tumors have been frequently linked with p53 mutations; about 50% of all tumors involve disruption of this gene [249,250]. The presence of the p53 gene in short lived organisms such as worms and flies, who do not develop cancers, indicate that tumor suppression is not the only function of p53 gene. According to recent studies, p53 and p73 play crucial roles in reproduction [251,252]. There is strong evidence that p53 has the complex role in the regulation of longevity of life in mice, flies, Caenorhabditis elegans, and humans [253,254,255], both longevity of lifespan and maturity age for reproduction are always coupled. A short form of p53 also known as p44 has been found associated with the body size and lifespan in mice. Regulated expression of p53 isoforms is responsible to maintain a balance between tissue regeneration and tumor suppression in mammals [256]. DNA damage is considered one of the main reasons behind short lifespan, p53 gene is responsible to maintain the integrity of DNA in species. In the genome of elephants 12 to 20 copies of p53 gene have been reported that are meant to reduce the chances of cancer and DNA damage [257]. (Role of p53 in the regulation of cell division or apoptosis under damaged or mutated conditions is described in the DNA repair section.
2.1.3. Sirtuin (SIRT1)
Sirtuin (SIRT) is considered an important factor to extend the longevity of lifespan by delaying cellular senescence [258]. SIRT1 is an important member of an NAD+ dependent family of enzymes that have multiple roles in the cells including cell cycle regulation, regulation of energy metabolism, gene silencing and gene regulation [259]. SIRT1 responsible for the deacetylation of tumor suppressor p53 protein, NF-κB and Ku70 (DNA repair factor), is the most well studied isozyme SIRT [260,261,262]. It also regulates FOXO family of transcription factors and STAT3 (signal transducer and activator of transcription 3) [263]. Sirtuin improves the cellular ability to sustain genome integrity by promoting the DNA repair process. It can enhance the ability of a cellular system and makes it resistant the oxidative stress [264]. Increased levels of sirtuin (SIRT2 and SIRT6) have shown prolongevity effect in Drosophila melanogaster, Saccharomyces cerevisiae, Caenorhabditis elegans, and mice [225,265,266]. Overexpression of brain-specific Sirt1 in transgenic mice prolonged their median lifespan by 9% in males and 16% in females [267]. The cellular levels of nicotinamide adenine dinucleotide (NAD+), a “cozymase” can promote DNA repair by reducing metabolic stress and improving mitochondrial function. An increased level of NAD+ in the mammalian and mice cells have shown an activation of SIRT1 and SIRT3 resulting in an increased oxidative metabolism eventually protecting against high fat diet induced problems [268]. The supplementation of precursor molecule of NAD+, NMN (nicotinamide mononucleotide) can reduce the DNA damage, heart failure, and effects of acute renal injury in a SIRT1-dependent manner [269,270]. NMN can help to maintain telomere length, decreases adipogenesis, and improves the osteogenesis in aged mice via SIRT1 activation [271,272]. Administration of NMN has reportedly increased the lifespan in aged mice via (PARP1)/SIRT1 axis [273]. SIRT1 activation reduce insulin resistance, enhances insulin sensitivity via PGC1-α (PPARγ co-activator 1α), implementing beneficial effects in obesity and diabetes type 2 [274,275]. Insulin sensitivity is enhanced by SIRT1 activation via reduction in the expression of proinflammatory genes and it attenuates the insulin resistance induced by tumor necrosis factor alpha (TNF-α) [259,276]. SIRT1 inhibits the production of proinflammatory cytokines via NF-kB [277], and STAT3 [278]. It also exhibits anti-apoptosis activity through p53 and FOXO regulation [279,280,281]. Activation of Sirt1 inhibits lipogenesis and promotes fatty acid β-oxidation [15,282,283] (Figure 1).
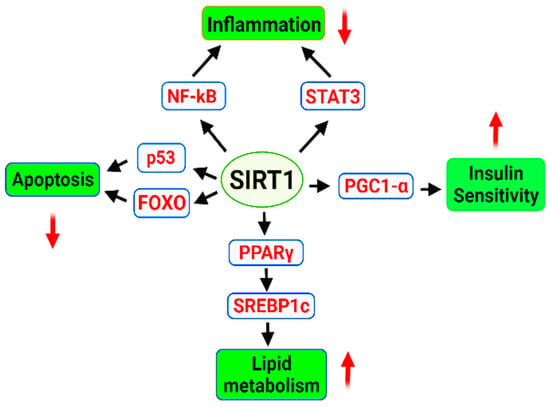
Figure 1.
Regulatory role of SRT1 signaling in the cell. SIRT1, an NAD-dependent deacetylase affects several downstream molecules and promotes life. Sirt1 based downregulation of apoptosis and inflammation leads to the extension of lifespan contrary to that enhanced lipid metabolism and insulin sensitivity are main factors that increase the lifespan. As the discussion included the sirt1 gene and we have a separate section for the associated pathways, Figure 1 was meant to indicate the central role of sirt1 gene in the cellular and metabolic pathways involved in the extended lifespan. Green colour indicates final process regulated, downward and upward arrows indicate downregulation and upregulation of process.
3. Genomic Instability and Oxidative Stress in Longevity of Lifespan
It has been a universal goal to understand the events and adopt the measures to slow down, stop or reverse the aging process [284]. The most important DNA modification associated with aging is DNA methylation. DNA methylation based biomarkers have answered many of the questions about the aging process and have described the role of epigenetics. As epigenetic changes in the human body are reversible, these biomarkers are therefore useful for the identification and validation of anti-aging interventions [285]. DNA methylation has been linked with human age and the aging process in man and other species. Measurement of changes in methylation (decrease/increase) at a few hundred specific CpG sites can estimate chronological age and mortality [286,287]. Studies have shown that an increased difference between chronological age and DNA methylation age is positively associated with chances of mortality [288,289,290]. Studies by Hannum and Horvath have shown that a difference of 5 years between these two age types can increase the chances of mortality by 11% to 21% [286,287]. An accelerated DNA methylation age significantly increases the risk of can, stroke and cardiovascular diseases [291,292]. The second genetic factor associated with aging is histone modifications. Alterations (decrease/increase) in the methylation of histones have been reported to regulate aging process in C. elegans [293]. High levels of trimethylation of H3K4 reduces lifespan of C. elegans [294]. Histone methylations have been linked with cellular pathways regulating the age and lifespan. The process of autophagy slows down with age, due to decrease in the ratio of cellular functional mitochondria and subsequent increase in the ROS, aggregation of insoluble or exhausted proteins in the cells. In such cases, avoiding the decrease in autophagy results in the extension of lifespan [295]. Another consequence of the aging process is accumulation of damaged DNA (mutated, irreparable, double stranded breaks) due to decreased efficiency of damaged DNA repair system (DDR system) in the old cells. Histone methylation promotes the DDR of cells and improve the life activity [296]. Acetylation of histones catalyzed mostly by NAD-dependent acetyltransferases is the third important genetic factor associated with the extension of lifespan. Histones are well established role in DNA packaging and regulation of gene expression [297]. Histone modifications are not only associated with rate of transcription but also regulate the precision of transcription process [298]. Acetylated histones are readily available to interact with transcription factors resulting in high levels of transcription [299]. Age associated decrease in histone acetylation is basis of decline in rate of transcription and metabolism leading to shortening of lifespan [300,301].
At present, the calorie restriction (CR) and calorie restriction mimetics such as rapamycin have been used as interventions to reduce the difference of methylation based age and chronological age [302]. CR interferes the DNA methylation by activation of DNA methyltransferase, resulting in hypermethylation and silencing of Ras and p16INK4a genes, both of these genes are associated with senescence [303,304]. CR presents an important anti-aging therapy to increase the lifespan of humans, non-human primates and rodents [305,306] (Figure 2).
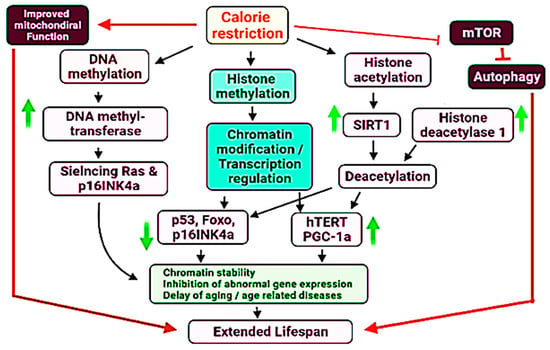
Figure 2.
Proposed mechanistic role of DNA methylation, histone methylation and histone acetylation in the regulation of lifespan in the human and other species. Upward and downward arrows indicate upregulation and downregulation of specific genes of processes. Some major aspects have been highlighted by specific colours.
Another critical factor associated with the survival of a cell is the maintenance of genomic DNA in its intact form. Damaged or mutated DNA accumulation in the multicellular organisms often leads to the onset of cancer or triggers the aging process [307,308,309]. DNA damage triggers aging process by blocking transcription, activating the signal transduction processes, regulating the DNA metabolism, altering the epigenome and finally by inducing apoptosis [310,311,312]. Normally, the DNA damage is recognized by genomic DNA followed by the induction of DNA repair mechanisms such as BER (base excision repair), and double standard breaks (DSBs). If the DNA damage signals continue, the cells select to avoid the replication of mutated or damaged genome rather they prefer to promote the processes leading cell cycle arrest of apoptosis [313,314]. Signaling cascades by the detection of damaged DNA are initiated by the activation of MRN (MRE11/RAD50/NBS1) complex subsequent activation of PIKKs (phosphatidylinositol 3-kinase-like kinases) ATM (ataxia-telangiectasia mutated), ATR (ATM-related kinase) [315,316,317]. ATM and ATR are activated by DNA DSBs and stalled replication forks respectively. Both of these activated factors operate in coordination with 53BP1 MDC1, TOPBP1, and BRCA1 that are sensor proteins responsible to bind the damaged DNA sites and for the recognition of DDR along [318]. TOPBP1 plays a critical role to preserve genome integrity during mitosis [319,320]. After the DNA damage recognition, the transducer kinase check points are phosphorylated and activated such as CHK2 (checkpoint kinase 1) and CHK1 (checkpoint kinase 1), followed by the activation of p53 [321,322] (major roles of p53 in the cellular physiology have already been described in the previous sections). Failure of proper DNA repair mechanism or in case of irreversible DNA damage, the cascades towards programmed cell death are initiated. Activated p53 is recognized as the master regulator of cell cycle, aging, apoptosis. P53 is required for cellular degeneration by genomic DNA damage not required for mitochondrial DNA damage response [323]. In fact a balance of antioxidant and prooxidant activities by p53 in response to oxidative stress play an important role in the longevity of lifespan [324] (Figure 3).
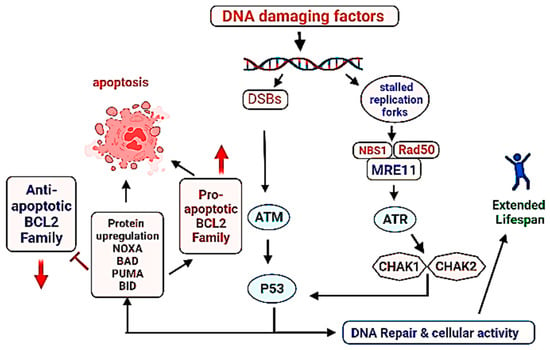
Figure 3.
Role of DNA damage repair system in the longevity of lifespan. Successful DNA repair leads to the continuation of life activity/cell division, contrary to that failure in the DNA repair promotes programmed cell death (apoptosis). Direction of arrows indicate upregulation or downregulation of a process.
Oxidative stress typically caused by reactive oxygen species (ROS), ultra-violet and ionizing radiations can damage cellular components, especially the lipids, proteins and DNA [325]. Genomic instability includes the events leading to permanent or temporary changes in the genome such as duplications, deletions, inversions and translocations in the chromosomes [326]. Genomic instability may lead to changes in the gene expression, enhance the changes of apoptosis, and it is known as a hall-mark of cancer initiation, age related neurodegenerative diseases [327,328,329]. In the human body, number and diversity of lymphocytes is regulated by the repeated hexanucleotides (TTAGGG) n of 10 kb to 15 kb length known as telomeres [330]. Shortening of telomeres stops the cell division and production of lymphocytes leading to subsequent loss of cellular ability in immune response [331].
Circadian rhythms also play important role in the mammalian cell biochemistry and physiology. Almost each cell has a mechanism to respond to the day night cycles which are mainly regulated by hypothalamus area of brain which is a master pacemaker to modulate cell signaling according to the photoperiods. The circadian desynchrony may lead to the neurodegenerative disorders and metabolic pathologies that ultimately impact the lifespan [332]. The aging process and longevity of lifespan has many well established risk factors including diabetes, obesity, problems of cardiovascular system due to the lack of physical activity. Several epigenetic factors also contribute to the longevity of lifespan [333].
4. DAF-16/FOXO (Forkhead Box Protein O1), TOR and JNK Pathways in Aging
DAF-16/FOXO (Forkhead Box Protein O1), TOR and JNK Pathways
DAF-16 is an orthologous gene coding for FoxO (fork head box transcription factors class O) that regulates the expression levels of genes associated with stress metabolism, aging development and immunity [334,335,336]. There are 19 subclasses of FOX transcription factor in mammals and 6 of them are found in humans [337]. DAF-16 activity is regulated by insulin/ IGF-1 (insulin like-growth factor 1) via IIS (insulin/IGF-1 signaling) pathway [338]. IIS cascade is triggered by the binding of insulin such as peptides (ILPs) to a tyrosine kinase receptor. There are 40 different types of ILPs in Caenorhabditis elegans and 10 members of ILPs are found in the human body. IGF-1 and IGF-2 behave similar to those found in C. elegans and interact with tyrosine kinase receptor [57,339,340]. These two transcription factors (DAF-16/FOXO and HLH-30) are co-expressed in most of the human tissues particularly in the neuron and intestinal cells [341,342]. For its activity, DAF-16/FOXO directly binds with HLH-30 (helix-loop-helix transcription factor HLH-30) to make a complex to two transcription factors, this binding is conserved in human [343,344]. Activation of DAF-16/FOXO by low IIS triggers the complex formation. DAF-16/FOXO is translocated to the nucleus, this action is mediated by HLH-30 under longevity promoting conditions. Both factors require each other for their action and participate in the coregulation of hundreds of genes associated with the longevity of lifespan [32,345]. The transcription factors DAF-16/FOXO and HLH-30 are not only associated with longevity of life but also play an important role in the stress resistance [346].
DAF-16/FOXO acts as a joining point for different signaling pathways leading to longevity of lifespan and management of stress. IIS is the basic pathway, highly conserved from C. elegans, Drosophila melanogaster and mammals [283,347,348]. A signaling cascade initiated by the interaction of insulin such as peptides (ILPs) to the insulin / IGF1 receptor known as (DAF2) results in the downstream activation or regulation of IRS (insulin receptor substrate), P13K (phosphoinositide 3-kinases), PDK (3-phosphoadenosine kinase), and AKT or PKB (protein kinase B), resulting in the inhibition of FOXO. The main consequences of this cascade include the promotion of glucose transport, protein synthesis, cell proliferation, cell differentiation, and inhibition of apoptosis that enhance the longevity. Any stressed conditions or food restrictions leading to the obstruction of IIS signaling pathway will subsequently enhance the DAF-16/FOXO transcriptional activity, regulation of downstream genes resulting to manage stress and increase in the longevity of lifespan (Figure 4).
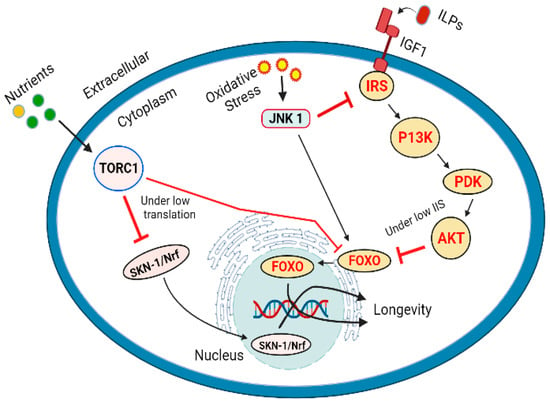
Figure 4.
Longevity of lifespan regulated by IIS pathway via DAF-16/FOXO, TOR pathway vis SKN1/Nrf and FOXO regulation, and by JNK under oxidative stress by phosphorylation and inactivation of IRS1 and activation of AKT.
Target of rapamycin (TOR) kinase which occurs in two distinct forms (TORC1 and TORC2) coded by the same gene. TOR participates to interact between the nutrients and growth promoting (anabolic) metabolic signals [349,350]. TOR pathway is activated by the presence of ample amounts of ATP, oxygen, and amino acids, it triggers the synthesis of nucleotides, lipids, promotes the levels of messenger RNAs and their subsequent translation [351]. TOR signaling is generally responsible for growth and development. However, it has also been linked with aging and diseases such as cancers, cardiovascular diseases, diabetes and neurodegenerative diseases [352,353]. Inhibition of TOR pathway can promote longevity by reducing mRNA translation [354,355,356]. Reduction in the protein synthesis reduces the overall burden on the cellular resources and machinery, it also promotes the expression of genes associated with cell protection. Genetic interference to inhibit the translation process has been linked with the longevity of life in C. elegans, the process has been associated with the inhibition of TOR pathway [357,358]. Transcription factor SKN1 (skinhead-1) and DAF-16 genes play an important role in the protection of cells by reducing translation. The similarity in their action suggests the regulation of these transcription factors by TOR pathway [359,360]. Reduced or inhibited TORC1 activity triggers SKN-1/Nrf and DAF-16/FoxO regulated reduction in the stress levels and increased lifespan [360]. The role of SKN-1 is interesting as it is considered as less important transcription factor to DAF-16. IIS inhibits both SKN-1 and DAF-16 by phosphorylation and translocation to the nucleus. However, TORC1 affects the SKN-1 present in the nucleus. A considerable number of SKN-1 molecules are found to occupy specific promoter sites under the conditions without significant stress [361]. Many isoforms of DAF-16 are accumulated in the nucleus when IIS is reduced [36,322,362], but only a single isoform DAF-16f is translocated to the nuclei by the inhibition of TORC1 indicating the most important isoform associated with longevity of life [36]. It has been reported that TORC1 and IIS influence DAF-16/FOXO and SKN1 by different mechanisms. In case of TORC1 induces longevity of life, SKN-1/Nrf are required and presence of DAF-16/FOXO is not obligatory [360] (Figure 4). Under heat stress conditions, JNK-1(Jun N-terminal kinase 1) also indorses the movement of DAF-16 into nucleus, it also facilitates the removal of binding partner 14-3-3 protein by phosphorylation [363,364]. In mammalian cells the FOXO4 can be directly phosphorylated, and its activity can be enhanced by JNK [365]. In mammals, the components of JNK also interact with insulin receptor substrate 1 (IRS-1). According to the reports JNK phosphorylated and inhibited IRS-1 and activated AKT1 [366] (Figure 4).
The well-known signaling pathways based on the NF-κB system that are associated with the human immune system are activated during aging. As the DNA damage in increased by the increased oxidative stress with age, it promotes the activity of NF-κB system that leads to adverse effects such as increase in apoptotic resistance, decrease in autophagy, and an enhanced inflammatory response. A number of inhibitors of NF-κB system are considered as longevity factors [367]. Nuclear receptors are also known to play an important role in the aging process. The NR4A subfamily of orphan nuclear receptors acts as nutrient sensors and promotes biogenesis, improves mitochondrial functioning, and contributes to DNA damage repair. These receptors are considered potential targets to slow down the aging process [368]. Nuclear receptor Nurr1 regulates the development of the dopaminergic phenotype from neuronal precursors. According to recent reports, a decrease in the Nurr1 expression leads to an enhanced inflammatory response resulting in the death of dopaminergic neurons. Hemizygosity of Nurr1 is necessary to protect against Parkinson’s disease [369].
5. Conclusions
The lifespan of living organisms is highly flexible and vulnerable to internal physiological factors and environmental conditions that govern the epigenetics. Expression and suppression levels of tens and hundreds of genes have been considered to play a significant contribution to the expected longevity of life. We have tabulated fifty such genes and a few have been elaborated for their influence in the regulation of aging process and lifespan. In addition to the contribution by gene expression regulatory systems, there is sufficient data with diverse evidence to support the critical role of DNA damage in the longevity of lifespan. The integrity of genomic DNA is an obligatory condition to maintain the cellular physiology and continuation of its transfer to the next generations from cell to organism levels. However, DNA is highly vulnerable to the internal and external stress conditions and chemical interferences. DNA modifications such as DNA methylation, acetylation of histones and physical damage to the DNA play important in the aging process and often lead to the unexpected outcomes in the lifespan. The efficient recovery of damaged DNA is crucial to the successful replication and inheritance of genome to the next generations. Failure to the proper DNA damage repair often leads to cell cycle arrest, apoptosis or cell death. The lifespan associated pathways regulate growth, essential physiology and reproduction of organisms. However, under adverse conditions, these pathways shift to protective and stress-bearing modes, eventually leading to an extended lifespan.
6. Future Perspective
Currently, research on the contribution of genes to the aging process, cellular stability, and longevity of lifespan is at initial stages. The data available is scattered, and the individual reports provide information about the contribution of selected either a gene or a group of similar genes and genetic mechanisms in the regulation of aging and lifespan, i.e., different reports are available representing the genes responsible for extended lifespan in humans, C. elegans and Drosophila. Genes regulating the cell cycle, apoptosis, managing the adverse effects of oxidative stress, and their association with life-threatening pathologies such as CVDs, diabetes, and neurodegenerative conditions have been reported but the mechanisms involved mostly remain unclear in the available reports. Large-scale genome-wide association studies (GWAS) in association with epigenetic approaches are required to demonstrate the coordination between different genes, their regulatory factors in the aging process and to reveal the mechanisms. Further studies for the identification of potential genetic targets to protect against aging-associated diseases are also required. Finally, the translation of these genetic findings into clinical practice poses a big challenge.
Author Contributions
Conceptualization, M.S.N., B.N.M., M.N.B.-J. and I.K.; writing—original draft preparation, M.S.N. and I.U.; writing—review and editing, critical revision, M.N.B.-J., S.J.G., F.A.A.-A., M.M.G., S.I.A., A.U. and S.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Vaupel, J.W.; Villavicencio, F.; Bergeron-Boucher, M.-P. Demographic perspectives on the rise of longevity. Proc. Natl. Acad. Sci. USA 2021, 118, e2019536118. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Song, Z.; Li, Y.; Yao, Y.; Fang, M.; Bai, C.; An, P.; Chen, H.; Chen, Z.; Tang, B. Integrated genetic analyses revealed novel human longevity loci and reduced risks of multiple diseases in a cohort study of 15,651 Chinese individuals. Aging Cell 2021, 20, e13323. [Google Scholar] [CrossRef] [PubMed]
- Van den Berg, N.; Rodríguez-Girondo, M.; van Dijk, I.; Mourits, R.; Mandemakers, K.; Janssens, A.; Beekman, M.; Smith, K.; Slagboom, P. Longevity defined as top 10% survivors and beyond is transmitted as a quantitative genetic trait. Nat Commun. 2019, 10, 35. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, J.K.; Elo, I.T.; Schupf, N.; Perls, T.T.; Stallard, E.; Yashin, A.I.; Christensen, K. The survival of spouses marrying into longevity-enriched families. J. Gerontol. Ser. A Biomed. Sci. Med. Sci. 2017, 72, 109–114. [Google Scholar] [CrossRef]
- Wijsman, C.A.; Rozing, M.P.; Streefland, T.C.; le Cessie, S.; Mooijaart, S.P.; Slagboom, P.E.; Westendorp, R.G.; Pijl, H.; van Heemst, D.; Leiden Longevity Study Group. Familial longevity is marked by enhanced insulin sensitivity. Aging Cell 2011, 10, 114–121. [Google Scholar] [CrossRef]
- Vaarhorst, A.A.; Beekman, M.; Suchiman, E.H.; Van Heemst, D.; Houwing-Duistermaat, J.J.; Westendorp, R.G.; Slagboom, P.E.; Heijmans, B.T. Lipid metabolism in long-lived families: The Leiden Longevity Study. Age 2011, 33, 219–227. [Google Scholar] [CrossRef]
- Andersen, S.L.; Sebastiani, P.; Dworkis, D.A.; Feldman, L.; Perls, T.T. Health span approximates life span among many supercentenarians: Compression of morbidity at the approximate limit of life span. J. Gerontol. Ser. A Biomed. Sci. Med. Sci. 2012, 67, 395–405. [Google Scholar] [CrossRef]
- Ash, A.S.; Kroll-Desrosiers, A.R.; Hoaglin, D.C.; Christensen, K.; Fang, H.; Perls, T.T. Are members of long-lived families healthier than their equally long-lived peers? Evidence from the long life family study. J. Gerontol. Ser. A Biomed. Sci. Med. Sci. 2015, 70, 971–976. [Google Scholar] [CrossRef]
- Slagboom, P.E.; van den Berg, N.; Deelen, J. Phenome and genome based studies into human ageing and longevity: An overview. Biochim. Biophys. Acta (BBA)—Mol. Basis Dis. 2018, 1864, 2742–2751. [Google Scholar] [CrossRef]
- Deelen, J.; Evans, D.S.; Arking, D.E.; Tesi, N.; Nygaard, M.; Liu, X.; Wojczynski, M.K.; Biggs, M.L.; van Der Spek, A.; Atzmon, G. A meta-analysis of genome-wide association studies identifies multiple longevity genes. Nat. Commun. 2019, 10, 3669. [Google Scholar] [CrossRef]
- Joshi, P.K.; Pirastu, N.; Kentistou, K.A.; Fischer, K.; Hofer, E.; Schraut, K.E.; Clark, D.W.; Nutile, T.; Barnes, C.L.; Timmers, P.R. Genome-wide meta-analysis associates HLA-DQA1/DRB1 and LPA and lifestyle factors with human longevity. Nat. Commun. 2017, 8, 910. [Google Scholar] [CrossRef] [PubMed]
- McDaid, A.F.; Joshi, P.K.; Porcu, E.; Komljenovic, A.; Li, H.; Sorrentino, V.; Litovchenko, M.; Bevers, R.P.; Rüeger, S.; Reymond, A. Bayesian association scan reveals loci associated with human lifespan and linked biomarkers. Nat. Commun. 2017, 8, 15842. [Google Scholar] [CrossRef] [PubMed]
- Vedder, O.; Moiron, M.; Bichet, C.; Bauch, C.; Verhulst, S.; Becker, P.H.; Bouwhuis, S. Telomere length is heritable and genetically correlated with lifespan in a wild bird. Mol. Ecol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Öngel, M.E.; Yıldız, C.; Akpınaroğlu, C.; Yilmaz, B.; Özilgen, M. Why women may live longer than men do? A telomere-length regulated and diet-based entropic assessment. Clin. Nutr. 2021, 40, 1186–1191. [Google Scholar] [CrossRef]
- Yu, Z.; Seim, I.; Yin, M.; Tian, R.; Sun, D.; Ren, W.; Yang, G.; Xu, S. Comparative analyses of aging-related genes in long-lived mammals provide insights into natural longevity. Innovation 2021, 2, 100108. [Google Scholar] [CrossRef]
- Gambini, J.; Gimeno-Mallench, L.; Olaso-Gonzalez, G.; Mastaloudis, A.; Traber, M.G.; Monleón, D.; Borrás, C.; Viña, J. Moderate Red Wine Consumption Increases the Expression of Longevity-Associated Genes in Controlled Human Populations and Extends Lifespan in Drosophila melanogaster. Antioxidants 2021, 10, 301. [Google Scholar] [CrossRef]
- Morris, B.J.; Chen, R.; Donlon, T.A.; Masaki, K.H.; Willcox, D.C.; Allsopp, R.C.; Willcox, B.J. Lifespan extension conferred by mitogen-activated protein kinase kinase kinase 5 (MAP3K5) longevity-associated gene variation is confined to at-risk men with a cardiometabolic disease. Aging 2021, 13, 7953. [Google Scholar] [CrossRef]
- Pawlikowska, L.; Hu, D.; Huntsman, S.; Sung, A.; Chu, C.; Chen, J.; Joyner, A.H.; Schork, N.J.; Hsueh, W.C.; Reiner, A.P. Association of common genetic variation in the insulin/IGF1 signaling pathway with human longevity. Aging Cell 2009, 8, 460–472. [Google Scholar] [CrossRef]
- Wilkinson, G.S.; Adams, D.M.; Haghani, A.; Lu, A.T.; Zoller, J.; Breeze, C.E.; Arnold, B.D.; Ball, H.C.; Carter, G.G.; Cooper, L.N. DNA methylation predicts age and provides insight into exceptional longevity of bats. Nat. Commun. 2021, 12, 1615. [Google Scholar] [CrossRef]
- Schumacher, B.; Pothof, J.; Vijg, J.; Hoeijmakers, J.H. The central role of DNA damage in the ageing process. Nature 2021, 592, 695–703. [Google Scholar] [CrossRef]
- Orkin, J.D.; Montague, M.J.; Tejada-Martinez, D.; de Manuel, M.; Del Campo, J.; Cheves Hernandez, S.; Di Fiore, A.; Fontsere, C.; Hodgson, J.A.; Janiak, M.C.; et al. The genomics of ecological flexibility, large brains, and long lives in capuchin monkeys revealed with fecalFACS. Proc. Natl. Acad. Sci. USA 2021, 118, e2010632118. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.P.; Demmitt, B.A.; Nath, R.D.; Brunet, A. The Genetics of Aging: A Vertebrate Perspective. Cell 2019, 177, 200–220. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.D.; Milman, S.; Lin, J.R.; Wierbowski, S.; Yu, H.; Barzilai, N.; Gorbunova, V.; Ladiges, W.C.; Niedernhofer, L.J.; Suh, Y.; et al. Genetics of extreme human longevity to guide drug discovery for healthy ageing. Nat. Metab. 2020, 2, 663–672. [Google Scholar] [CrossRef] [PubMed]
- Ang, L.S.; Cruz, R.P.; Hendel, A.; Granville, D.J. Apolipoprotein E, an important player in longevity and age-related diseases. Exp. Gerontol. 2008, 43, 615–622. [Google Scholar] [CrossRef] [PubMed]
- Raghavachari, N. The Impact of Apolipoprotein E Genetic Variability in Health and Life Span. J. Gerontol. Ser. A 2020, 75, 1855–1857. [Google Scholar] [CrossRef]
- Strittmatter, W.J.; Hill, C.B. Molecular biology of apolipoprotein E. Curr. Opin. Lipidol. 2002, 13, 119–123. [Google Scholar] [CrossRef]
- Mazzone, T. Apolipoprotein E secretion by macrophages: Its potential physiological functions. Curr. Opin. Lipidol. 1996, 7, 303–307. [Google Scholar] [CrossRef]
- Feng, Z.; Lin, M.; Wu, R. The Regulation of Aging and Longevity: A New and Complex Role of p53. Genes Cancer 2011, 2, 443–452. [Google Scholar] [CrossRef]
- Zhao, Y.; Wu, L.; Yue, X.; Zhang, C.; Wang, J.; Li, J.; Sun, X.; Zhu, Y.; Feng, Z.; Hu, W. A polymorphism in the tumor suppressor p53 affects aging and longevity in mouse models. Elife 2018, 7, e34701. [Google Scholar] [CrossRef]
- Brady, C.A.; Attardi, L.D. p53 at a glance. J. Cell Sci. 2010, 123, 2527–2532. [Google Scholar] [CrossRef]
- Leibiger, I.B.; Berggren, P.-O. Sirt1: A metabolic master switch that modulates lifespan. Nat. Med. 2006, 12, 34–36. [Google Scholar] [CrossRef]
- Ng, F.; Wijaya, L.; Tang, B.L. SIRT1 in the brain—connections with aging-associated disorders and lifespan. Front. Cell. Neurosci. 2015, 9, 64. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhou, M.; Ge, Y.; Wang, X. SIRT1 and aging related signaling pathways. Mech. Ageing Dev. 2020, 187, 111215. [Google Scholar] [CrossRef] [PubMed]
- Nogueiras, R.; Habegger, K.M.; Chaudhary, N.; Finan, B.; Banks, A.S.; Dietrich, M.O.; Horvath, T.L.; Sinclair, D.A.; Pfluger, P.T.; Tschöp, M.H. Sirtuin 1 and sirtuin 3: Physiological modulators of metabolism. Physiol. Rev. 2012. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Yuan, Z.; Li, Y.; Ling, H.; Izumi, V.; Fang, B.; Fukasawa, K.; Koomen, J.; Chen, J.; Seto, E. Ubiquitinated sirtuin 1 (SIRT1) function is modulated during DNA damage-induced cell death and survival. J. Biol. Chem. 2015, 290, 8904–8912. [Google Scholar] [CrossRef]
- Kwon, E.S.; Narasimhan, S.D.; Yen, K.; Tissenbaum, H.A. A new DAF-16 isoform regulates longevity. Nature 2010, 466, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Chen, W.D.; Wang, Y.D. DAF-16/FOXO Transcription Factor in Aging and Longevity. Front. Pharmacol. 2017, 8, 548. [Google Scholar] [CrossRef]
- Furuyama, T.; Kitayama, K.; Shimoda, Y.; Ogawa, M.; Sone, K.; Yoshida-Araki, K.; Hisatsune, H.; Nishikawa, S.I.; Nakayama, K.; Nakayama, K.; et al. Abnormal angiogenesis in Foxo1 (Fkhr)-deficient mice. J. Biol. Chem. 2004, 279, 34741–34749. [Google Scholar] [CrossRef]
- Joshi, P.K.; Fischer, K.; Schraut, K.E.; Campbell, H.; Esko, T.; Wilson, J.F. Variants near CHRNA3/5 and APOE have age-and sex-related effects on human lifespan. Nat. Commun. 2016, 7, 11174. [Google Scholar] [CrossRef]
- Haller, G.; Kapoor, M.; Budde, J.; Xuei, X.; Edenberg, H.; Nurnberger, J.; Kramer, J.; Brooks, A.; Tischfield, J.; Almasy, L.; et al. Rare missense variants in CHRNB3 and CHRNA3 are associated with risk of alcohol and cocaine dependence. Hum. Mol. Genet. 2014, 23, 810–819. [Google Scholar] [CrossRef][Green Version]
- Song, W.; Ren, D.; Li, W.; Jiang, L.; Cho, K.W.; Huang, P.; Fan, C.; Song, Y.; Liu, Y.; Rui, L. SH2B regulation of growth, metabolism, and longevity in both insects and mammals. Cell Metab. 2010, 11, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Kuo, C.-L.; Joaquim, M.; Kuchel, G.A.; Ferrucci, L.; Harries, L.W.; Pilling, L.C.; Melzer, D. The longevity-associated SH2B3 (LNK) genetic variant: Selected aging phenotypes in 379, 758 subjects. J. Gerontol. Ser. A 2020, 75, 1656–1662. [Google Scholar] [CrossRef] [PubMed]
- Maures, T.J.; Kurzer, J.H.; Carter-Su, C. SH2B1 (SH2-B) and JAK2: A multifunctional adaptor protein and kinase made for each other. Trends Endocrinol. Metab. 2007, 18, 38–45. [Google Scholar] [CrossRef]
- Gery, S.; Koeffler, H.P. Role of the adaptor protein LNK in normal and malignant hematopoiesis. Oncogene 2013, 32, 3111–3118. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.; Yang, H.; White, M.F.; Rui, L. Disruption of the SH2-B gene causes age-dependent insulin resistance and glucose intolerance. Mol. Cell. Biol. 2004, 24, 7435–7443. [Google Scholar] [CrossRef] [PubMed]
- Baker, D.J.; Jin, F.; Van Deursen, J.M. The yin and yang of the Cdkn2a locus in senescence and aging. Cell Cycle 2008, 7, 2795–2802. [Google Scholar] [CrossRef] [PubMed]
- Sándor, S.; Tátrai, K.; Czeibert, K.; Egyed, B.; Kubinyi, E. CDKN2A gene expression as a potential aging biomarker in dogs. Front. Vet. Sci. 2021, 8, 348. [Google Scholar] [CrossRef]
- Pal, A.; Potjer, T.P.; Thomsen, S.K.; Ng, H.J.; Barrett, A.; Scharfmann, R.; James, T.J.; Bishop, D.T.; Karpe, F.; Godsland, I.F.; et al. Loss-of-function mutations in the cell-cycle control gene CDKN2A impact on glucose homeostasis in humans. Diabetes 2016, 65, 527–533. [Google Scholar] [CrossRef]
- Kahoul, Y.; Oger, F.; Montaigne, J.; Froguel, P.; Breton, C.; Annicotte, J.S. Emerging Roles for the INK4a/ARF (CDKN2A) Locus in Adipose Tissue: Implications for Obesity and Type 2 Diabetes. Biomolecules 2020, 10, 1350. [Google Scholar] [CrossRef]
- Rabhi, N.; Hannou, S.A.; Gromada, X.; Salas, E.; Yao, X.; Oger, F.; Carney, C.; Lopez-Mejia, I.C.; Durand, E.; Rabearivelo, I.; et al. Cdkn2a deficiency promotes adipose tissue browning. Mol. Metab. 2018, 8, 65–76. [Google Scholar] [CrossRef]
- Pegoli, G.; Milan, M.; Manti, P.G.; Bianchi, A.; Lucini, F.; Santarelli, P.; Bearzi, C.; Rizzi, R.; Lanzuolo, C. Role of Cdkn2a in the Emery–Dreifuss Muscular Dystrophy Cardiac Phenotype. Biomolecules 2021, 11, 538. [Google Scholar] [CrossRef] [PubMed]
- Bacalini, M.G.; Deelen, J.; Pirazzini, C.; De Cecco, M.; Giuliani, C.; Lanzarini, C.; Ravaioli, F.; Marasco, E.; Van Heemst, D.; Suchiman, H.E.D. Systemic age-associated DNA hypermethylation of ELOVL2 gene: In vivo and in vitro evidences of a cell replication process. J. Gerontol. Ser. A Biomed. Sci. Med. Sci. 2017, 72, 1015–1023. [Google Scholar] [CrossRef] [PubMed]
- Chao, D.L.; Skowronska-Krawczyk, D. ELOVL2: Not just a biomarker of aging. Transl. Med. Aging 2020, 4, 78–80. [Google Scholar] [CrossRef] [PubMed]
- Pauter, A.M.; Olsson, P.; Asadi, A.; Herslöf, B.; Csikasz, R.I.; Zadravec, D.; Jacobsson, A. Elovl2 ablation demonstrates that systemic DHA is endogenously produced and is essential for lipid homeostasis in mice [S]. J. Lipid Res. 2014, 55, 718–728. [Google Scholar] [CrossRef]
- Bellini, L.; Campana, M.; Rouch, C.; Chacinska, M.; Bugliani, M.; Meneyrol, K.; Hainault, I.; Lenoir, V.; Denom, J.; Véret, J.; et al. Protective role of the ELOVL2/docosahexaenoic acid axis in glucolipotoxicity-induced apoptosis in rodent beta cells and human islets. Diabetologia 2018, 61, 1780–1793. [Google Scholar] [CrossRef]
- Kuningas, M.; Slagboom, P.E.; Westendorp, R.G.; van Heemst, D. Impact of genetic variations in the WRN gene on age related pathologies and mortality. Mech. Ageing Dev. 2006, 127, 307–313. [Google Scholar] [CrossRef]
- Ding, S.-L.; Yu, J.-C.; Chen, S.-T.; Hsu, G.-C.; Shen, C.-Y. Genetic variation in the premature aging gene WRN: A case-control study on breast cancer susceptibility. Cancer Epidemiol. Prev. Biomark. 2007, 16, 263–269. [Google Scholar] [CrossRef]
- Szekely, A.M.; Bleichert, F.; Nümann, A.; Van Komen, S.; Manasanch, E.; Ben Nasr, A.; Canaan, A.; Weissman, S.M. Werner protein protects nonproliferating cells from oxidative DNA damage. Mol. Cell. Biol. 2005, 25, 10492–10506. [Google Scholar] [CrossRef]
- Ozgenc, A.; Loeb, L.A. Current advances in unraveling the function of the Werner syndrome protein. Mutat. Res./Fundam. Mol. Mech. Mutagenesis 2005, 577, 237–251. [Google Scholar] [CrossRef]
- Opresko, P.L.; Cheng, W.H.; von Kobbe, C.; Harrigan, J.A.; Bohr, V.A. Werner syndrome and the function of the Werner protein; what they can teach us about the molecular aging process. Carcinogenesis 2003, 24, 791–802. [Google Scholar] [CrossRef]
- Lescai, F.; Marchegiani, F.; Franceschi, C. PON1 is a longevity gene: Results of a meta-analysis. Ageing Res. Rev. 2009, 8, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Erdman, V.V.N.T.; Tuktarova, I.A.; Timasheva, Y.R.; Viktorova, T.V. The PON1 gene polymorphic marker rs662 is associated with longevity in Bashkirs. Биoмика 2020, 12, 435–441. [Google Scholar] [CrossRef]
- Lacinski, M.; Skorupski, W.; Cieslinski, A.; Sokolowska, J.; Trzeciak, W.H. OF the paraoxonase-1 (PON1) protein in humans. Cell. Mol. Biol. 2004, 50, 885–893. [Google Scholar]
- Mackness, M.I.; Durrington, P.N.; Mackness, B. The role of paraoxonase 1 activity in cardiovascular disease. Am. J. Cardiovasc. Drugs 2004, 4, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Van Raamsdonk, J.M.; Hekimi, S. Deletion of the mitochondrial superoxide dismutase sod-2 extends lifespan in Caenorhabditis elegans. PLoS Genet. 2009, 5, e1000361. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Yolitz, J.; Alberico, T.; Sun, X.; Zou, S. Lifespan extension by cranberry supplementation partially requires SOD2 and is life stage independent. Exp. Gerontol. 2014, 50, 57–63. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Afolayan, A.J.; Teng, R.J.; Eis, A.; Rana, U.; Broniowska, K.A.; Corbett, J.A.; Pritchard, K.; Konduri, G.G. Inducible HSP70 regulates superoxide dismutase-2 and mitochondrial oxidative stress in the endothelial cells from developing lungs. Am. J. Physiol. Lung Cell. Mol. Physiol. 2014, 306, L351–L360. [Google Scholar] [CrossRef]
- Saxena, P.; Selvaraj, K.; Khare, S.K.; Chaudhary, N. Superoxide dismutase as multipotent therapeutic antioxidant enzyme: Role in human diseases. Biotechnol. Lett. 2021, 3, 1–22. [Google Scholar] [CrossRef]
- Conneely, K.N.; Capell, B.C.; Erdos, M.R.; Sebastiani, P.; Solovieff, N.; Swift, A.J.; Baldwin, C.T.; Budagov, T.; Barzilai, N.; Atzmon, G.; et al. Human longevity and common variations in the LMNA gene: A meta-analysis. Aging Cell 2012, 11, 475–481. [Google Scholar] [CrossRef]
- Lopez-Mejia, I.C.; Vautrot, V.; De Toledo, M.; Behm-Ansmant, I.; Bourgeois, C.F.; Navarro, C.L.; Osorio, F.G.; Freije, J.M.; Stévenin, J.; De Sandre-Giovannoli, A.; et al. A conserved splicing mechanism of the LMNA gene controls premature aging. Hum. Mol. Genet. 2011, 20, 4540–4555. [Google Scholar] [CrossRef]
- Cenni, V.; Capanni, C.; Mattioli, E.; Schena, E.; Squarzoni, S.; Bacalini, M.G.; Garagnani, P.; Salvioli, S.; Franceschi, C.; Lattanzi, G. Lamin A involvement in ageing processes. Ageing Res. Rev. 2020, 62, 101073. [Google Scholar] [CrossRef] [PubMed]
- Carmosino, M.; Torretta, S.; Procino, G.; Gerbino, A.; Forleo, C.; Favale, S.; Svelto, M. Role of nuclear Lamin A/C in cardiomyocyte functions. Biol. Cell 2014, 106, 346–358. [Google Scholar] [CrossRef] [PubMed]
- Soerensen, M.; Dato, S.; Tan, Q.; Thinggaard, M.; Kleindorp, R.; Beekman, M.; Suchiman, H.E.; Jacobsen, R.; McGue, M.; Stevnsner, T.; et al. Evidence from case-control and longitudinal studies supports associations of genetic variation in APOE, CETP, and IL6 with human longevity. Age 2013, 35, 487–500. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.K.; Gong, Y.Y.; Xie, L.; Yang, Y.; Xu, L.Y.; Zhang, Y.P. Association study of promoter polymorphisms in the CETP gene with longevity in the Han Chinese population. Mol. Biol. Rep. 2014, 41, 325–329. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; De, A. Pathophysiology and inhibition of cholesteryl ester transfer protein for prevention of cardiovascular diseases: An update. Drug Discov. Today 2021, 26, 1759–1764. [Google Scholar] [CrossRef]
- Yamashita, S.; Hirano, K.I.; Sakai, N.; Matsuzawa, Y. Molecular biology and pathophysiological aspects of plasma cholesteryl ester transfer protein. Biochim. Biophys. Acta (BBA)—Mol. Cell Biol. Lipids 2000, 1529, 257–275. [Google Scholar] [CrossRef]
- Novelli, V.; Viviani Anselmi, C.; Roncarati, R.; Guffanti, G.; Malovini, A.; Piluso, G.; Puca, A.A. Lack of replication of genetic associations with human longevity. Biogerontology 2008, 9, 85–92. [Google Scholar] [CrossRef]
- Muenchhoff, J.; Song, F.; Poljak, A.; Crawford, J.D.; Mather, K.A.; Kochan, N.A.; Yang, Z.; Trollor, J.N.; Reppermund, S.; Maston, K.; et al. Plasma apolipoproteins and physical and cognitive health in very old individuals. Neurobiol. Aging 2017, 55, 49–60. [Google Scholar] [CrossRef]
- Buckner, T.; Shao, B.; Eckel, R.H.; Heinecke, J.W.; Bornfeldt, K.E.; Snell-Bergeon, J. Association of apolipoprotein C3 with insulin resistance and coronary artery calcium in patients with type 1 diabetes. J. Clin. Lipidol. 2021, 15, 235–242. [Google Scholar] [CrossRef]
- Liu, C.C.; Kanekiyo, T.; Xu, H.; Bu, G. Apolipoprotein E and Alzheimer disease: Risk, mechanisms and therapy. Nat. Rev. Neurol. 2013, 9, 106–118. [Google Scholar] [CrossRef]
- Huffman, D.M.; Deelen, J.; Ye, K.; Bergman, A.; Slagboom, E.P.; Barzilai, N.; Atzmon, G. Distinguishing between longevity and buffered-deleterious genotypes for exceptional human longevity: The case of the MTP gene. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2012, 67, 1153–1160. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.M.; Rava, P.; Walsh, M.; Rana, M.; Iqbal, J. Multiple functions of microsomal triglyceride transfer protein. Nutr. Metab. 2012, 9, 14. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, J.; Jahangir, Z.; Al-Qarni, A.A. Microsomal triglyceride transfer protein: From lipid metabolism to metabolic diseases. In Lipid Transfer in Lipoprotein Metabolism and Cardiovascular Disease; Springer: Singapore, 2020; pp. 37–52. [Google Scholar]
- Xie, S.; Ni, J.; Guo, H.; Luu, V.; Wang, Y.; Zhao, J.J.; Roberts, T.M. The role of the PIK3CA gene in the development and aging of the brain. Sci. Rep. 2021, 11, 291. [Google Scholar] [CrossRef] [PubMed]
- Ditlevsen, D.K.; Køhler, L.B.; Pedersen, M.V.; Risell, M.; Kolkova, K.; Meyer, M.; Berezin, V.; Bock, E. The role of phosphatidylinositol 3-kinase in neural cell adhesion molecule-mediated neuronal differentiation and survival. J. Neurochem. 2003, 84, 546–556. [Google Scholar] [CrossRef] [PubMed]
- Xing, M. Genetic alterations in the phosphatidylinositol-3 kinase/Akt pathway in thyroid cancer. Thyroid 2010, 20, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Ihle, N.T.; Powis, G. Take your PIK: Phosphatidylinositol 3-kinase inhibitors race through the clinic and toward cancer therapy. Mol. Cancer Ther. 2009, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Pietsch, K.; Saul, N.; Menzel, R.; Stürzenbaum, S.R.; Steinberg, C.E. Quercetin mediated lifespan extension in Caenorhabditis elegans is modulated by age-1, daf-2, sek-1 and unc-43. Biogerontology 2009, 10, 565–578. [Google Scholar] [CrossRef]
- Ayyadevara, S.; Dandapat, A.; Singh, S.P.; Benes, H.; Zimniak, L.; Shmookler Reis, R.J.; Zimniak, P. Lifespan extension in hypomorphic daf-2 mutants of Caenorhabditis elegans is partially mediated by glutathione transferase CeGSTP2-2. Aging Cell 2005, 4, 299–307. [Google Scholar] [CrossRef]
- Laron, Z. Insulin-like growth factor 1 (IGF-1): A growth hormone. Mol. Pathol. 2001, 54, 311. [Google Scholar] [CrossRef]
- Larsson, O.; Girnita, A.; Girnita, L. Role of insulin-like growth factor 1 receptor signalling in cancer. Br. J. Cancer 2005, 92, 2097–2101. [Google Scholar] [CrossRef]
- Amer, M.; Elhefnawi, M.; El-Ahwany, E.; Awad, A.F.; Gawad, N.A.; Zada, S.; Tawab, F.M. Hsa-miR-195 targets PCMT1 in hepatocellular carcinoma that increases tumor life span. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2014, 35, 11301–11309. [Google Scholar] [CrossRef] [PubMed]
- Furuchi, T.; Sakurako, K.; Katane, M.; Sekine, M.; Homma, H. The role of protein L-isoaspartyl/D-aspartyl O-methyltransferase (PIMT) in intracellular signal transduction. Chem. Biodivers. 2010, 7, 1337–1348. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Ikegami, T.; Ogawara, M.; Suzuki, Y.I.; Takahashi, M.; Morio, H.; Shirasawa, T. Transgenic expression of the protein-L-isoaspartyl methyltransferase (PIMT) gene in the brain rescues mice from the fatal epilepsy of PIMT deficiency. J. Neurosci. Res. 2002, 69, 341–352. [Google Scholar] [CrossRef]
- Van Heemst, D. Insulin, IGF-1 and longevity. Aging Dis. 2010, 1, 147–157. [Google Scholar] [PubMed]
- López-Otín, C.; Galluzzi, L.; Freije, J.M.P.; Madeo, F.; Kroemer, G. Metabolic Control of Longevity. Cell 2016, 166, 802–821. [Google Scholar] [CrossRef] [PubMed]
- Waters, M.J.; Kaye, P.L. The role of growth hormone in fetal development. Growth Horm. IGF Res. 2002, 1, 137–146. [Google Scholar] [CrossRef]
- Vijayakumar, A.; Yakar, S.; Le Roith, D. The intricate role of growth hormone in metabolism. Front. Endocrinol. 2011, 2, 32. [Google Scholar] [CrossRef]
- Bartke, A. Growth hormone and aging: Updated review. World J. Men’s Health 2019, 37, 19–30. [Google Scholar] [CrossRef]
- Mahesh, S.; Kaskel, F. Growth hormone axis in chronic kidney disease. Pediatr. Nephrol. 2008, 23, 41–48. [Google Scholar] [CrossRef]
- Yokoyama, J.S.; Sturm, V.E.; Bonham, L.W.; Klein, E.; Arfanakis, K.; Yu, L.; Coppola, G.; Kramer, J.H.; Bennett, D.A.; Miller, B.L.; et al. Variation in longevity gene KLOTHO is associated with greater cortical volumes. Ann. Clin. Transl. Neurol. 2015, 2, 215–230. [Google Scholar] [CrossRef]
- Di Bona, D.; Accardi, G.; Virruso, C.; Candore, G.; Caruso, C. Association of Klotho polymorphisms with healthy aging: A systematic review and meta-analysis. Rejuvenation Res. 2014, 17, 212–216. [Google Scholar] [CrossRef] [PubMed]
- Dias, G.P.; Murphy, T.; Stangl, D.; Ahmet, S.; Morisse, B.; Nix, A.; Aimone, L.J.; Aimone, J.B.; Kuro, O.M.; Gage, F.H.; et al. Intermittent fasting enhances long-term memory consolidation, adult hippocampal neurogenesis, and expression of longevity gene Klotho. Mol. Psychiatry 2021, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, M.; Köhler, L.; Kovarova, M.; Lerche, S.; Schulte, C.; Wurster, I.; Machetanz, G.; Deuschle, C.; Hauser, A.K.; Gasser, T.; et al. The longevity gene Klotho and its cerebrospinal fluid protein profiles as a modifier for Parkinson’s disease. Eur. J. Neurol. 2021, 28, 1557–1565. [Google Scholar] [CrossRef] [PubMed]
- Bian, A.; Xing, C.; Hu, M.C. Alpha Klotho and phosphate homeostasis. J. Endocrinol. Investig. 2014, 37, 1121–1126. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Hwang, K.H.; Park, K.S.; Kong, I.D.; Cha, S.K. Biological role of anti-aging protein Klotho. J. Lifestyle Med. 2015, 5, 1. [Google Scholar] [CrossRef]
- Mencke, R.; Hillebrands, J.L. NIGRAM consortium. The role of the anti-ageing protein Klotho in vascular physiology and pathophysiology. Ageing Res. Rev. 2017, 35, 124–146. [Google Scholar] [CrossRef]
- Saar-Kovrov, V.; Donners, M.M.; Van Der Vorst, E.P. Shedding of Klotho: Functional Implications in Chronic Kidney Disease and Associated Vascular Disease. Front. Cardiovasc. Med. 2021, 7, 407. [Google Scholar] [CrossRef]
- Mair, W.; Morantte, I.; Rodrigues, A.P.; Manning, G.; Montminy, M.; Shaw, R.J.; Dillin, A. Lifespan extension induced by AMPK and calcineurin is mediated by CRTC-1 and CREB. Nature 2011, 470, 404–408. [Google Scholar] [CrossRef]
- Chen, Y.C.; Chen, H.J.; Tseng, W.C.; Hsu, J.M.; Huang, T.T.; Chen, C.H.; Pan, C.L. A C. elegans Thermosensory Circuit Regulates Longevity through crh-1/CREB-Dependent flp-6 Neuropeptide Signaling. Dev. Cell 2016, 39, 209–223. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K.; Karelina, K.; Obrietan, K. CREB: A multifaceted regulator of neuronal plasticity and protection. J. Neurochem. 2011, 116, 1–9. [Google Scholar] [CrossRef]
- Lee, A.H. The role of CREB-H transcription factor in triglyceride metabolism. Curr. Opin. Lipidol. 2012, 23, 141. [Google Scholar] [CrossRef] [PubMed]
- Amidfar, M.; de Oliveira, J.; Kucharska, E.; Budni, J.; Kim, Y.K. The role of CREB and BDNF in neurobiology and treatment of Alzheimer’s disease. Life Sci. 2020, 257, 118020. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Chen, H.; Wang, J.; Liu, G.; Cui, S.W.; Kang, J.; Jiang, Y.; Wang, H. Naringenin prolongs lifespan and delays aging mediated by IIS and MAPK in Caenorhabditis elegans. Food Funct. 2021, 12, 12127–12141. [Google Scholar] [CrossRef] [PubMed]
- Matilainen, O.; Ribeiro, A.R.S.; Verbeeren, J.; Cetinbas, M.; Sood, H.; Sadreyev, R.I.; Garcia, S. Loss of muscleblind splicing factor shortens Caenorhabditis elegans lifespan by reducing the activity of p38 MAPK/PMK-1 and transcription factors ATF-7 and Nrf/SKN-1. Genetics 2021, 219, iyab114. [Google Scholar] [CrossRef]
- Salojin, K.V.; Owusu, I.B.; Millerchip, K.A.; Potter, M.; Platt, K.A.; Oravecz, T. Essential role of MAPK phosphatase-1 in the negative control of innate immune responses. J. Immunol. 2006, 176, 1899–1907. [Google Scholar] [CrossRef]
- De Zelicourt, A.; Colcombet, J.; Hirt, H. The role of MAPK modules and ABA during abiotic stress signaling. Trends Plant Sci. 2016, 21, 677–685. [Google Scholar] [CrossRef]
- Zhu, X.; Lee, H.G.; Raina, A.K.; Perry, G.; Smith, M.A. The role of mitogen-activated protein kinase pathways in Alzheimer’s disease. Neurosignals 2002, 11, 270–281. [Google Scholar] [CrossRef]
- Bernardo, B.C.; Weeks, K.L.; Pretorius, L.; McMullen, J.R. Molecular distinction between physiological and pathological cardiac hypertrophy: Experimental findings and therapeutic strategies. Pharmacol. Ther. 2010, 128, 191–227. [Google Scholar] [CrossRef]
- Akhoon, B.A.; Rathor, L.; Pandey, R. Withanolide A extends the lifespan in human EGFR-driven cancerous Caenorhabditis elegans. Exp. Gerontol. 2018, 104, 113–117. [Google Scholar] [CrossRef]
- Kunugi, H.; Mohammed Ali, A. Royal Jelly and Its Components Promote Healthy Aging and Longevity: From Animal Models to Humans. Int. J. Mol. Sci. 2019, 20, 4662. [Google Scholar] [CrossRef]
- Romano, R.; Bucci, C. Role of EGFR in the Nervous System. Cells 2020, 9, 1887. [Google Scholar] [CrossRef]
- Breitbart, H.; Etkovitz, N. Role and regulation of EGFR in actin remodeling in sperm capacitation and the acrosome reaction. Asian J. Androl. 2011, 13, 106. [Google Scholar] [CrossRef] [PubMed]
- Sigismund, S.; Avanzato, D.; Lanzetti, L. Emerging functions of the EGFR in cancer. Mol. Oncol. 2018, 12, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Salminen, A.; Ojala, J.; Huuskonen, J.; Kauppinen, A.; Suuronen, T.; Kaarniranta, K. Interaction of aging-associated signaling cascades: Inhibition of NF-kappaB signaling by longevity factors FoxOs and SIRT1. Cell. Mol. Life Sci. CMLS 2008, 65, 1049–1058. [Google Scholar] [CrossRef] [PubMed]
- Ryu, S.; Han, J.; Norden-Krichmar, T.M.; Zhang, Q.; Lee, S.; Zhang, Z.; Atzmon, G.; Niedernhofer, L.J.; Robbins, P.D.; Barzilai, N.; et al. Genetic signature of human longevity in PKC and NF-κB signaling. Aging Cell 2021, 20, e13362. [Google Scholar] [CrossRef]
- Tripathi, P.; Aggarwal, A. NF-kB transcription factor: A key player in the generation of immune response. Curr. Sci. 2006, 25, 519–531. [Google Scholar]
- Caviedes, A.; Lafourcade, C.; Soto, C.; Wyneken, U. BDNF/NF-κB signaling in the neurobiology of depression. Curr. Pharm. Des. 2017, 23, 3154–3163. [Google Scholar] [CrossRef]
- Coto, E.; Reguero, J.R.; Avanzas, P.; Pascual, I.; Martín, M.; Hevia, S.; Morís, C.; Díaz-Molina, B.; Lambert, J.L.; Alonso, B.; et al. Gene variants in the NF-KB pathway (NFKB1, NFKBIA, NFKBIZ) and risk for early-onset coronary artery disease. Immunol. Lett. 2019, 208, 39–43. [Google Scholar] [CrossRef]
- Zinatizadeh, M.R.; Schock, B.; Chalbatani, G.M.; Zarandi, P.K.; Jalali, S.A.; Miri, S.R. The Nuclear Factor Kappa B (NF-kB) signaling in cancer development and immune diseases. Genes Dis. 2021, 8, 287–297. [Google Scholar] [CrossRef]
- Wong, C.O.; Karagas, N.E.; Jung, J.; Wang, Q.; Rousseau, M.A.; Chao, Y.; Insolera, R.; Soppina, P.; Collins, C.A.; Zhou, Y.; et al. Regulation of longevity by depolarization-induced activation of PLC-β-IP(3)R signaling in neurons. Proc. Natl. Acad. Sci. USA 2021, 118, 3118. [Google Scholar] [CrossRef]
- Bill, C.A.; Vines, C.M. Phospholipase, C. Calcium Signal 2020, 215–242. [Google Scholar]
- Rusciano, I.; Marvi, M.V.; Obeng, E.O.; Mongiorgi, S.; Ramazzotti, G.; Follo, M.Y.; Zoli, M.; Morandi, L.; Asioli, S.; Fabbri, V.P.; et al. Location-dependent role of phospholipase C signaling in the brain: Physiology and pathology. Adv. Biol. Regul. 2021, 79, 100771. [Google Scholar] [CrossRef] [PubMed]
- Lattanzio, R.; Piantelli, M.; Falasca, M. Role of phospholipase C in cell invasion and metastasis. Adv. Biol. Regul. 2013, 53, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Salmon, A.B.; Kim, G.; Liu, C.; Wren, J.D.; Georgescu, C.; Richardson, A.; Levine, R.L. Effects of transgenic methionine sulfoxide reductase A (MsrA) expression on lifespan and age-dependent changes in metabolic function in mice. Redox Biol. 2016, 10, 251–256. [Google Scholar] [CrossRef] [PubMed]
- Ruan, H.; Tang, X.D.; Chen, M.L.; Joiner, M.A.; Sun, G.; Brot, N.; Weissbach, H.; Heinemann, S.H.; Iverson, L.; Wu, C.F.; et al. High-quality life extension by the enzyme peptide methionine sulfoxide reductase. Proc. Natl. Acad. Sci. USA 2002, 99, 2748–2753. [Google Scholar] [CrossRef]
- Zhao, H.; Sun, J.; Deschamps, A.M.; Kim, G.; Liu, C.; Murphy, E.; Levine, R.L. Myristoylated methionine sulfoxide reductase A protects the heart from ischemia-reperfusion injury. Am. J. Physiol. Heart Circ. Physiol. 2011, 301, H1513–H1518. [Google Scholar] [CrossRef]
- Jiang, B.; Moskovitz, J. The functions of the mammalian methionine sulfoxide reductase system and related diseases. Antioxidants 2018, 7, 122. [Google Scholar] [CrossRef]
- Ewald, C.Y.; Hourihan, J.M.; Bland, M.S.; Obieglo, C.; Katic, I.; Moronetti Mazzeo, L.E.; Alcedo, J.; Blackwell, T.K.; Hynes, N.E. NADPH oxidase-mediated redox signaling promotes oxidative stress resistance and longevity through memo-1 in C. elegans. eLife 2017, 6, 19493. [Google Scholar] [CrossRef]
- Schotanus, M.D.; Van Otterloo, E. Finding MEMO—Emerging Evidence for MEMO1′s Function in Development and Disease. Genes 2020, 11, 1316. [Google Scholar] [CrossRef]
- Hooten, N.N.; Fitzpatrick, M.; Kompaniez, K.; Jacob, K.D.; Moore, B.R.; Nagle, J.; Barnes, J.; Lohani, A.; Evans, M. Coordination of DNA repair by NEIL1 and PARP-1: A possible link to aging. Aging 2012, 4, 674–685. [Google Scholar] [CrossRef]
- Han, D.; Schomacher, L.; Schüle, K.M.; Mallick, M.; Musheev, M.U.; Karaulanov, E.; Krebs, L.; Von Seggern, A.; Niehrs, C. Author response: NEIL1 and NEIL2 DNA glycosylases protect neural crest development against mitochondrial oxidative stress. eLife 2019, 8, e49044. [Google Scholar] [CrossRef] [PubMed]
- Kwiatkowski, D.; Czarny, P.; Toma, M.; Korycinska, A.; Sowinska, K.; Galecki, P.; Bachurska, A.; Bielecka-Kowalska, A.; Szemraj, J.; Maes, M.; et al. Association between Single-Nucleotide Polymorphisms of the hOGG1,NEIL1,APEX1, FEN1,LIG1, and LIG3 Genes and Alzheimer’s Disease Risk. Neuropsychobiology 2016, 73, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Argmann, C.; Dobrin, R.; Heikkinen, S.; Auburtin, A.; Pouilly, L.; Cock, T.-A.; Koutnikova, H.; Zhu, J.; Schadt, E.E.; Auwerx, J. Pparγ2 Is a Key Driver of Longevity in the Mouse. PLoS Genet. 2009, 5, e1000752. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Ma, X.; Verma, N.; Perie, L.; Pendse, J.; Shamloo, S.; Josephson, A.M.; Wang, D.; Qiu, J.; Guo, M.; et al. PPARγ agonists delay age-associated metabolic disease and extend longevity. Aging Cell 2020, 19, e13267. [Google Scholar] [CrossRef]
- Ge, K.; Guermah, M.; Yuan, C.-X.; Ito, M.; Wallberg, A.E.; Spiegelman, B.M.; Roeder, R.G. Transcription coactivator TRAP220 is required for PPARγ2-stimulated adipogenesis. Nature 2002, 417, 563–567. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, J.; Ding, J. Association of the PPARγ2 Pro12Ala polymorphism with increased risk of cardiovascular diseases. Genet. Mol. Res. 2015, 14, 18662–18674. [Google Scholar] [CrossRef]
- Cattie, D.J.; Richardson, C.E.; Reddy, K.C.; Ness-Cohn, E.M.; Droste, R.; Thompson, M.K.; Gilbert, W.V.; Kim, D.H. Mutations in Nonessential eIF3k and eIF3l Genes Confer Lifespan Extension and Enhanced Resistance to ER Stress in Caenorhabditis elegans. PLoS Genet. 2016, 12, e1006326. [Google Scholar] [CrossRef]
- Lin, Y.M.; Chen, Y.R.; Lin, J.R.; Wang, W.J.; Inoko, A.; Inagaki, M.; Wu, Y.C.; Chen, R.H. eIF3k regulates apoptosis in epithelial cells by releasing caspase 3 from keratin-containing inclusions. J. Cell Sci. 2008, 121, 2382–2393. [Google Scholar] [CrossRef]
- Wang, D.; Jia, Y.; Zheng, W.; Li, C.; Cui, W. Overexpression of eIF3D in lung adenocarcinoma is a new independent prognostic marker of poor survival. Dis. Markers 2019, 2019, 6019637. [Google Scholar] [CrossRef]
- Qian, M.; Liu, Z.; Peng, L.; Tang, X.; Meng, F.; Ao, Y.; Zhou, M.; Wang, M.; Cao, X.; Qin, B.; et al. Boosting ATM activity alleviates aging and extends lifespan in a mouse model of progeria. eLife 2018, 7, e34836. [Google Scholar] [CrossRef]
- Shiloh, Y.; Ziv, Y. The ATM protein kinase: Regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 2013, 14, 197–210. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; He, Y.; Hao, Q.; Chen, S.; Yang, M.; Leng, S.X.; Yue, J.; Dong, B. The association of single nucleotide polymorphism rs189037C> T in ATM gene with coronary artery disease in Chinese Han populations: A case control study. Medicine 2018, 97, e9747. [Google Scholar] [CrossRef] [PubMed]
- Pause, F.G.; Wacker, P.; Sappino, P. ATM gene and lymphoid malignancies. Leukemia 2003, 18, 238–242. [Google Scholar] [CrossRef]
- Fernández, Á.F.; Sebti, S.; Wei, Y.; Zou, Z.; Shi, M.; McMillan, K.L.; He, C.; Ting, T.; Liu, Y.; Chiang, W.-C.; et al. Disruption of the beclin 1–BCL2 autophagy regulatory complex promotes longevity in mice. Nature 2018, 558, 136–140. [Google Scholar] [CrossRef] [PubMed]
- Hardwick, J.M.; Soane, L. Multiple Functions of BCL-2 Family Proteins. Cold Spring Harb. Perspect. Biol. 2013, 5, a008722. [Google Scholar] [CrossRef]
- Sassone, J.; Maraschi, A.M.; Sassone, F.; Silani, V.; Ciammola, A. Defining the role of the Bcl-2 family proteins in Huntington’s disease. Cell Death Dis. 2013, 4, e772. [Google Scholar] [CrossRef]
- Wang, L.; Yang, L.; Debidda, M.; Witte, D.; Zheng, Y. Cdc42 GTPase-activating protein deficiency promotes genomic instability and premature aging-like phenotypes. Proc. Natl. Acad. Sci. USA 2007, 104, 1248–1253. [Google Scholar] [CrossRef]
- Florian, M.C.; Leins, H.; Gobs, M.; Han, Y.; Marka, G.; Soller, K.; Vollmer, A.; Sakk, V.; Nattamai, K.J.; Rayes, A.; et al. Inhibition of Cdc42 activity extends lifespan and decreases circulating inflammatory cytokines in aged female C57BL/6 mice. Aging Cell 2020, 19, e13208. [Google Scholar] [CrossRef]
- Florian, M.C.; Dörr, K.; Niebel, A.; Daria, D.; Schrezenmeier, H.; Rojewski, M.; Filippi, M.-D.; Hasenberg, A.; Gunzer, M.; Scharffetter-Kochanek, K.; et al. Cdc42 Activity Regulates Hematopoietic Stem Cell Aging and Rejuvenation. Cell Stem Cell 2012, 10, 520–530. [Google Scholar] [CrossRef]
- Druso, J.E.; Endo, M.; Lin, M.-C.J.; Peng, X.; Antonyak, M.A.; Meller, S.; Cerione, R.A. An Essential Role for Cdc42 in the Functioning of the Adult Mammary Gland. J. Biol. Chem. 2016, 291, 8886–8895. [Google Scholar] [CrossRef]
- Geiger, H.; Zheng, Y. Cdc42 and aging of hematopoietic stem cells. Curr. Opin. Hematol. 2013, 20, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Streeper, R.S.; Grueter, C.A.; Salomonis, N.; Cases, S.; Levin, M.C.; Koliwad, S.K.; Zhou, P.; Hirschey, M.; Verdin, E.; Farese, R.V. Deficiency of the lipid synthesis enzyme, DGAT1, extends longevity in mice. Aging 2012, 4, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.A.; Stolzing, A. The role of lipid metabolism in aging, lifespan regulation, and age-related disease. Aging Cell 2019, 18, e13048. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, T.; Sasaki, E.; Kakinuma, C.; Yano, T.; Miura, S.; Ezaki, O. Increased Very Low Density Lipoprotein Secretion and Gonadal Fat Mass in Mice Overexpressing Liver DGAT1. J. Biol. Chem. 2005, 280, 21506–21514. [Google Scholar] [CrossRef] [PubMed]
- Haas, J.; Winter, H.S.; Lim, E.; Kirby, A.; Blumenstiel, B.; DeFelice, M.; Gabriel, S.; Jalas, C.; Branski, D.; Grueter, C. DGAT1 mutation is linked to a congenital diarrheal disorder. J. Clin. Investig. 2012, 122, 4680–4684. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, S.M.; Kim, S.K. The GATA transcription factor/ MTA-1 homolog egr-1 promotes longevity and stress resistance in C aenorhabditis elegans. Aging Cell 2013, 13, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Tazearslan, C.; Huang, J.; Barzilai, N.; Suh, Y. Impaired IGF1R signaling in cells expressing longevity-associated human IGF1R alleles. Aging Cell 2011, 10, 551–554. [Google Scholar] [CrossRef]
- Sun, L.Y.; Spong, A.; Swindell, W.R.; Fang, Y.; Hill, C.; Huber, J.A.; Boehm, J.D.; Westbrook, R.; Salvatori, R.; Bartke, A. Growth hormone-releasing hormone disruption extends lifespan and regulates response to caloric restriction in mice. eLife 2013, 2, e01098. [Google Scholar] [CrossRef]
- Li, T.-T.; Liu, M.-R.; Pei, D.-S. Friend or foe, the role of EGR-1 in cancer. Med. Oncol. 2019, 37, 7. [Google Scholar] [CrossRef]
- Kuro-O, M. Aging and FGF23-klotho system. Vitam. Horm. 2021, 115, 317–332. [Google Scholar] [CrossRef]
- Shimada, T.; Kakitani, M.; Yamazaki, Y.; Hasegawa, H.; Takeuchi, Y.; Fujita, T.; Fukumoto, S.; Tomizuka, K.; Yamashita, T. Targeted ablation of Fgf23 demonstrates an essential physiological role of FGF23 in phosphate and vitamin D metabolism. J. Clin. Investig. 2004, 113, 561–568. [Google Scholar] [CrossRef] [PubMed]
- Quarles, L.D. Role of FGF23 in vitamin D and phosphate metabolism: Implications in chronic kidney disease. Exp. Cell Res. 2012, 318, 1040–1048. [Google Scholar] [CrossRef] [PubMed]
- Salminen, A.; Kaarniranta, K.; Kauppinen, A. Regulation of longevity by FGF21: Interaction between energy metabolism and stress responses. Ageing Res. Rev. 2017, 37, 79–93. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Nie, Y.; Cao, J.; Luo, M.; Yan, M.; Chen, Z.; He, B. The Roles and Pharmacological Effects of FGF21 in Preventing Aging-Associated Metabolic Diseases. Front. Cardiovasc. Med. 2021, 8, 221. [Google Scholar] [CrossRef] [PubMed]
- Fujii, N.; Uta, S.; Kobayashi, M.; Sato, T.; Okita, N.; Higami, Y. Impact of aging and caloric restriction on fibroblast growth factor 21 signaling in rat white adipose tissue. Exp. Gerontol. 2019, 118, 55–64. [Google Scholar] [CrossRef]
- Fisher, F.M.; Maratos-Flier, E. Understanding the Physiology of FGF21. Annu. Rev. Physiol. 2016, 78, 223–241. [Google Scholar] [CrossRef]
- Cheng, P.; Zhang, F.; Yu, L.; Lin, X.; He, L.; Li, X.; Lu, X.; Yan, X.; Tan, Y.; Zhang, C. Physiological and Pharmacological Roles of FGF21 in Cardiovascular Diseases. J. Diabetes Res. 2016, 2016, 1540267. [Google Scholar] [CrossRef]
- Torres, G.G.; Nygaard, M.; Caliebe, A.; Blanché, H.; Chantalat, S.; Galan, P.; Lieb, W.; Christiansen, L.; Deleuze, J.-F.; Christensen, K. Exome-Wide Association Study Identifies FN3KRP and PGP as New Candidate Longevity Genes. J. Gerontol. Ser. A Boil. Sci. Med Sci. 2021, 76, 786–795. [Google Scholar] [CrossRef]
- Szwergold, B. A Hypothesis: Fructosamine-3-Kinase-Related-Protein (FN3KRP) Catalyzes Deglycation of Maillard Intermediates Directly Downstream from Fructosamines. Rejuvenation Res. 2021, 24, 310–318. [Google Scholar] [CrossRef]
- Possik, E.; Madiraju, S.M.; Prentki, M. Glycerol-3-phosphate phosphatase/PGP: Role in intermediary metabolism and target for cardiometabolic diseases. Biochimie 2017, 143, 18–28. [Google Scholar] [CrossRef]
- Selman, C.; Partridge, L.; Withers, D. Replication of Extended Lifespan Phenotype in Mice with Deletion of Insulin Receptor Substrate 1. PLoS ONE 2011, 6, e16144. [Google Scholar] [CrossRef] [PubMed]
- Selman, C.; Lingard, S.; Choudhury, A.I.; Batterham, R.L.; Claret, M.; Clements, M.; Ramadani, F.; Okkenhaug, K.; Schuster, E.; Blanc, E.; et al. Evidence for lifespan extension and delayed age–related biomarkers in insulin receptor substrate 1 null mice. FASEB J. 2008, 22, 807–818. [Google Scholar] [CrossRef] [PubMed]
- Liberman, Z.; Eldar-Finkelman, H. Serine 332 Phosphorylation of Insulin Receptor Substrate-1 by Glycogen Synthase Kinase-3 Attenuates Insulin Signaling. J. Biol. Chem. 2005, 280, 4422–4428. [Google Scholar] [CrossRef] [PubMed]
- Baroni, M.G.; D’Andrea, M.P.; Montali, A.; Pannitteri, G.; Barillà, F.; Campagna, F.; Mazzei, E.; Lovari, S.; Seccareccia, F.; Campa, P.P.; et al. A Common Mutation of the Insulin Receptor Substrate-1 Gene Is A Risk Factor for Coronary Artery Disease. Arter. Thromb. Vasc. Biol. 1999, 19, 2975–2980. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yarchoan, M.; Toledo, J.; Lee, E.B.; Arvanitakis, Z.; Kazi, H.; Han, L.-Y.; Louneva, N.; Lee, V.M.-Y.; Kim, S.F.; Trojanowski, J.Q.; et al. Abnormal serine phosphorylation of insulin receptor substrate 1 is associated with tau pathology in Alzheimer’s disease and tauopathies. Acta Neuropathol. 2014, 128, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Gu, M.; Shen, L.; Bai, L.; Gao, J.; Marshall, C.; Wu, T.; Ding, J.; Miao, D.; Xiao, M. Heterozygous knockout of the Bmi-1 gene causes an early onset of phenotypes associated with brain aging. Age 2013, 36, 129–139. [Google Scholar] [CrossRef]
- Lee, J.Y.; Yu, K.-R.; Kim, H.-S.; Kang, I.; Kim, J.-J.; Lee, B.-C.; Choi, S.W.; Shin, J.-H.; Seo, Y.; Kang, K.-S. BMI1 inhibits senescence and enhances the immunomodulatory properties of human mesenchymal stem cells via the direct suppression of MKP-1/DUSP1. Aging 2016, 8, 1670–1689. [Google Scholar] [CrossRef]
- Abdouh, M.; Chatoo, W.; El Hajjar, J.; David, J.; Ferreira, J.; Bernier, G. Bmi1 Is Down-Regulated in the Aging Brain and Displays Antioxidant and Protective Activities in Neurons. PLoS ONE 2012, 7, e31870. [Google Scholar] [CrossRef]
- Sahasrabuddhe, A.A. BMI1: A Biomarker of Hematologic Malignancies. Biomarkers Cancer 2016, 8, BIC.S33376-75. [Google Scholar] [CrossRef]
- Bhattacharya, R.; Mustafi, S.B.; Street, M.; Dey, A.; Dwivedi, S.K. Bmi-1: At the crossroads of physiological and pathological biology. Genes Dis. 2015, 2, 225–239. [Google Scholar] [CrossRef]
- Edrey, Y.H.; Casper, D.; Huchon, D.; Mele, J.; Gelfond, J.A.; Kristan, D.M.; Nevo, E.; Buffenstein, R. Sustained high levels of neuregulin-1 in the longest-lived rodents; a key determinant of rodent longevity. Aging Cell 2011, 11, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Spivak, D.; Zhekalov, A.; Nyrov, V.; Shapovalov, P.; Spivak, I. Creativity and Its Genetic Foundations. In International Conference on Professional Culture of the Specialist of the Future; Springer: Cham, Switzerland, 2021; pp. 72–86. [Google Scholar]
- Dang, R.; Guo, Y.; Zhu, Y.; Yang, R.; Cai, H.; Jiang, P. Chronic administration of calcitriol enhanced neuregulin-1/ErbB signaling in rat myocardium. Pharm. Int. J. Pharm. Sci. 2016, 71, 192–195. [Google Scholar]
- Cheret, C.; Willem, M.; Fricker, F.R.; Wende, H.; Wulf-Goldenberg, A.; Tahirovic, S.; Nave, K.-A.; Saftig, P.; Haass, C.; Garratt, A.N.; et al. Bace1 and Neuregulin-1 cooperate to control formation and maintenance of muscle spindles. EMBO J. 2013, 32, 2015–2028. [Google Scholar] [CrossRef] [PubMed]
- Harrison, P.J.; Law, A.J. Neuregulin 1 and SchizophreniA Genetics, Gene Expression, and Neurobiology. Biol. Psychiatry 2006, 60, 132–140. [Google Scholar] [CrossRef]
- Erdman, V.; Nasibullin, T.R.; Tuktarova, I.A.; Somova, R.S.; Mustafina, O.E. Association Analysis of Polymorphic Gene Variants in the JAK/STAT Signaling Pathway with Aging and Longevity. Russ. J. Genet. 2019, 55, 728–737. [Google Scholar] [CrossRef]
- Hofmann, P.; Sommer, J.; Theodorou, K.; Kirchhof, L.; Fischer, A.; Li, Y.; Perisic, L.; Hedin, U.; Maegdefessel, L.; Dimmeler, S.; et al. Long non-coding RNA H19 regulates endothelial cell aging via inhibition of STAT3 signalling. Cardiovasc. Res. 2018, 115, 230–242. [Google Scholar] [CrossRef]
- Awasthi, N.; Liongue, C.; Ward, A.C. STAT proteins: A kaleidoscope of canonical and non-canonical functions in immunity and cancer. J. Hematol. Oncol. 2021, 14, 198. [Google Scholar] [CrossRef]
- Lorenzini, T.; Dotta, L.; Giacomelli, M.; Vairo, D.; Badolato, R. STAT mutations as program switchers: Turning primary immuno-deficiencies into autoimmune diseases. J. Leukoc. Biol. 2017, 101, 29–38. [Google Scholar] [CrossRef]
- Gu, Y.; Mohammad, I.S.; Liu, Z. Overview of the STAT-3 signaling pathway in cancer and the development of specific inhibitors. Oncol. Lett. 2020, 19, 2585–2594. [Google Scholar] [CrossRef]
- Yamasaki, L.; Bronson, R.; Williams, B.O.; Dyson, N.J.; Harlow, E.; Jacks, T. Loss of E2F-1 reduces tumorigenesis and extends the lifespan of Rb1 (+/−) mice. Nat. Genet. 1998, 18, 360–364. [Google Scholar] [CrossRef]
- Xie, Q.; Peng, S.; Tao, L.; Ruan, H.; Yang, Y.; Li, T.-M.; Adams, U.; Meng, S.; Bi, X.; Dong, M.-Q.; et al. E2F Transcription Factor 1 Regulates Cellular and Organismal Senescence by Inhibiting Forkhead Box O Transcription Factors. J. Biol. Chem. 2014, 289, 34205–34213. [Google Scholar] [CrossRef] [PubMed]
- Warg, L.A.; Oakes, J.L.; Burton, R.; Neidermyer, A.J.; Rutledge, H.R.; Groshong, S.; Schwartz, D.A.; Yang, I.V. The role of the E2F1 tran-scription factor in the innate immune response to systemic LPS. Am. J. Physiol. Lung Cell. Mol. Physiol. 2012, 303, L391–L400. [Google Scholar] [CrossRef] [PubMed]
- Denechaud, P.-D.; Fajas, L.; Giralt, A. E2F1, a Novel Regulator of Metabolism. Front. Endocrinol. 2017, 8, 311. [Google Scholar] [CrossRef] [PubMed]
- Mandigo, A.C.; Yuan, W.; Xu, K.; Gallagher, P.; Pang, A.; Guan, Y.F.; Shafi, A.A.; Thangavel, C.; Sheehan, B.; Bogdan, D.; et al. RB/E2F1 as a Master Regulator of Cancer Cell Metabolism in Advanced Disease. Cancer Discov. 2021, 11, 2334–2353. [Google Scholar] [CrossRef]
- Grunewald, M.; Kumar, S.; Sharife, H.; Volinsky, E.; Gileles-Hillel, A.; Licht, T.; Permyakova, A.; Hinden, L.; Azar, S.; Friedmann, Y.; et al. Counteracting age-related VEGF signaling insufficiency promotes healthy aging and extends life span. Science 2021, 373, eabc8479. [Google Scholar] [CrossRef]
- Del Bo, R.; Ghezzi, S.; Scarlato, M.; Albani, D.; Galimberti, D.; Lucca, U.; Tettamanti, M.; Scarpini, E.; Forloni, G.; Bresolin, N.; et al. Role of VEGF gene variability in longevity: A lesson from the Italian population. Neurobiol. Aging 2008, 29, 1917–1922. [Google Scholar] [CrossRef]
- Matsumoto, K.; Ema, M. Roles of VEGF-A signalling in development, regeneration, and tumours. J. Biochem. 2014, 156, 1–10. [Google Scholar] [CrossRef]
- Tyshkovskiy, A.; Bozaykut, P.; Borodinova, A.A.; Gerashchenko, M.V.; Ables, G.P.; Garratt, M.; Khaitovich, P.; Clish, C.; Miller, R.A.; Gladyshev, V.N. Identification and Application of Gene Expression Signatures Associated with Lifespan Extension. Cell Metab. 2019, 30, 573–593.e8. [Google Scholar] [CrossRef]
- Steinbaugh, M.; Sun, L.Y.; Bartke, A.; Miller, R.A. Activation of genes involved in xenobiotic metabolism is a shared signature of mouse models with extended lifespan. Am. J. Physiol. Metab. 2012, 303, E488–E495. [Google Scholar] [CrossRef]
- Crocco, P.; Montesanto, A.; Dato, S.; Geracitano, S.; Iannone, F.; Passarino, G.; Rose, G. Inter-individual variability in xenobi-otic-metabolizing enzymes: Implications for human aging and longevity. Genes 2019, 10, 403. [Google Scholar] [CrossRef]
- Shore, D.E.; Ruvkun, G. A cytoprotective perspective on longevity regulation. Trends Cell Biol. 2013, 23, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, J.W.; Zhao, X.; De Cecco, M.; Peterson, A.L.; Pagliaroli, L.; Manivannan, J.; Hubbard, G.B.; Ikeno, Y.; Zhang, Y.; Feng, B.; et al. Reduced Expression of MYC Increases Longevity and Enhances Healthspan. Cell 2015, 160, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Carroll, P.A.; Eisenman, R.N. Growing old with Myc. Cell 2015, 160, 365–366. [Google Scholar] [CrossRef]
- Neumann, B.; Segel, M.; Ghosh, T.; Zhao, C.; Tourlomousis, P.; Young, A.; Förster, S.; Sharma, A.; Chen, C.Z.-Y.; Cubillos, J.F.; et al. Myc determines the functional age state of oligodendrocyte progenitor cells. Nat. Aging 2021, 1, 826–837. [Google Scholar] [CrossRef]
- Marinkovic, D.; Marinkovic, T. The new role for an old guy: MYC as an immunoplayer. J. Cell. Physiol. 2020, 236, 3234–3243. [Google Scholar] [CrossRef] [PubMed]
- Spinetti, G.; Sangalli, E.; Specchia, C.; Villa, F.; Spinelli, C.C.; Pipolo, R.; Carrizzo, A.; Greco, S.; Voellenkle, C.; Vecchione, C.; et al. The expression of the BPIFB4 and CXCR4 associates with sustained health in long-living individuals from Cilento-Italy. Aging 2017, 9, 370–380. [Google Scholar] [CrossRef]
- Moll, N.M.; Ransohoff, R.M. CXCL12 and CXCR4 in bone marrow physiology. Expert Rev. Hematol. 2010, 3, 315–322. [Google Scholar] [CrossRef]
- Zlotnik, A. New insights on the role of CXCR4 in cancer metastasis. J. Pathol. A J. Pathol. Soc. Great Br. Irel. 2008, 215, 211–213. [Google Scholar] [CrossRef]
- Tissenbaum, H.A.; Guarente, L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 2001, 410, 227–230. [Google Scholar] [CrossRef]
- Whitaker, R.; Faulkner, S.; Miyokawa, R.; Burhenn, L.; Henriksen, M.; Wood, J.G.; Helfand, S.L. Increased expression of Drosophila Sir 2 extends life span in a dose-dependent manner. Aging 2013, 5, 682–691. [Google Scholar] [CrossRef]
- Hoffmann, J.; Romey, R.; Fink, C.; Yong, L.; Roeder, T. Overexpression of Sir2 in the adult fat body is sufficient to extend lifespan of male and female Drosophila. Aging 2013, 5, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Turdi, S.; Li, Q.; Lopez, F.L.; Ren, J. Catalase alleviates cardiomyocyte dysfunction in diabetes: Role of Akt, Forkhead transcriptional factor and silent information regulator 2. Life Sci. 2007, 81, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Hattori, Y.; Okamoto, Y.; Maki, T.; Yamamoto, Y.; Oishi, N.; Yamahara, K.; Nagatsuka, K.; Takahashi, R.; Kalaria, R.N.; Fukuyama, H.; et al. Silent Information Regulator 2 Homolog 1 Counters Cerebral Hypoperfusion Injury by Deacetylating Endothelial Nitric Oxide Synthase. Stroke 2014, 45, 3403–3411. [Google Scholar] [CrossRef] [PubMed]
- Yoon, D.S.; Cha, D.S.; Choi, Y.; Lee, J.W.; Lee, M.H. MPK-1/ERK is required for the full activity of resveratrol in extended lifespan and reproduction. Aging Cell 2019, 18, e12867. [Google Scholar] [CrossRef]
- Chung, E.; Kondo, M. Role of Ras/Raf/MEK/ERK signaling in physiological hematopoiesis and leukemia development. Immunol. Res. 2010, 49, 248–268. [Google Scholar] [CrossRef]
- Kim, H.; Wu, X.; Lee, J. SLC31 (CTR) family of copper transporters in health and disease. Mol. Asp. Med. 2013, 34, 561–570. [Google Scholar] [CrossRef]
- Mahley, R.W.; Rall, S.C., Jr. Apolipoprotein E: Far more than a lipid transport protein. Annu. Rev. Genom. Hum. Genet. 2000, 1, 507–537. [Google Scholar] [CrossRef]
- Fullerton, S.M.; Clark, A.; Weiss, K.M.; Nickerson, D.A.; Taylor, S.L.; Stengård, J.H.; Salomaa, V.; Vartiainen, E.; Perola, M.; Boerwinkle, E.; et al. Apolipoprotein E Variation at the Sequence Haplotype Level: Implications for the Origin and Maintenance of a Major Human Polymorphism. Am. J. Hum. Genet. 2000, 67, 881–900. [Google Scholar] [CrossRef]
- Stengård, J.H.; Clark, A.; Weiss, K.M.; Kardia, S.; Nickerson, D.A.; Salomaa, V.; Ehnholm, C.; Boerwinkle, E.; Sing, C.F. Contributions of 18 Additional DNA Sequence Variations in the Gene Encoding Apolipoprotein E to Explaining Variation in Quantitative Measures of Lipid Metabolism. Am. J. Hum. Genet. 2002, 71, 501–517. [Google Scholar] [CrossRef]
- Swertfeger, D.; Hui, D.Y. Apolipoprotein E Receptor Binding VersusHeparan Sulfate Proteoglycan Binding in Its Regulation of Smooth Muscle Cell Migration and Proliferation. J. Biol. Chem. 2001, 276, 25043–25048. [Google Scholar] [CrossRef]
- Tudorache, I.F.; Trusca, V.G.; Gafencu, A.V. Apolipoprotein E-a multifunctional protein with implications in various pathol-ogies as a result of its structural features. Comput. Struct. Biotechnol. J. 2017, 15, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Gerdes, L.U.; Gerdes, C.; Hansen, P.S.; Klausen, I.C.; Faergeman, O.; Dyerberg, J. The apolipoprotein E polymorphism in Greenland Inuit in its global perspective. Qual. Life Res. 1996, 98, 546–550. [Google Scholar] [CrossRef] [PubMed]
- Ewbank, D.C. The APOE gene and differences in life expectancy in Europe. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2004, 59, B16–B20. [Google Scholar] [CrossRef]
- Song, Y.; Stampfer, M.J.; Liu, S. Meta-Analysis: Apolipoprotein E Genotypes and Risk for Coronary Heart Disease. Ann. Intern. Med. 2004, 141, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Bertram, L.; McQueen, M.B.; Mullin, K.; Blacker, D.; Tanzi, R.E. Systematic meta-analyses of Alzheimer disease genetic asso-ciation studies: The AlzGene database. Nat. Genet. 2007, 39, 17–23. [Google Scholar] [CrossRef]
- Kulminski, A.M.; Arbeev, K.G.; Culminskaya, I.; Arbeeva, L.; Ukraintseva, S.V.; Stallard, E.; Christensen, K.; Schupf, N.; Province, M.A.; Yashin, A.I. Age, Gender, and Cancer but Not Neurodegenerative and Cardiovascular Diseases Strongly Modulate Systemic Effect of the Apolipoprotein E4 Allele on Lifespan. PLoS Genet. 2014, 10, e1004141. [Google Scholar] [CrossRef]
- Finch, C.E. Evolution of the human lifespan and diseases of aging: Roles of infection, inflammation, and nutrition. Proc. Natl. Acad. Sci. USA 2009, 107, 1718–1724. [Google Scholar] [CrossRef]
- Finch, C.E. Evolution of the human lifespan, past, present, and future: Phases in the evolution of human life expectancy in relation to the inflammatory load. Proc. Am. Philos. Soc. USA 2012, 156, 9–44. [Google Scholar]
- Finch, C.E.; Stanford, C.B. Meat-Adaptive Genes and the Evolution of Slower Aging in Humans. Q. Rev. Biol. 2004, 79, 3–50. [Google Scholar] [CrossRef]
- Raichlen, D.A.; Alexander, G.E. Exercise, APOE genotype, and the evolution of the human lifespan. Trends Neurosci. 2014, 37, 247–255. [Google Scholar] [CrossRef]
- Shinohara, M.; Suzuki, K.; Bu, G.; Sato, N. Interaction Between APOE Genotype and Diabetes in Longevity. J. Alzheimer’s Dis. 2021, 82, 719–726. [Google Scholar] [CrossRef] [PubMed]
- Hawkes, K.; O’Connell, J.F. How old is human longevity? J. Hum. Evol. 2005, 49, 650–653. [Google Scholar] [CrossRef] [PubMed]
- Makkar, S.R.; Lipnicki, D.M.; Crawford, J.D.; Kochan, N.A.; Castro-Costa, E.; Lima-Costa, M.F.; Diniz, B.S.; Brayne, C.; Stephan, B.; Matthews, F.; et al. APOE ε4 and the Influence of Sex, Age, Vascular Risk Factors, and Ethnicity on Cognitive Decline. J. Gerontol. Ser. A Boil. Sci. Med Sci. 2020, 75, 1863–1873. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, T.; Nishimoto, Y.; Abe, Y.; Takayama, M.; Hirose, N.; Okano, H.; Arai, Y. Sex-Specific Effects of Apolipoprotein ε4 Allele on Mortality in Very Old and Centenarian Japanese Men. J. Gerontol. Ser. A Boil. Sci. Med Sci. 2019, 75, 1874–1879. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, V.S.; Deo, P.; Chua, A.; Thomas, P.; Fenech, M. Shorter telomere length in carriers of APOE-ε4 and high plasma concentration of glucose, glyoxal and other advanced glycation end products (AGEs). J. Gerontol. Ser. A 2020, 75, 1894–1898. [Google Scholar] [CrossRef] [PubMed]
- Garatachea, N.; Emanuele, E.; Calero, M.; Fuku, N.; Arai, Y.; Abe, Y.; Murakami, H.; Miyachi, M.; Yvert, T.; Verde, Z.; et al. ApoE gene and exceptional longevity: Insights from three independent cohorts. Exp. Gerontol. 2014, 53, 16–23. [Google Scholar] [CrossRef]
- Bourdon, J.-C. p53 and its isoforms in cancer. Br. J. Cancer 2007, 97, 277–282. [Google Scholar] [CrossRef]
- Levine, A.J. p53: 800 million years of evolution and 40 years of discovery. Nat. Cancer 2020, 20, 471–480. [Google Scholar] [CrossRef]
- Hu, W.; Feng, Z.; Teresky, A.K.; Levine, A.J. p53 regulates maternal reproduction through LIF. Nature 2007, 450, 721–724. [Google Scholar] [CrossRef]
- Levine, A.J.; Tomasini, R.; McKeon, F.D.; Mak, T.W.; Melino, G. The p53 family: Guardians of maternal reproduction. Nat. Rev. Mol. Cell Biol. 2011, 12, 259–265. [Google Scholar] [CrossRef]
- van Heemst, D.; Mooijaart, S.; Beekman, M.; Schreuder, J.; de Craen, A.J.; Brandt, B.W.; Slagboom, P.; Westendorp, R.G. Variation in the human TP53 gene affects old age survival and cancer mortality. Exp. Gerontol. 2004, 40, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Ørsted, D.D.; Bojesen, S.E.; Tybjærg-Hansen, A.; Nordestgaard, B.G. Tumor suppressor p53 Arg72Pro polymorphism and longevity, cancer survival, and risk of cancer in the general population. J. Cell Biol. 2007, 177, i14. [Google Scholar] [CrossRef]
- Boyd-Kirkup, J.D.; Green, C.D.; Wu, G.; Wang, D.; Han, J.D. Epigenomics and the regulation of aging. Epigenomics 2013, 5, 205–227. [Google Scholar] [CrossRef] [PubMed]
- Maier, B.; Gluba, W.; Bernier, B.; Turner, T.; Mohammad, K.; Guise, T.; Sutherland, A.; Thorner, M.; Scrable, H. Modulation of mammalian life span by the short isoform of p53. Genes Dev. 2004, 18, 306–319. [Google Scholar] [CrossRef]
- Sulak, M.; Fong, L.; Mika, K.; Chigurupati, S.; Yon, L.; Mongan, N.; Emes, R.; Lynch, V.J. Correction: TP53 copy number expansion is associated with the evolution of increased body size and an enhanced DNA damage response in elephants. eLife 2016, 5, e11994. [Google Scholar] [CrossRef]
- Lee, S.-H.; Lee, J.-H.; Lee, H.-Y.; Min, A.K.-J. Sirtuin signaling in cellular senescence and aging. BMB Rep. 2019, 52, 24–34. [Google Scholar] [CrossRef]
- Yoshizaki, T.; Schenk, S.; Imamura, T.; Babendure, J.L.; Sonoda, N.; Bae, E.J.; Oh, D.Y.; Lu, M.; Milne, J.C.; Westphal, C.; et al. SIRT1 inhibits inflammatory pathways in macrophages and modulates insulin sensitivity. Am. J. Physiol. Metab. 2010, 298, E419–E428. [Google Scholar] [CrossRef]
- Yeung, F.; Hoberg, J.E.; Ramsey, C.S.; Keller, M.D.; Jones, D.R.; Frye, R.A.; Mayo, M.W. Modulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004, 23, 2369–2380. [Google Scholar] [CrossRef]
- Cohen, H.Y.; Miller, C.; Bitterman, K.J.; Wall, N.R.; Hekking, B.; Kessler, B.; Howitz, K.T.; Gorospe, M.; de Cabo, R.; Sinclair, D.A. Calorie Restriction Promotes Mammalian Cell Survival by Inducing the SIRT1 Deacetylase. Science 2004, 305, 390–392. [Google Scholar] [CrossRef]
- Wang, Y.; He, J.; Liao, M.; Hu, M.; Li, W.; Ouyang, H.; Wang, X.; Ye, T.; Zhang, Y.; Ouyang, L. An overview of Sirtuins as potential therapeutic target: Structure, function and modulators. Eur. J. Med. Chem. 2018, 161, 48–77. [Google Scholar] [CrossRef]
- Bernier, M.; Paul, R.K.; Martin-Montalvo, A.; Scheibye-Knudsen, M.; Song, S.; He, H.-J.; Armour, S.M.; Hubbard, B.; Bohr, V.A.; Wang, L.; et al. Negative Regulation of STAT3 Protein-mediated Cellular Respiration by SIRT1 Protein. J. Biol. Chem. 2011, 286, 19270–19279. [Google Scholar] [CrossRef] [PubMed]
- Brunet, A.; Sweeney, L.B.; Sturgill, J.F.; Chua, K.F.; Greer, P.L.; Lin, Y.; Tran, H.; Ross, S.E.; Mostoslavsky, R.; Cohen, H.Y.; et al. Stress-Dependent Regulation of FOXO Transcription Factors by the SIRT1 Deacetylase. Science 2004, 303, 2011–2015. [Google Scholar] [CrossRef] [PubMed]
- Rogina, B.; Helfand, S.L. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc. Natl. Acad. Sci. USA 2004, 101, 15998–16003. [Google Scholar] [CrossRef] [PubMed]
- Kanfi, Y.; Naiman, S.; Amir, G.; Peshti, V.; Zinman, G.; Nahum, L.; Bar-Joseph, Z.; Cohen, H.Y. The sirtuin SIRT6 regulates lifespan in male mice. Nature 2012, 483, 218–221. [Google Scholar] [CrossRef]
- Satoh, A.; Brace, C.S.; Rensing, N.; Cliften, P.; Wozniak, D.F.; Herzog, E.; Yamada, K.A.; Imai, S.-I. Sirt1 Extends Life Span and Delays Aging in Mice through the Regulation of Nk2 Homeobox 1 in the DMH and LH. Cell Metab. 2013, 18, 416–430. [Google Scholar] [CrossRef]
- Canto, C.; Houtkooper, R.H.; Pirinen, E.; Youn, D.Y.; Oosterveer, M.H.; Cen, Y.; Fernandez-Marcos, P.J.; Yamamoto, H.; Andreux, P.A.; Cettour-Rose, P.; et al. The NAD+ Precursor Nicotinamide Riboside Enhances Oxidative Metabolism and Protects against High-Fat Diet-Induced Obesity. Cell Metab. 2012, 15, 838–847. [Google Scholar] [CrossRef]
- Mills, K.F.; Yoshida, S.; Stein, L.R.; Grozio, A.; Kubota, S.; Sasaki, Y.; Redpath, P.; Migaud, M.E.; Apte, R.S.; Uchida, K.; et al. Long-Term Administration of Nicotinamide Mononucleotide Mitigates Age-Associated Physiological Decline in Mice. Cell Metab. 2016, 24, 795–806. [Google Scholar] [CrossRef]
- Guan, Y.; Wang, S.R.; Huang, X.Z.; Xie, Q.H.; Xu, Y.Y.; Shang, D.; Hao, C.M. Nicotinamide Mononucleotide, an NAD+ Precursor, Rescues Age-Associated Susceptibility to AKI in a Sirtuin 1–Dependent Manner. J. Am. Soc. Nephrol. 2017, 28, 2337–2352. [Google Scholar] [CrossRef]
- Amano, H.; Chaudhury, A.; Rodriguez-Aguayo, C.; Lu, L.; Akhanov, V.; Catic, A.; Popov, Y.V.; Verdin, E.; Johnson, H.; Stossi, F.; et al. Telomere Dysfunction Induces Sirtuin Repression that Drives Telomere-Dependent Disease. Cell Metab. 2019, 29, 1274–1290.e9. [Google Scholar] [CrossRef]
- Pi, C.; Yang, Y.; Sun, Y.; Wang, H.; Sun, H.; Ma, M.; Lin, L.; Shi, Y.; Li, Y.; Li, Y.; et al. Nicotinamide phosphoribosyltransferase postpones rat bone marrow mesenchymal stem cell senescence by mediating NAD+–Sirt1 signaling. Aging 2019, 11, 3505–3522. [Google Scholar] [CrossRef]
- Mendelsohn, A.R.; Larrick, J.W. The NAD+/PARP1/SIRT1 Axis in Aging. Rejuvenation Res. 2017, 20, 244–247. [Google Scholar] [CrossRef] [PubMed]
- Liang, F.; Chen, R.; Nakagawa, A.; Nishizawa, M.; Tsuda, S.; Wang, H.; Koya, D. Low-Frequency Electroacupuncture Improves Insulin Sensitivity in Obese Diabetic Mice through Activation of SIRT1/PGC-1αin Skeletal Muscle. Evid.-Based Complement. Altern. Med. 2011, 2011, 735297. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; An, H.J.; Kim, D.H.; Lee, B.; Lee, H.J.; Ullah, S.; Kim, S.J.; Jeong, H.O.; Moon, K.M.; Lee, E.K.; et al. Novel SIRT1 activator MHY2233 improves glucose tolerance and reduces hepatic lipid accumulation in db/db mice. Bioorgan. Med. Chem. Lett. 2018, 28, 684–688. [Google Scholar] [CrossRef]
- Yoshizaki, T.; Milne, J.C.; Imamura, T.; Schenk, S.; Sonoda, N.; Babendure, J.L.; Lu, J.-C.; Smith, J.J.; Jirousek, M.R.; Olefsky, J.M. SIRT1 Exerts Anti-Inflammatory Effects and Improves Insulin Sensitivity in Adipocytes. Mol. Cell. Biol. 2009, 29, 1363–1374. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.-Y.; Kao, C.-L.; Ma, H.-I.; Hung, C.-H.; Wang, C.-T.; Liu, D.-H.; Chen, P.-Y.; Tsai, K.-L. SIRT1-related inhibition of pro-inflammatory responses and oxidative stress are involved in the mechanism of nonspecific low back pain relief after exercise through modulation of Toll-like receptor 4. J. Biochem. 2015, 158, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Xu, J.; Xiong, X.; Deng, Y. Salidroside inhibits MAPK, NF-κB, and STAT3 pathways in psoriasis-associated oxidative stress via SIRT1 activation. Redox Rep. 2019, 24, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Hori, Y.S.; Kuno, A.; Hosoda, R.; Horio, Y. Regulation of FOXOs and p53 by SIRT1 Modulators under Oxidative Stress. PLoS ONE 2013, 8, e73875. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Yao, Z.; Zhang, S.; Zhang, W.; Zhou, W. Upregulation of SIRT1 inhibits H2O2-induced osteoblast apoptosis via FoxO1/β-catenin pathway. Mol. Med. Rep. 2018, 17, 6681–6690. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, S.; Zhao, D.; Zhang, X.; Xia, C.; Wang, T.; Zhang, M.; Liu, T.; Huang, W.; Wu, B. SIRT1 inhibits apoptosis in in vivo and in vitro models of spinal cord injury via microRNA-494. Int. J. Mol. Med. 2019, 43, 1758–1768. [Google Scholar] [CrossRef]
- Ren, R.; Wang, Z.; Wu, M.; Wang, H. Emerging Roles of SIRT1 in Alcoholic Liver Disease. Int. J. Biol. Sci. 2020, 16, 3174–3183. [Google Scholar] [CrossRef]
- Yu, M.; Zhang, H.; Wang, B.; Zhang, Y.; Zheng, X.; Shao, B.; Zhuge, Q.; Jin, K. Key Signaling Pathways in Aging and Potential Interventions for Healthy Aging. Cells 2021, 10, 660. [Google Scholar] [CrossRef] [PubMed]
- Warner, H.R. The Future of Aging Interventions: Current Status of Efforts to Measure and Modulate the Biological Rate of Aging. J. Gerontol. Ser. A Boil. Sci. Med Sci. 2004, 59, B692–B696. [Google Scholar] [CrossRef] [PubMed]
- Jylhävä, J.; Pedersen, N.L.; Hägg, S. Biological Age Predictors. eBioMedicine 2017, 21, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Hannum, G.; Guinney, J.; Zhao, L.; Zhang, L.; Hughes, G.; Sadda, S.; Klotzle, B.; Bibikova, M.; Fan, J.-B.; Gao, Y.; et al. Genome-wide Methylation Profiles Reveal Quantitative Views of Human Aging Rates. Mol. Cell 2012, 49, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 2013, 14, R115. [Google Scholar] [CrossRef] [PubMed]
- Levine, M.E.; Lu, A.T.; Quach, A.; Chen, B.H.; Assimes, T.L.; Bandinelli, S.; Hou, L.; Baccarelli, A.A.; Stewart, J.D.; Li, Y.; et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging 2018, 10, 573–591. [Google Scholar] [CrossRef]
- Lu, A.T.; Quach, A.; Wilson, J.G.; Reiner, A.P.; Aviv, A.; Raj, K.; Hou, L.; Baccarelli, A.A.; Li, Y.; Stewart, J.D.; et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging 2019, 11, 303–327. [Google Scholar] [CrossRef]
- Fransquet, P.D.; Wrigglesworth, J.; Woods, R.L.; Ernst, M.E.; Ryan, J. The epigenetic clock as a predictor of disease and mortality risk: A systematic review and meta-analysis. Clin. Epigenet. 2019, 11, 62. [Google Scholar] [CrossRef]
- Dugué, P.-A.; Bassett, J.K.; Joo, J.E.; Jung, C.-H.; Wong, E.M.; Moreno-Betancur, M.; Schmidt, D.; Makalic, E.; Li, S.; Severi, G.; et al. DNA methylation-based biological aging and cancer risk and survival: Pooled analysis of seven prospective studies. Int. J. Cancer 2017, 142, 1611–1619. [Google Scholar] [CrossRef]
- Zheng, C.; Li, L.; Xu, R. Association of Epigenetic Clock with Consensus Molecular Subtypes and Overall Survival of Colorectal Cancer. Cancer Epidemiol. Biomark. Prev. 2019, 28, 1720–1724. [Google Scholar] [CrossRef]
- Ni, Z.; Ebata, A.; Alipanahiramandi, E.; Lee, S.S. Two SET domain containing genes link epigenetic changes and aging in Caenorhabditis elegans. Aging Cell 2011, 11, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Greer, E.L.; Maures, T.J.; Hauswirth, A.G.; Green, E.M.; Leeman, D.S.; Maro, G.S.; Han, S.; Banko, M.R.; Gozani, O.; Brunet, A. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature 2010, 466, 383–387. [Google Scholar] [CrossRef] [PubMed]
- Lionaki, E.; Markaki, M.; Tavernarakis, N. Autophagy and ageing: Insights from invertebrate model organisms. Ageing Res. Rev. 2013, 12, 413–428. [Google Scholar] [CrossRef] [PubMed]
- Wysocki, R.; Javaheri, A.; Allard, S.; Sha, F.; Cote, J.; Kron, S.J. Role of Dot1-Dependent Histone H3 Methylation in G 1 and S Phase DNA Damage Checkpoint Functions of Rad9. Mol. Cell. Biol. 2005, 25, 8430–8443. [Google Scholar] [CrossRef] [PubMed]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.C.A.; Choate, L.A.; Rice, E.J.; Miller, D.C.; Chu, T.; Chou, S.P.; Kingsley, N.B.; Petersen, J.L.; Finno, C.J.; Bellone, R.R. Accurate imputation of histone modifications using transcription. bioRxiv 2020. [Google Scholar] [CrossRef]
- Downey, M.; Baetz, K. Building a KATalogue of acetyllysine targeting and function. Brief. Funct. Genom. 2015, 15, 109–118. [Google Scholar] [CrossRef]
- Benayoun, B.; Pollina, E.A.; Brunet, A. Epigenetic regulation of ageing: Linking environmental inputs to genomic stability. Nat. Rev. Mol. Cell Biol. 2015, 16, 593–610. [Google Scholar] [CrossRef]
- Peleg, S.; Feller, C.; Ladurner, A.; Imhof, A. The Metabolic Impact on Histone Acetylation and Transcription in Ageing. Trends Biochem. Sci. 2016, 41, 700–711. [Google Scholar] [CrossRef]
- Soukas, A.A.; Hao, H.; Wu, L. Metformin as Anti-Aging Therapy: Is It for Everyone? Trends Endocrinol. Metab. 2019, 30, 745–755. [Google Scholar] [CrossRef]
- DiMauro, T. Ras-Induced Senescence and its Physiological Relevance in Cancer. Curr. Cancer Drug Targets 2010, 10, 869–876. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tollefsbol, T.O. p16INK4a Suppression by Glucose Restriction Contributes to Human Cellular Lifespan Extension through SIRT1-Mediated Epigenetic and Genetic Mechanisms. PLoS ONE 2011, 6, e17421. [Google Scholar] [CrossRef] [PubMed]
- Everitt, A.V.; Le Couteur, D. Life Extension by Calorie Restriction in Humans. Ann. N. Y. Acad. Sci. 2007, 1114, 428–433. [Google Scholar] [CrossRef] [PubMed]
- Giacomello, E.; Toniolo, L. The Potential of Calorie Restriction and Calorie Restriction Mimetics in Delaying Aging: Focus on Experimental Models. Nutrients 2021, 13, 2346. [Google Scholar] [CrossRef] [PubMed]
- Freitas, A.; de Magalhaes, J.P. A review and appraisal of the DNA damage theory of ageing. Mutat. Res. Mutat. Res. 2011, 728, 12–22. [Google Scholar] [CrossRef]
- Roca-Bayerri, C.; Robertson, F.; Pyle, A.; Hudson, G.; Payne, B.A.I. Mitochondrial DNA Damage and Brain Aging in Human Immunodeficiency Virus. Clin. Infect. Dis. 2020, 73, e466–e473. [Google Scholar] [CrossRef]
- Vijg, J. From DNA damage to mutations: All roads lead to aging. Ageing Res. Rev. 2021, 68, 101316. [Google Scholar] [CrossRef]
- Oberdoerffer, P.; Michan, S.; McVay, M.; Mostoslavsky, R.; Vann, J.; Park, S.-K.; Hartlerode, A.; Stegmuller, J.; Hafner, A.; Loerch, P.; et al. SIRT1 Redistribution on Chromatin Promotes Genomic Stability but Alters Gene Expression during Aging. Cell 2008, 135, 907–918. [Google Scholar] [CrossRef]
- Kajitani, G.S.; Nascimento, L.L.D.S.; Neves, M.R.D.C.; Leandro, G.D.S.; Garcia, C.C.M.; Menck, C.F.M. Transcription blockage by DNA damage in nucleotide excision repair-related neurological dysfunctions. Semin. Cell Dev. Biol. 2020, 114, 20–35. [Google Scholar] [CrossRef]
- Roos, W.P.; Kaina, B. DNA damage-induced cell death: From specific DNA lesions to the DNA damage response and apoptosis. Cancer Lett. 2013, 332, 237–248. [Google Scholar] [CrossRef]
- Cheng, C.-H.; Ma, H.-L.; Deng, Y.-Q.; Feng, J.; Jie, Y.-K.; Guo, Z.-X. Oxidative stress, cell cycle arrest, DNA damage and apoptosis in the mud crab (Scylla paramamosain) induced by cadmium exposure. Chemosphere 2020, 263, 128277. [Google Scholar] [CrossRef] [PubMed]
- Sadoughi, F.; Hallajzadeh, J.; Asemi, Z.; Mansournia, M.A.; Alemi, F.; Yousefi, B. Signaling pathways involved in cell cycle arrest during the DNA breaks. DNA Repair 2021, 98, 103047. [Google Scholar] [CrossRef] [PubMed]
- Jansma, M.; Hopfner, K.-P. Structural basis of the (in)activity of the apical DNA damage response kinases ATM, ATR and DNA-PKcs. Prog. Biophys. Mol. Biol. 2020, 163, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.M.; Zhang, X. Roles of ATM and ATR in DNA double strand breaks and replication stress. Prog. Biophys. Mol. Biol. 2020, 161, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Shibata, A.; Jeggo, P.A. ATM’s Role in the Repair of DNA Double-Strand Breaks. Genes 2021, 12, 1370. [Google Scholar] [CrossRef] [PubMed]
- Eliezer, Y.; Argaman, L.; Rhie, A.; Doherty, A.J.; Goldberg, M. The Direct Interaction between 53BP1 and MDC1 Is Required for the Recruitment of 53BP1 to Sites of Damage. J. Biol. Chem. 2009, 284, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Day, M.; Oliver, A.W.; Pearl, L.H. Phosphorylation-dependent assembly of DNA damage response systems and the central roles of TOPBP1. DNA Repair 2021, 108, 103232. [Google Scholar] [CrossRef]
- Bagge, J.; Oestergaard, V.H.; Lisby, M. Functions of TopBP1 in preserving genome integrity during mitosis. Semin. Cell Dev. Biol. 2020, 113, 57–64. [Google Scholar] [CrossRef]
- Shieh, S.Y.; Ahn, J.; Tamai, K.; Taya, Y.; Prives, C. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phos-phorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 2000, 14, 289–300. [Google Scholar] [CrossRef]
- Jiang, S.; Deng, N.; Zheng, B.; Li, T.; Liu, R.H. Rhodiola extract promotes longevity and stress resistance of Caenorhabditis elegans via DAF-16 and SKN-1. Food Funct. 2021, 12, 4471–4483. [Google Scholar] [CrossRef]
- Geden, M.J.; Romero, S.E.; Deshmukh, M. p53 is required for nuclear but not mitochondrial DNA damage-induced degeneration. Cell Death Dis. 2021, 12, 104. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Xu, Y. p53, oxidative stress, and aging. Antioxid. Redox Signal. 2011, 15, 1669–1678. [Google Scholar] [CrossRef] [PubMed]
- Kawanishi, S.; Hiraku, Y.; Oikawa, S. Mechanism of guanine-specific DNA damage by oxidative stress and its role in carcinogenesis and aging. Mutat. Res. Mutat. Res. 2001, 488, 65–76. [Google Scholar] [CrossRef]
- Valdiglesias, V.; Bonassi, S.; Dell’Armi, V.; Settanni, S.; Celi, M.; Mastropaolo, S.; Antocicco, M.; Fini, M.; Onder, G. Micronucleus frequency in peripheral blood lymphocytes and frailty status in elderly. A lack of association with clinical features. Mutat. Res. Mol. Mech. Mutagen. 2015, 780, 47–54. [Google Scholar] [CrossRef]
- Thomas, P.; Fenech, M. A review of genome mutation and Alzheimer’s disease. Mutagenesis 2006, 22, 15–33. [Google Scholar] [CrossRef][Green Version]
- Coppedè, F.; Migliore, L. DNA repair in premature aging disorders and neurodegeneration. Curr. Aging Sci. 2010, 3, 3–19. [Google Scholar] [CrossRef]
- Migliore, L.; Coppedè, F.; Fenech, M.; Thomas, P. Association of micronucleus frequency with neurodegenerative diseases. Mutagenesis 2010, 26, 85–92. [Google Scholar] [CrossRef]
- Weng, N.-P. Aging of the Immune System: How Much Can the Adaptive Immune System Adapt? Immunity 2006, 24, 495–499. [Google Scholar] [CrossRef]
- Reed, J.R.; Vukmanovic-Stejic, M.; Fletcher, J.M.; Soares, M.V.D.; Cook, J.E.; Orteu, C.H.; Jackson, S.E.; Birch, K.E.; Foster, G.R.; Salmon, M.; et al. Telomere Erosion in Memory T Cells Induced by Telomerase Inhibition at the Site of Antigenic Challenge In Vivo. J. Exp. Med. 2004, 199, 1433–1443. [Google Scholar] [CrossRef]
- Liu, F.; Chang, H.-C. Physiological links of circadian clock and biological clock of aging. Protein Cell 2017, 8, 477–488. [Google Scholar] [CrossRef]
- Kucia, M.; Ratajczak, M.Z. Plausible Links Between Metabolic Networks, Stem Cells, and Longevity. Stem Cells 2019, 1201, 355–388. [Google Scholar] [CrossRef]
- Li, Y.; Ma, Z.; Jiang, S.; Hu, W.; Li, T.; Di, S.; Wang, D.; Yang, Y. A global perspective on FOXO1 in lipid metabolism and lipid-related diseases. Prog. Lipid Res. 2017, 66, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Graves, D.T.; Milovanova, T.N. Mucosal Immunity and the FOXO1 Transcription Factors. Front. Immunol. 2019, 10, 2530. [Google Scholar] [CrossRef] [PubMed]
- Delpoux, A.; Marcel, N.; Michelini, R.H.; Katayama, C.D.; Allison, K.A.; Glass, C.K.; Quiñones-Parra, S.M.; Murre, C.; Loh, L.; Kedzierska, K.; et al. FOXO1 constrains activation and regulates senescence in CD8 T cells. Cell Rep. 2021, 34, 108674. [Google Scholar] [CrossRef] [PubMed]
- Hannenhalli, S.; Kaestner, K.H. The evolution of Fox genes and their role in development and disease. Nat. Rev. Genet. 2009, 10, 233–240. [Google Scholar] [CrossRef]
- Zečić, A.; Braeckman, B.P. DAF-16/FoxO in Caenorhabditis elegans and Its Role in Metabolic Remodeling. Cells 2020, 9, 109. [Google Scholar] [CrossRef]
- Menu, E.; Jernberg-Wiklund, H.; Stromberg, T.; De Raeve, H.; Girnita, L.; Larsson, O.; Axelson, M.; Asosingh, K.; Nilsson, K.; Van Camp, B.; et al. Inhibiting the IGF-1 receptor tyrosine kinase with the cyclolignan PPP: An in vitro and in vivo study in the 5T33MM mouse model. Blood 2006, 107, 655–660. [Google Scholar] [CrossRef]
- Zemva, J.; Schubert, M. The role of neuronal insulin/insulin-like growth factor-1 signaling for the pathogenesis of Alzheimer’s disease: Possible therapeutic implications. CNS Neurol. Disord. Drug Targets 2014, 13, 322–337. [Google Scholar] [CrossRef]
- Libina, N.; Berman, J.R.; Kenyon, C. Tissue-Specific Activities of C. elegans DAF-16 in the Regulation of Lifespan. Cell 2003, 115, 489–502. [Google Scholar] [CrossRef]
- Dimov, I.; Maduro, M.F. The C. elegans intestine: Organogenesis, digestion, and physiology. Cell Tissue Res. 2019, 377, 383–396. [Google Scholar] [CrossRef]
- Lin, X.-X.; Sen, I.; Janssens, G.E.; Zhou, X.; Fonslow, B.R.; Edgar, D.; Stroustrup, N.; Swoboda, P.; Yates, J.R.; Ruvkun, G.; et al. DAF-16/FOXO and HLH-30/TFEB function as combinatorial transcription factors to promote stress resistance and longevity. Nat. Commun. 2018, 9, 4400. [Google Scholar] [CrossRef] [PubMed]
- Sen, I.; Zhou, X.; Chernobrovkin, A.; Puerta-Cavanzo, N.; Kanno, T.; Salignon, J.; Stoehr, A.; Lin, X.-X.; Baskaner, B.; Brandenburg, S.; et al. DAF-16/FOXO requires Protein Phosphatase 4 to initiate transcription of stress resistance and longevity promoting genes. Nat. Commun. 2020, 11, 138. [Google Scholar] [CrossRef] [PubMed]
- Schuster, E.; McElwee, J.J.; Tullet, J.; Doonan, R.; Matthijssens, F.; Reece-Hoyes, J.S.; Hope, I.; Vanfleteren, J.R.; Thornton, J.; Gems, D. DamID in C. elegans reveals longevity-associated targets of DAF-16/FoxO. Mol. Syst. Biol. 2010, 6, 399. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Dai, Y.; Tang, H.; Pang, S. SKN-1 Is a Negative Regulator of DAF-16 and Somatic Stress Resistance in Caenorhabditis elegans. G3: Genes Genomes Genet. 2020, 10, 1707–1712. [Google Scholar] [CrossRef] [PubMed]
- Li, W.-J.; Wang, C.-W.; Tao, L.; Yan, Y.-H.; Zhang, M.-J.; Liu, Z.-X.; Li, Y.-X.; Zhao, H.-Q.; Li, X.-M.; He, X.-D.; et al. Insulin signaling regulates longevity through protein phosphorylation in Caenorhabditis elegans. Nat. Commun. 2021, 12, 4568. [Google Scholar] [CrossRef]
- Zhang, C.; Seong, K.M.; Sun, W.; Mittapalli, O.; Qiu, B.; Clark, J.M.; Pittendrigh, B.R. The insulin signaling pathway in Drosophila melanogaster: A nexus revealing an “Achilles’ heel” in DDT resistance. Pestic. Biochem. Physiol. 2020, 171, 104727. [Google Scholar] [CrossRef]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976, Erratum in Cell 2017, 169, 361–371. [Google Scholar] [CrossRef]
- Burkart, G.M.; Brandizzi, F. A Tour of TOR Complex Signaling in Plants. Trends Biochem. Sci. 2020, 46, 417–428. [Google Scholar] [CrossRef]
- Wu, Y.; Shi, L.; Li, L.; Fu, L.; Liu, Y.; Xiong, Y.; Sheen, J. Integration of nutrient, energy, light, and hormone signalling via TOR in plants. J. Exp. Bot. 2019, 70, 2227–2238. [Google Scholar] [CrossRef]
- Beauchamp, E.M.; Platanias, L.C. The evolution of the TOR pathway and its role in cancer. Oncogene 2012, 32, 3923–3932. [Google Scholar] [CrossRef]
- Lushchak, O.; Strilbytska, O.; Piskovatska, V.; Storey, K.B.; Koliada, A.; Vaiserman, A. The role of the TOR pathway in mediating the link between nutrition and longevity. Mech. Ageing Dev. 2017, 164, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Daneshgar, N.; Rabinovitch, P.; Dai, D.-F. TOR Signaling Pathway in Cardiac Aging and Heart Failure. Biomolecules 2021, 11, 168. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Troya, S.; Pérez-Pérez, M.E.; Pérez-Martín, M.; Moes, S.; Jeno, P.; Florencio, F.J.; Crespo, J.L. Inhibition of Protein Synthesis by TOR Inactivation Revealed a Conserved Regulatory Mechanism of the BiP Chaperone in Chlamydomonas. Plant Physiol. 2011, 157, 730–741. [Google Scholar] [CrossRef] [PubMed]
- Suginohara, T.; Wakabayashi, K.; Ato, S.; Ogasawara, R. Effect of 2-deoxyglucose-mediated inhibition of glycolysis on the regulation of mTOR signaling and protein synthesis before and after high-intensity muscle contraction. Metabolism 2020, 114, 154419. [Google Scholar] [CrossRef]
- Wu, G.; Yan, Q.; Jones, J.A.; Tang, Y.J.; Fong, S.S.; Koffas, M.A. Metabolic Burden: Cornerstones in Synthetic Biology and Metabolic Engineering Applications. Trends Biotechnol. 2016, 34, 652–664. [Google Scholar] [CrossRef]
- Yee, Z.; Lim, S.H.Y.; Ng, L.F.; Gruber, J. Inhibition of mTOR decreases insoluble proteins burden by reducing translation in C. elegans. Biogerontology 2020, 22, 101–118. [Google Scholar] [CrossRef]
- Kenyon, C.J. The genetics of ageing. Nature 2010, 464, 504–512. [Google Scholar] [CrossRef]
- Robida-Stubbs, S.; Glover-Cutter, K.; Lamming, D.W.; Mizunuma, M.; Narasimhan, S.D.; Neumann-Haefelin, E.; Sabatini, D.M.; Blackwell, T.K. TOR Signaling and Rapamycin Influence Longevity by Regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab. 2012, 15, 713–724. [Google Scholar] [CrossRef]
- Niu, W.; Lu, Z.J.; Zhong, M.; Sarov, M.; Murray, J.I.; Brdlik, C.M.; Janette, J.; Chen, C.; Alves, P.; Preston, E.; et al. Diverse transcription factor binding features revealed by genome-wide ChIP-seq in C. elegans. Genome Res. 2010, 21, 245–254. [Google Scholar] [CrossRef]
- Somogyvari, M.; Gecse, E.; Sőti, C. DAF-21/Hsp90 is required for C. elegans longevity by ensuring DAF-16/FOXO isoform A function. Sci. Rep. 2018, 8, 12048. [Google Scholar] [CrossRef]
- Oh, S.W.; Mukhopadhyay, A.; Svrzikapa, N.; Jiang, F.; Davis, R.J.; Tissenbaum, H.A. JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proc. Natl. Acad. Sci. USA 2005, 102, 4494–4499. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Zhang, H.; Lin, Y.; Li, J.; Pober, J.S.; Min, W. RIP1-mediated AIP1 Phosphorylation at a 14-3-3-binding Site Is Critical for Tumor Necrosis Factor-induced ASK1-JNK/p38 Activation. J. Biol. Chem. 2007, 282, 14788–14796. [Google Scholar] [CrossRef] [PubMed]
- Soh, R.; Hardy, A.; Nieden, N.I.Z. The FOXO signaling axis displays conjoined functions in redox homeostasis and stemness. Free. Radic. Biol. Med. 2021, 169, 224–237. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.H.; Sasaki, T.; Chao, M.V. JNK-interacting Protein 1 Promotes Akt1 Activation. J. Biol. Chem. 2003, 278, 29830–29836. [Google Scholar] [CrossRef]
- Salminen, A.; Kaarniranta, K. Genetics vs. entropy: Longevity factors suppress the NF-κB-driven entropic aging process. Ageing Res. Rev. 2010, 9, 298–314. [Google Scholar] [CrossRef]
- Paillasse, M.R.; de Medina, P. The NR4A nuclear receptors as potential targets for anti-aging interventions. Med. Hypotheses 2015, 84, 135–140. [Google Scholar] [CrossRef]
- Argyrofthalmidou, M.; Spathis, A.D.; Maniati, M.; Poula, A.; Katsianou, M.A.; Sotiriou, E.; Manousaki, M.; Perier, C.; Papapanagiotou, I.; Papadopoulou-Daifoti, Z.; et al. Nurr1 repression mediates cardinal features of Parkinson’s disease in α-synuclein transgenic mice. Hum. Mol. Genet. 2021, 30, 1469–1483. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).