Simultaneous Improvement of Grain Yield and Quality through Manipulating Two Type C G Protein Gamma Subunits in Rice
Abstract
:1. Introduction
2. Results
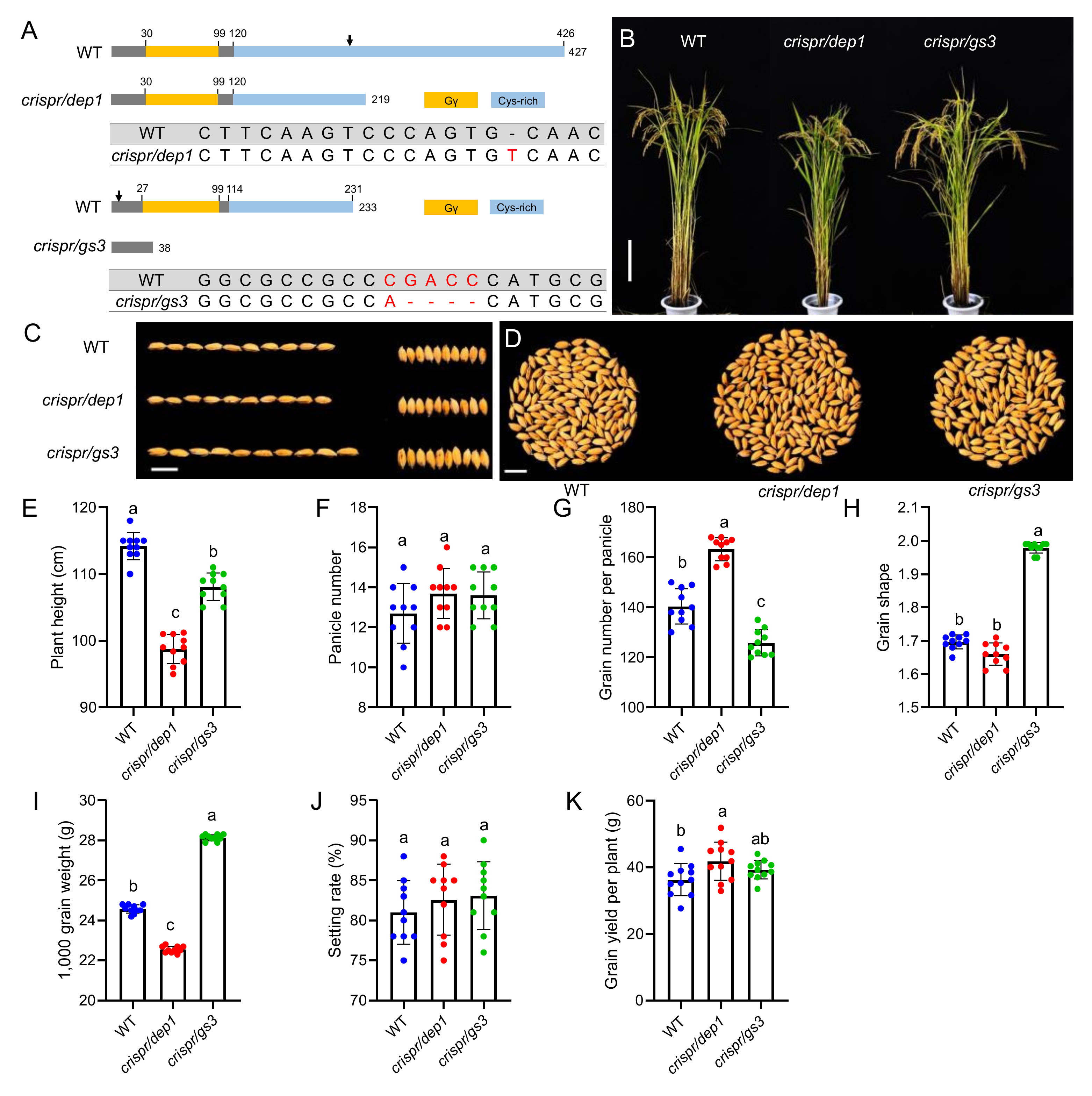
2.1. Construction of Two Type C G Protein Gamma Subunit Mutants in Rice
2.2. The Grain Changes of DEP1 and GS3 Mutants
2.3. The Quality Traits of WT and the DEP1 and GS3 Mutants
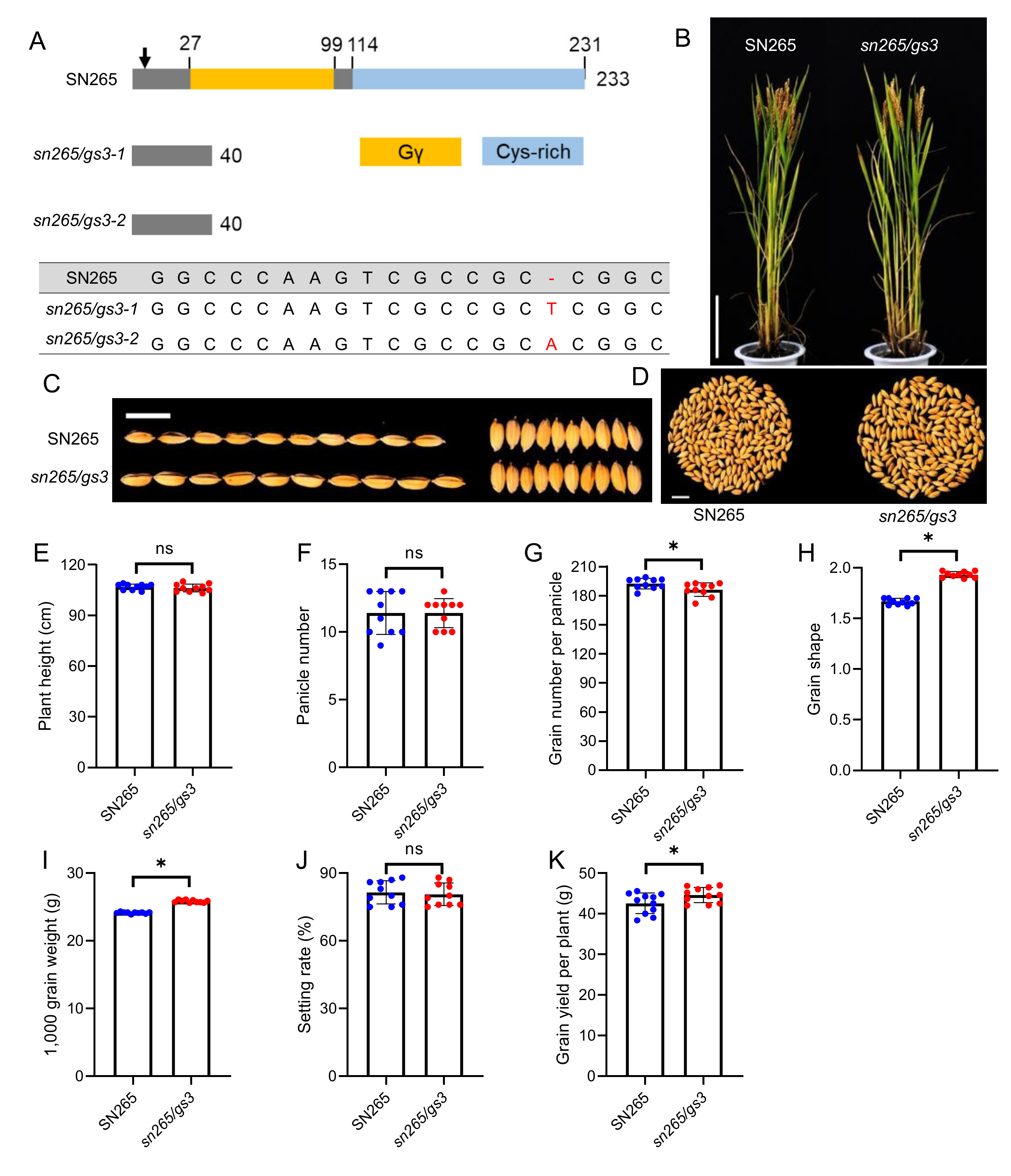
2.4. Molecular Design Breeding of a “Super Rice” Variety
2.5. The Quality Traits of the Improved Variety
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Vector Construction and Plant Transformation
4.3. Microscopy Observations
4.4. Quality Traits Measurement
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Muthayya, S.; Sugimoto, J.D.; Montgomery, S.; Maberly, G.F. An overview of global rice production, supply, trade, and consumption. Ann. N. Y. Acad. Sci. 2014, 1324, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Fei, C.; Xu, Q.; Xu, Z.; Chen, W. Effect of Rice Breeding Process on Improvement of Yield and Quality in China. Rice Sci. 2020, 27, 363–367. [Google Scholar]
- Urano, D.; Miura, K.; Wu, Q.; Iwasaki, Y.; Jackson, D.; Jones, A.M. Plant Morphology of Heterotrimeric G protein Mutants. Plant Cell Physiol. 2016, 57, 437–445. [Google Scholar] [CrossRef]
- Temple, B.R.; Jones, A.M. The plant heterotrimeric G-protein complex. Annu. Rev. Plant Biol. 2007, 58, 249–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, A.M.; Assmann, S.M. Plants: The latest model system for G-protein research. EMBO Rep. 2004, 5, 572–578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Q.; Zhao, M.; Wu, K.; Fu, X.; Liu, Q. Emerging Insights into Heterotrimeric G Protein Signaling in Plants. J. Genet. Genom. 2016, 43, 495–502. [Google Scholar] [CrossRef]
- Ferrero-Serrano, A.; Assmann, S.M. The alpha-subunit of the rice heterotrimeric G protein, RGA1, regulates drought tolerance during the vegetative phase in the dwarf rice mutant d1. J. Exp. Bot. 2016, 67, 3433–3443. [Google Scholar] [CrossRef] [Green Version]
- Jangam, A.P.; Pathak, R.R.; Raghuram, N. Microarray Analysis of Rice d1 (RGA1) Mutant Reveals the Potential Role of G-Protein Alpha Subunit in Regulating Multiple Abiotic Stresses Such as Drought, Salinity, Heat, and Cold. Front. Plant Sci. 2016, 7, 11. [Google Scholar] [CrossRef]
- Utsunomiya, Y.; Samejima, C.; Takayanagi, Y.; Izawa, Y.; Yoshida, T.; Sawada, Y.; Fujisawa, Y.; Kato, H.; Iwasaki, Y. Suppression of the rice heterotrimeric G protein beta-subunit gene, RGB1, causes dwarfism and browning of internodes and lamina joint regions. Plant J. 2011, 67, 907–916. [Google Scholar] [CrossRef]
- Miao, J.; Yang, Z.; Zhang, D.; Wang, Y.; Xu, M.; Zhou, L.; Wang, J.; Wu, S.; Yao, Y.; Du, X.; et al. Mutation of RGG2, which encodes a type B heterotrimeric G protein gamma subunit, increases grain size and yield production in rice. Plant Biotechnol. J. 2019, 17, 650–664. [Google Scholar] [CrossRef] [Green Version]
- Trusov, Y.; Chakravorty, D.; Botella, J.R. Diversity of heterotrimeric G-protein γ subunits in plants. BMC Res. Notes 2012, 5, 608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Qian, Q.; Liu, Z.; Sun, H.; He, S.; Luo, D.; Xia, G.; Chu, C.; Li, J.; Fu, X. Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 2009, 41, 494–497. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Zhao, M.; Zhang, Q.; Xu, Z.; Xu, Q. The DENSE AND ERECT PANICLE 1 (DEP1) gene offering the potential in the breeding of high-yielding rice. Breed. Sci. 2016, 66, 659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Botella, J.R. Can heterotrimeric G proteins help to feed the world? Trends Plant Sci. 2012, 17, 563–568. [Google Scholar] [CrossRef]
- Zhao, M.; Sun, J.; Xiao, Z.; Cheng, F.; Xu, H.; Tang, L.; Chen, W.; Xu, Z.; Xu, Q. Variations in DENSE AND ERECT PANICLE 1 (DEP1) contribute to the diversity of the panicle trait in high-yielding japonica rice varieties in northern China. Breed. Sci. 2016, 66, 16058. [Google Scholar] [CrossRef] [Green Version]
- Fan, C.; Xing, Y.; Mao, H.; Lu, T.; Han, B.; Xu, C.; Li, X.; Zhang, Q. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 2006, 112, 1164–1171. [Google Scholar] [CrossRef]
- Mao, H.; Sun, S.; Yao, J.; Wang, C.; Yu, S.; Xu, C.; Li, X.; Zhang, Q. Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. USA 2010, 107, 19579–19584. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Tao, Q.; Miao, J.; Yang, Z.; Gu, M.; Liang, G.; Zhou, Y. Evaluation of differential qPE9-1/DEP1 protein domains in rice grain length and weight variation. Rice 2019, 12, 5. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Li, X.; Zhou, Z.; Wu, P.; Fang, M.; Pan, X.; Lin, Q.; Luo, W.; Wu, G.; Li, H. Reassessment of the Four Yield-related Genes Gn1a, DEP1, GS3, and IPA1 in Rice Using a CRISPR/Cas9 System. Front. Plant Sci. 2016, 7, 377. [Google Scholar] [CrossRef] [Green Version]
- Sun, S.; Wang, L.; Mao, H.; Shao, L.; Li, X.; Xiao, J.; Ouyang, Y.; Zhang, Q. A G-protein pathway determines grain size in rice. Nat. Commun. 2018, 9, 851. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Zhu, Z.; Chern, M.; Yin, J.; Yang, C.; Ran, L.; Cheng, M.; He, M.; Wang, K.; Wang, J. A Natural Allele of a Transcription Factor in Rice Confers Broad-Spectrum Blast Resistance. Cell 2017, 170, 114–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef]
- Nishimura, A.; Aichi, I.; Matsuoka, M. A protocol for Agrobacterium-mediated transformation in rice. Nat. Protoc. 2006, 1, 2796. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.F.; Li, J.X.; Yu, S.B.; Xing, Y.Z.; Xu, C.G.; Zhang, Q. The three important traits for cooking and eating quality of rice grains are controlled by a single locus in an elite rice hybrid, Shanyou 63. Theor. Appl. Genet. 1999, 99, 642–648. [Google Scholar] [CrossRef]
- Zhang, C.-Q.; HU, B.; Zhu, K.-Z.; Zhang, H.; Leng, Y.-L.; Tang, S.-Z.; Gu, M.-H.; Liu, Q.-Q. QTL Mapping for Rice RVA Properties Using High-Throughput Re-sequenced Chromosome Segment Substitution Lines. Rice Sci. 2013, 20, 407–414. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, L.; Wang, X.; Yu, Z.; Cui, X.; Xu, Q. Simultaneous Improvement of Grain Yield and Quality through Manipulating Two Type C G Protein Gamma Subunits in Rice. Int. J. Mol. Sci. 2022, 23, 1463. https://doi.org/10.3390/ijms23031463
Wu L, Wang X, Yu Z, Cui X, Xu Q. Simultaneous Improvement of Grain Yield and Quality through Manipulating Two Type C G Protein Gamma Subunits in Rice. International Journal of Molecular Sciences. 2022; 23(3):1463. https://doi.org/10.3390/ijms23031463
Chicago/Turabian StyleWu, Lian, Xiaodong Wang, Zhiwen Yu, Xin Cui, and Quan Xu. 2022. "Simultaneous Improvement of Grain Yield and Quality through Manipulating Two Type C G Protein Gamma Subunits in Rice" International Journal of Molecular Sciences 23, no. 3: 1463. https://doi.org/10.3390/ijms23031463
APA StyleWu, L., Wang, X., Yu, Z., Cui, X., & Xu, Q. (2022). Simultaneous Improvement of Grain Yield and Quality through Manipulating Two Type C G Protein Gamma Subunits in Rice. International Journal of Molecular Sciences, 23(3), 1463. https://doi.org/10.3390/ijms23031463





