A Systematic Review of Expression and Immunogenicity of Human Endogenous Retroviral Proteins in Cancer and Discussion of Therapeutic Approaches
Abstract
1. Introduction
2. Materials and Methods
2.1. Information Sources
2.2. Eligibility Criteria and Data Management
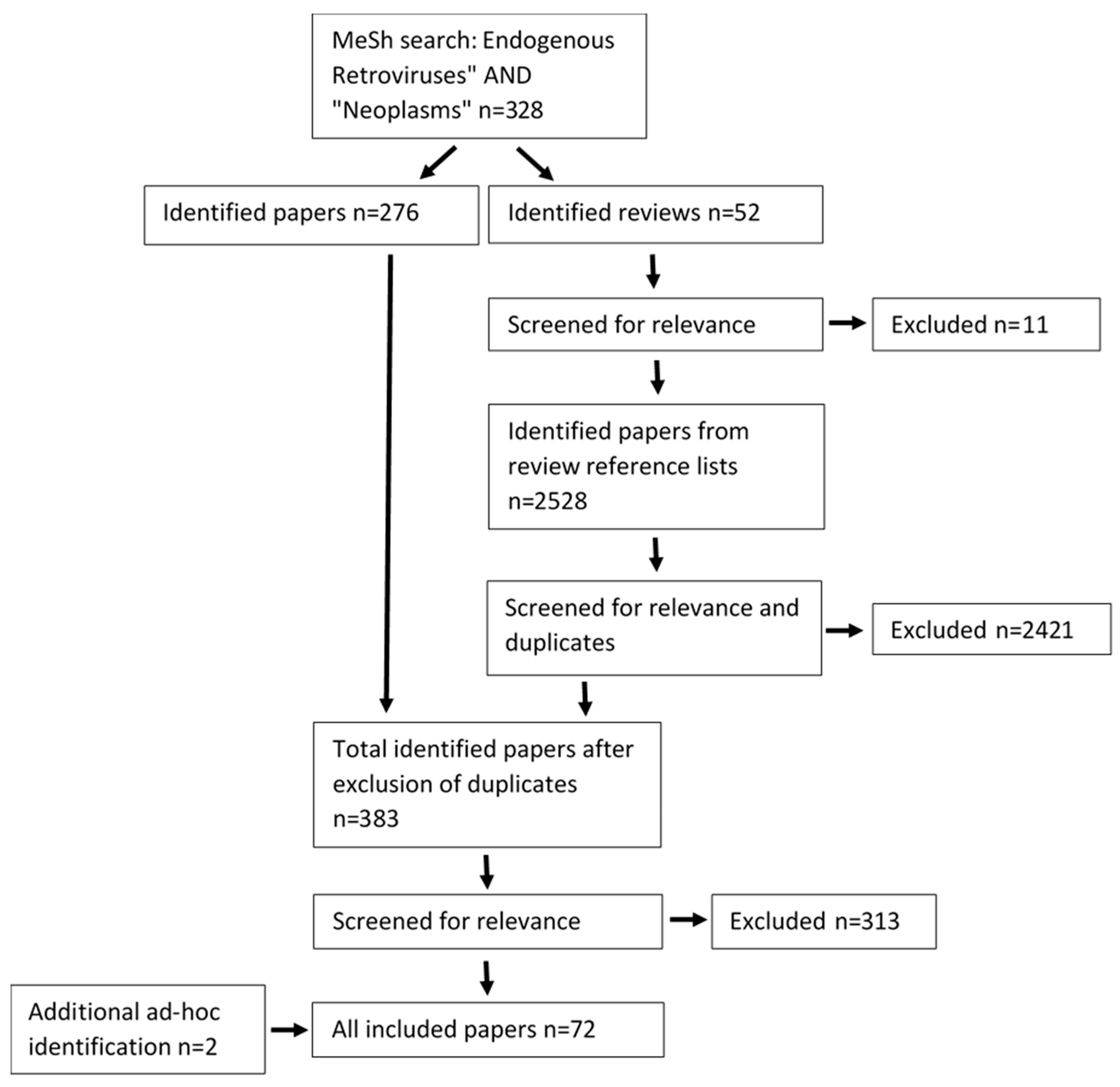
2.3. Search Results
3. Results and Discussion
3.1. Germ Cell Cancers
| Indication | HERV | Tissue | Cell Line | Controls | Case/Control | Conclusion | Reference |
|---|---|---|---|---|---|---|---|
| Teratocarcinoma | HERV-K | - | NCCIT | - | - | Expression Tumor function | [63] |
| Teratocarcinoma | HERV-K | - | Tera-1 | - | - | Expression Tumor function | [64] |
| Teratocarcinoma | HERV-K | - | Tera-1, Tera-2, NCCIT, 2102Ep, PA-1 | - | - | Expression | [65] |
| Teratocarcinoma | HERV-K | Serum | GH | - | Unclear | Expression Seroreactivity | [59] |
| Teratocarcinoma | HERV-K | - | GH, Tera-2, MRC-5 | - | - | Expression | [58] |
| Teratocarcinoma | HERV-K | - | Tera-1, Tera-2 | - | - | Expression | [56] |
| Teratocarcinoma | HERV-K | Tumor | Tera-1, PA-1 | - | 8/0 | Expression Tumor function | [61] |
| Testicular cancer | HERV-K | PBMCs | - | Healthy controls | 26/18 | T cell activity | [67] |
| Testicular cancer | HERV-K | Serum Tumor | Tera-1 | - | 57/0, 8/0 | Seroreactivity | [55] |
| Multiple GCT | HERV-K | Serum | - | Serum | Unclear | Seroreactivity Prognostic | [66] |
| Multiple GCT | HERV-K | Serum | - | - | 52/84 | Seroreactivity | [60] |
| Multiple GCT | HERV-K | Serum | - | - | 49/15 | Seroreactivity | [57] |
| Unspecified GCT | HERV-K | Serum | - | - | 18/0 | Tumor function Seroreactivity | [62] |
3.2. Neurological Cancers
3.3. Skin Cancers
| Indication | HERV | Tissue | Cell Line | Controls | Case/Control | Conclusion | Reference |
|---|---|---|---|---|---|---|---|
| CTCL | HERV-W | - | Mac-1, Mac-2A, MyLa | - | - | Expression Tumor function | [80] |
| CTCL | HERV-W | Tumor | Non-malignant skin lesions | 26/5 | Expression | [79] | |
| Melanoma | HERV-H | - | Hs294T | - | - | Expression Tumor function | [81] |
| Melanoma | HERV-K | Tumor | A-375-SM | Healthy controls | 220/55 | Expression Therapeutic | [77] |
| Melanoma | HERV-K | Tumor | - | Naevi | 35/38 | Expression Tumor function | [42] |
| Melanoma | HERV-K | - | TVM-A12, TVM-A197 | - | - | Expression Tumor function | [73] |
| Melanoma | HERV-K | Serum | - | Healthy controls | 312/70 | Expression Prognostic | [76] |
| Melanoma | HERV-K | - | A-375 | - | - | Expression Tumor function | [74] |
| Melanoma | HERV-K | - | SK-MEL-28 | - | - | Expression | [72] |
| Melanoma | HERV-K | Tumor | SK-MEL-28 | - | 53/0 | Expression | [71] |
| Melanoma | HERV-K | Serum | - | Healthy controls | 81/95 | Seroreactivity | [75] |
| Melanoma | HERV-K | - | A-375 | - | - | Expression Tumor function Therapeutic | [78] |
| Melanoma | HERV-K | PBMCs | - | - | 1/0 | T cell activity | [82] |
| Melanoma | HERV-K | Primary tumor and metastases | SK-MEL-28, SK-MEL-1 | Naevi | Unclear | Expression | [70] |
3.4. Hematological Cancers
| Indication | HERV | Tissue | Cell Line | Controls | Case/Control | Conclusion | Reference |
|---|---|---|---|---|---|---|---|
| AML, MDS, CMML | HERV-K HERV-H HERV-W HERV-FRD HERV-E | PBMCs Bone marrow | - | Healthy controls | 22/27 | T cell activity | [83] |
| AML, ALL | HERV-W | - | FA-AML1, MOLT-4 | - | - | Expression Tumor function | [86] |
| AML, CML, ALL Multiple myeloma | HERV-K | Blood | K562, K562/adr, KCL-22, KCL-22M, KG-1, HL-60, NB4, Kasumi-1, Jurkat, Molt-4, H9, Raji, KM3, 8226 | Healthy controls | 50/22 | Expression Tumor function | [43] |
| CLL, CML, ALL, AML, AMLL, NHL | HERV-W | Blood | - | - | 57/20 | Expression Prognostic | [87] |
| B cell lymphoma | HERV-K | - | JVM2, REC1 | - | - | Expression | [85] |
| Large cell lymphoma, mantle cell lymphoma | HERV-K | Blood | Healthy controls | 3/3 | Expression | [84] | |
| CML, ET | HERV-K | Blood | - | Healthy controls | 2/2 | Expression | [21] |
| ALL and lymphomas | HERV-K | Serum | - | - | 81/0 227/0 | Seroreactivity | [55] |
3.5. Prostate Cancer
| Indication | HERV | Tissue | Cell Line | Controls | Case/Control | Conclusion | Reference |
|---|---|---|---|---|---|---|---|
| Adenocarcinoma | HERV-K | - | LNCaP | - | - | Expression Tumor function Therapeutics | [90] |
| Adenocarcinoma | HERV-K | Tumor Blood | CWR22, 22Rv1, PC-3 and DU145 | Healthy controls | 377/0 294/135 | Expression Diagnostic | [46] |
| Adenocarcinoma | HERV-K | - | PC-3 | - | - | Expression Tumor function Therapeutic | [78] |
| Prostate cancer | HERV-K | Tumor | - | Adjacent | 18/18 | Expression Tumor function | [88] |
| Prostate cancer | HERV-K | Serum | - | Serum | Unclear | Expression Diagnostic | [92] |
| Prostate cancer | HERV-K | Tumor Serum | Vcap, LNcap, PC-3 | Healthy controls | 188/22 483/148 284 * | Tumor function Seroreactivity Prognostic | [91] |
3.6. Mammary Cancer
| Indication | HERV | Tissue | Cell Line | Controls | Case/Control | Conclusion | Reference |
|---|---|---|---|---|---|---|---|
| DCIS and IDC | HERV-K | Tumor | MDA-MB-231, Hs578T, MCF-7, SKBR3 and 293TN | - | - | Expression Tumor function | [98] |
| DCIS and IDC | HERV-K | Serum | - | Healthy controls | 49/13 | Seroreactivity Diagnostic | [96] |
| DCIS and IDC | HERV-K | Tumor | MDA-MB-231, SKBR3, MDA-MB-453, T47D, and ZR-75-1 | Adjacent | 2/2 | Expression Therapeutic | [95] |
| DCIS and IDC | HERV-K | Tumor | - | Adjacent Healthy controls | 110/30 195/0 | Expression Prognostic | [97] |
| DCIS, IDC, IMPC, SCC | HERV-K | Tumor | Healthy controls | 119/63 | Expression T cell activity | [94] | |
| Breast cancer | HERV-K | Tumor | Adjacent | 110/85 | Expression Prognostic | [22] | |
| Adenocarcinoma | HERV-K | - | T47D | - | - | Expression Tumor function | [102] |
| Breast cancer | HERV-K | Serum | - | - | 103/0 | Seroreactivity | [55] |
3.7. Gastrointestinal Cancers
| Indication | HERV | Tissue | Cell Line | Controls | Case/Control | Conclusion | Reference |
|---|---|---|---|---|---|---|---|
| Colorectal cancer | HERV-K | - | DLD-1 and HCT116 | - | - | Expression Tumor function | [44] |
| Colorectal cancer | HERV-K HERV-W | - | Caco-2, SK-CO-1 | - | - | Expression Tumor function | [103] |
| Colorectal cancer | HERV-H | Tumor | - | - | 13/13 | Expression | [104] |
| Colorectal cancer | HERV-W HERV-FRD HERV-3 | Tumor | HCT8 | - | 20/0 | Expression Tumor function Therapeutic | [50] |
| Colorectal cancer | HERV-H | - | Colo320 and HCT116 | - | - | Expression Tumor function | [81] |
| Colorectal cancer | HERV-W | Tumor | - | - | 140/0 | Expression Prognostic | [105] |
| Colorectal cancer | HERV-K | - | HT29 | - | - | Expression Tumor function Therapeutic | [78] |
3.8. Gynecological Cancers
| Indication | HERV | Tissue | Cell Line | Controls | Case/Control | Conclusion | Reference |
|---|---|---|---|---|---|---|---|
| Endometrial carcinoma | HERV-W HERV-FRD ERV-3 | Tumor | RL95-2 | 7 adjacent/22 age matched | 38/29 | Expression Tumor function | [112] |
| Endometrial carcinoma | HERV-W | Tumor | - | 12 adjacent/12 age matched | 24/24 | Expression | [107] |
| Ovarian | HERV-W HERV-FRD HERV-V ERV-3 | Tumor | SKOV3 OVCAR | - | 10/0 | Expression Tumor function Therapeutic | [106] |
| Ovarian | HERV-K ERV-3 | Tumor | - | Healthy controls | 70/10 | Expression Tumor function | [113] |
| Ovarian | HEMO | Serum Tumor | HeLa | - | Unclear | Expression Tumor function | [109] |
| Ovarian | HERV-K | Tumor cysts | - | Adjacent | 89/89 | T cell activity eroreactivity | [99] |
| Ovarian | HERV-W ERV-3 | - | A2780 | - | - | Expression Tumor function Therapeutic | [48] |
| Ovarian | HERV-K ERV-3 | Tumor Serum | SKOV3, OVCA 430, OVCA 433, OVCA 420, OVCAR3, DOV 13 and OVCA 429, T29, T72 and T80 | Healthy controls | 553/3 20/20 | Seroreactivity | [100] |
| Choriocarcinoma | HERV-W HERV-FRD HERV-K | - | BeWo, JEG | - | - | T cell activity | [110] |
| Choriocarcinoma | HERV-W | - | JAR, JEG-3 | Normal trophoblast | - | Expression | [108] |
| Choriocarcinoma | HERV-E | - | JEG-3, JAR, BeWo, HeLa | Normal trophoblast | Expression | [111] |
3.9. Urological Cancers
| Indication | HERV | Tissue | Cell Line | Controls | Case/Control | Conclusion | Reference |
|---|---|---|---|---|---|---|---|
| Urothelial cell carcinoma | HERV-W | Tumor | - | Adjacent | 82/82 | Expression Tumor function Prognostic | [114] |
| ccRCC | HERV-K | Tumor | MZ1257RC | - | 288/0 | Expression Tumor function Prognostic | [119] |
| ccRCC | HERV-E | Tumoral T cells | - | Healthy controls | 4/4 | T cell activity | [10] |
| ccRCC | HERV-E | PBMCs | - | - | 7/0 | T cell activity Therapeutic | [117] |
| ccRCC | HERV-E | Tumor | - | - | 67/17 | Expression Tumor function | [116] |
| ccRCC | HERV-E | PBMCs | Multiple RCC cell lines | - | 1 | T cell activity | [115] |
3.10. Lung Cancers
3.11. Pancreatic Cancers
3.12. Endocrine Cancers
3.13. Sarcomas
3.14. Discussion
3.14.1. Underexplored HERVs in the Published Literature
3.14.2. Historical Context
3.14.3. Biological Function in Cancer Development and Progression
3.14.4. Immunotherapeutic Prospects
3.14.5. Antiretrovirals
3.14.6. Diagnostic and Prognostic Use of HERVs
3.15. Limitations and Bias
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bagchi, S.; Yuan, R.; Engleman, E.G. Immune Checkpoint Inhibitors for the Treatment of Cancer: Clinical Impact and Mechanisms of Response and Resistance. Annu. Rev. Pathol. 2021, 16, 223–249. [Google Scholar] [CrossRef] [PubMed]
- Robert, C. A decade of immune-checkpoint inhibitors in cancer therapy. Nat. Commun. 2020, 11, 3801. [Google Scholar] [CrossRef] [PubMed]
- Binnewies, M.; Roberts, E.W.; Kersten, K.; Chan, V.; Fearon, D.F.; Merad, M.; Coussens, L.M.; Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Hedrick, C.C.; et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018, 24, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Syn, N.L.; Teng, M.W.L.L.; Mok, T.S.K.K.; Soo, R.A. De-novo and acquired resistance to immune checkpoint targeting. Lancet Oncol. 2017, 18, e731–e741. [Google Scholar] [CrossRef]
- Snyder, A.; Makarov, V.; Merghoub, T.; Yuan, J.; Zaretsky, J.M.; Desrichard, A.; Walsh, L.A.; Postow, M.A.; Wong, P.; Ho, T.S.; et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 2014, 371, 2189–2199. [Google Scholar] [CrossRef] [PubMed]
- Spranger, S.; Dai, D.; Horton, B.; Gajewski, T.F. Tumor-Residing Batf3 Dendritic Cells Are Required for Effector T Cell Trafficking and Adoptive T Cell Therapy. Cancer Cell 2017, 31, 711–723.e4. [Google Scholar] [CrossRef]
- Gros, A.; Parkhurst, M.R.; Tran, E.; Pasetto, A.; Robbins, P.F.; Ilyas, S.; Prickett, T.D.; Gartner, J.J.; Crystal, J.S.; Roberts, I.M.; et al. Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat. Med. 2016, 22, 433–438. [Google Scholar] [CrossRef]
- Maleki Vareki, S. High and low mutational burden tumors versus immunologically hot and cold tumors and response to immune checkpoint inhibitors. J. Immunother. Cancer 2018, 6, 157. [Google Scholar] [CrossRef]
- Ficial, M.; Jegede, O.A.; Sant’Angelo, M.; Hou, Y.; Flaifel, A.; Pignon, J.-C.C.; Braun, D.A.; Wind-Rotolo, M.; Sticco-Ivins, M.A.; Catalano, P.J.; et al. Expression of T-Cell exhaustion molecules and human endogenous retroviruses as predictive biomarkers for response to nivolumab in metastatic clear cell renal cell carcinoma. Clin. Cancer Res. 2021, 27, 1371–1380. [Google Scholar] [CrossRef]
- Smith, C.C.; Beckermann, K.E.; Bortone, D.S.; De Cubas, A.A.; Bixby, L.M.; Lee, S.J.; Panda, A.; Ganesan, S.; Bhanot, G.; Wallen, E.M.; et al. Endogenous retroviral signatures predict immunotherapy response in clear cell renal cell carcinoma. J. Clin. Investig. 2018, 128, 4804–4820. [Google Scholar] [CrossRef]
- Lu, T.; Zhang, Z.; Zhu, J.; Wang, Y.; Jiang, P.; Xiao, X.; Bernatchez, C.; Heymach, J.V.; Gibbons, D.L.; Wang, J.; et al. Deep learning-based prediction of the T cell receptor–antigen binding specificity. Nat. Mach. Intell. 2021, 3, 864–875. [Google Scholar] [CrossRef]
- Szpakowski, S.; Sun, X.; Lage, J.M.; Dyer, A.; Rubinstein, J.; Kowalski, D.; Sasaki, C.; Costa, J.; Lizardi, P.M. Loss of epigenetic silencing in tumors preferentially affects primate-specific retroelements. Gene 2009, 448, 151–167. [Google Scholar] [CrossRef] [PubMed]
- Grandi, N.; Tramontano, E. Human Endogenous Retroviruses Are Ancient Acquired Elements Still Shaping Innate Immune Responses. Front. Immunol. 2018, 9, 2039. [Google Scholar] [CrossRef] [PubMed]
- Petrizzo, A.; Ragone, C.; Cavalluzzo, B.; Mauriello, A.; Manolio, C.; Tagliamonte, M.; Buonaguro, L. Human Endogenous Retrovirus Reactivation: Implications for Cancer Immunotherapy. Cancers 2021, 13, 1999. [Google Scholar] [CrossRef] [PubMed]
- Vitiello, G.A.F.; Ferreira, W.A.S.; Cordeiro de Lima, V.C.; Medina, T.d.S. Antiviral Responses in Cancer: Boosting Antitumor Immunity Through Activation of Interferon Pathway in the Tumor Microenvironment. Front. Immunol. 2021, 12, 782852. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef]
- Griffiths, D.J. Endogenous retroviruses in the human genome sequence. Genome Biol. 2001, 2, 1–5. [Google Scholar] [CrossRef]
- Contreras-Galindo, R.; Kaplan, M.H.; Dube, D.; Gonzalez-Hernandez, M.J.; Chan, S.; Meng, F.; Dai, M.; Omenn, G.S.; Gitlin, S.D.; Markovitz, D.M. Human Endogenous Retrovirus Type K (HERV-K) Particles Package and Transmit HERV-K–Related Sequences. J. Virol. 2015, 89, 7187–7201. [Google Scholar] [CrossRef]
- Vargiu, L.; Rodriguez-Tomé, P.; Sperber, G.O.; Cadeddu, M.; Grandi, N.; Blikstad, V.; Tramontano, E.; Blomberg, J. Classification and characterization of human endogenous retroviruses mosaic forms are common. Retrovirology 2016, 13, 7. [Google Scholar] [CrossRef]
- Fuchs, N.V.; Loewer, S.; Daley, G.Q.; Izsvák, Z.; Löwer, J.; Löwer, R. Human endogenous retrovirus K (HML-2) RNA and protein expression is a marker for human embryonic and induced pluripotent stem cells. Retrovirology 2013, 10, 115. [Google Scholar] [CrossRef]
- Morgan, D.; Brodsky, I. Human endogenous retrovirus (HERV-K) particles in megakaryocytes cultured from essential thrombocythemia peripheral blood stem cells. Exp. Hematol. 2004, 32, 520–525. [Google Scholar] [CrossRef] [PubMed]
- Golan, M.; Hizi, A.; Resau, J.H.; Yaal-Hahoshen, N.; Reichman, H.; Keydar, I.; Tsarfaty, I. Human Endogenous Retrovirus (HERV-K) Reverse Transcriptase as a Breast Cancer Prognostic Marker. Neoplasia 2008, 10, 521–533. [Google Scholar] [CrossRef] [PubMed]
- Bergallo, M.; Montanari, P.; Mareschi, K.; Merlino, C.; Berger, M.; Bini, I.; Daprà, V.; Galliano, I.; Fagioli, F. Expression of the pol gene of human endogenous retroviruses HERV-K and -W in leukemia patients. Arch. Virol. 2017, 162, 3639–3644. [Google Scholar] [CrossRef] [PubMed]
- Blond, J.-L.; Lavillette, D.; Cheynet, V.; Bouton, O.; Oriol, G.; Chapel-Fernandes, S.; Mandrand, B.; Mallet, F.; Cosset, F.-L. An Envelope Glycoprotein of the Human Endogenous Retrovirus HERV-W Is Expressed in the Human Placenta and Fuses Cells Expressing the Type D Mammalian Retrovirus Receptor. J. Virol. 2000, 74, 3321–3329. [Google Scholar] [CrossRef]
- Huang, Q.; Li, J.; Wang, F.; Oliver, M.T.; Tipton, T.; Gao, Y.; Jiang, S.W. Syncytin-1 modulates placental trophoblast cell proliferation by promoting G1/S transition. Cell. Signal. 2013, 25, 1027–1035. [Google Scholar] [CrossRef]
- Singh, A.M.; Dalton, S. The Cell Cycle and Myc Intersect with Mechanisms that Regulate Pluripotency and Reprogramming. Cell Stem Cell 2009, 5, 141–149. [Google Scholar] [CrossRef]
- Argaw-Denboba, A.; Balestrieri, E.; Serafino, A.; Cipriani, C.; Bucci, I.; Sorrentino, R.; Sciamanna, I.; Gambacurta, A.; Sinibaldi-Vallebona, P.; Matteucci, C. HERV-K activation is strictly required to sustain CD133+ melanoma cells with stemness features. J. Exp. Clin. Cancer Res. 2017, 36, 20. [Google Scholar] [CrossRef]
- Mikhalkevich, N.; O’Carroll, I.P.; Tkavc, R.; Lund, K.; Sukumar, G.; Dalgard, C.L.; Johnson, K.R.; Li, W.; Wang, T.; Nath, A.; et al. Response of human macrophages to gamma radiation is mediated via expression of endogenous retroviruses. PLoS Pathog. 2021, 17, e1009305. [Google Scholar] [CrossRef]
- Hurme, M.; Pawelec, G. Human endogenous retroviruses and ageing. Immun. Ageing 2021, 18, 14. [Google Scholar] [CrossRef]
- Tokuyama, M.; Gunn, B.M.; Venkataraman, A.; Kong, Y.; Kang, I.; Rakib, T.; Townsend, M.J.; Costenbader, K.H.; Alter, G.; Iwasaki, A. Antibodies against human endogenous retrovirus K102 envelope activate neutrophils in systemic lupus erythematosus. J. Exp. Med. 2021, 218, e20191766. [Google Scholar] [CrossRef]
- Talotta, R.; Atzeni, F.; Laska, M.J. The contribution of HERV-E clone 4-1 and other HERV-E members to the pathogenesis of rheumatic autoimmune diseases. APMIS 2020, 128, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Dolei, A.; Ibba, G.; Piu, C.; Serra, C. Expression of HERV genes as possible biomarker and target in neurodegenerative diseases. Int. J. Mol. Sci. 2019, 20, 3706. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Lee, M.-H.H.; Henderson, L.; Tyagi, R.; Bachani, M.; Steiner, J.; Campanac, E.; Hoffman, D.A.; Von Geldern, G.; Johnson, K.; et al. Human endogenous retrovirus-K contributes to motor neuron disease. Sci. Transl. Med. 2015, 7, 307ra153. [Google Scholar] [CrossRef] [PubMed]
- Grandi, N.; Tramontano, E. HERV Envelope Proteins: Physiological Role and Pathogenic Potential in Cancer and Autoimmunity. Front. Microbiol. 2018, 9, 462. [Google Scholar] [CrossRef] [PubMed]
- Jansz, N.; Faulkner, G.J. Endogenous retroviruses in the origins and treatment of cancer. Genome Biol. 2021, 22, 147. [Google Scholar] [CrossRef] [PubMed]
- Hurst, T.; Magiorkinis, G. Epigenetic Control of Human Endogenous Retrovirus Expression: Focus on Regulation of Long-Terminal Repeats (LTRs). Viruses 2017, 9, 130. [Google Scholar] [CrossRef] [PubMed]
- Goering, W.; Ribarska, T.; Schulz, W.A. Selective changes of retroelement expression in human prostate cancer. Carcinogenesis 2011, 32, 1484–1492. [Google Scholar] [CrossRef]
- Mareschi, K.; Montanari, P.; Rassu, M.; Galliano, I.; Daprà, V.; Adamini, A.; Castiglia, S.; Fagioli, F.; Bergallo, M. Human Endogenous Retrovirus-H and K Expression in Human Mesenchymal Stem Cells as Potential Markers of Stemness. Intervirology 2019, 62, 9–14. [Google Scholar] [CrossRef]
- Nguyen, T.D.; Davis, J.; Eugenio, R.A.; Liu, Y. Female Sex Hormones Activate Human Endogenous Retrovirus Type K Through the OCT4 Transcription Factor in T47D Breast Cancer Cells. AIDS Res. Hum. Retrovir. 2019, 35, 348–356. [Google Scholar] [CrossRef]
- Kitsou, K.; Kotanidou, A.; Paraskevis, D.; Karamitros, T.; Katzourakis, A.; Tedder, R.; Hurst, T.; Sapounas, S.; Kotsinas, A.; Gorgoulis, V.; et al. Upregulation of Human Endogenous Retroviruses in Bronchoalveolar Lavage Fluid of COVID-19 Patients. Microbiol. Spectr. 2021, 9, e01260-21. [Google Scholar] [CrossRef]
- Xiaoqian, L.; Zunpeng, L.; Liang, S.; Jie, R.; Zeming, W.; Xiaoyu, J.; Qianzhao, J.; Qianran, W.; Yanling, F.; Yusheng, C.; et al. Resurrection of human endogenous retroviruses during aging reinforces senescence. bioRxiv 2021. [Google Scholar] [CrossRef]
- Dong, J.; Huang, G.; Li, Z.; Wan, X.; Wang, Y.; Dong, J. Human endogenous retroviral K element encodes fusogenic activity in melanoma cells. J. Carcinog. 2013, 12, 5. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Chen, T.; Meng, Z.; Gan, Y.; Wang, X.; Xu, F.; Gu, Y.; Xu, X.; Tang, J.; Zhou, H.; et al. The viral oncogene Np9 acts as a critical molecular switch for co-activating β-catenin, ERK, Akt and Notch1 and promoting the growth of human leukemia stem/progenitor cells. Leukemia 2013, 27, 1469–1478. [Google Scholar] [CrossRef]
- Ko, E.-J.J.; Ock, M.-S.S.; Choi, Y.-H.H.; Iovanna, J.L.; Mun, S.; Han, K.; Kim, H.-S.S.; Cha, H.-J.J. Human Endogenous Retrovirus (HERV)-K env Gene Knockout Affects Tumorigenic Characteristics of nupr1 Gene in DLD-1 Colorectal Cancer Cells. Int. J. Mol. Sci. 2021, 22, 3941. [Google Scholar] [CrossRef] [PubMed]
- Johanning, G.L.; Malouf, G.G.; Zheng, X.; Esteva, F.J.; Weinstein, J.N.; Wang-Johanning, F.; Su, X. Expression of human endogenous retrovirus-K is strongly associated with the basal-like breast cancer phenotype. Sci. Rep. 2017, 7, 41960. [Google Scholar] [CrossRef]
- Wallace, T.A.; Downey, R.F.; Seufert, C.J.; Schetter, A.; Dorsey, T.H.; Johnson, C.A.; Goldman, R.; Loffredo, C.A.; Yan, P.; Sullivan, F.J.; et al. Elevated HERV-K mRNA expression in PBMC is associated with a prostate cancer diagnosis particularly in older men and smokers. Carcinogenesis 2014, 35, 2074–2083. [Google Scholar] [CrossRef]
- Mangeney, M.; Pothlichet, J.; Renard, M.; Ducos, B.; Heidmann, T. Endogenous Retrovirus Expression Is Required for Murine Melanoma Tumor Growth In vivo. Cancer Res. 2005, 65, 2588–2591. [Google Scholar] [CrossRef]
- Chiappinelli, K.B.; Strissel, P.L.; Desrichard, A.; Li, H.; Henke, C.; Akman, B.; Hein, A.; Rote, N.S.; Cope, L.M.; Snyder, A.; et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell 2015, 162, 974–986. [Google Scholar] [CrossRef]
- Roulois, D.; Loo Yau, H.; Singhania, R.; Wang, Y.; Danesh, A.; Shen, S.Y.; Han, H.; Liang, G.; Jones, P.A.; Pugh, T.J.; et al. DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts. Cell 2015, 162, 961–973. [Google Scholar] [CrossRef]
- Díaz-Carballo, D.; Acikelli, A.H.; Klein, J.; Jastrow, H.; Dammann, P.; Wyganowski, T.; Guemues, C.; Gustmann, S.; Bardenheuer, W.; Malak, S.; et al. Therapeutic potential of antiviral drugs targeting chemorefractory colorectal adenocarcinoma cells overexpressing endogenous retroviral elements. J. Exp. Clin. Cancer Res. 2015, 34, 81. [Google Scholar] [CrossRef]
- Kraus, B.; Fischer, K.; Sliva, K.; Schnierle, B.S. Vaccination directed against the human endogenous retrovirus-K (HERV-K) gag protein slows HERV-K gag expressing cell growth in a murine model system. Virol. J. 2014, 11, 58. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Grabski, D.F.; Hu, Y.; Sharma, M.; Rasmussen, S.K. Close to the Bedside: A Systematic Review of Endogenous Retroviruses and Their Impact in Oncology. J. Surg. Res. 2019, 240, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Mayer, J.; Blomberg, J.; Seal, R.L. A revised nomenclature for transcribed human endogenous retroviral loci. Mob. DNA 2011, 2, 7. [Google Scholar] [CrossRef] [PubMed]
- Gifford, R.J.; Blomberg, J.; Coffin, J.M.; Fan, H.; Heidmann, T.; Mayer, J.; Stoye, J.; Tristem, M.; Johnson, W.E. Nomenclature for endogenous retrovirus (ERV) loci. Retrovirology 2018, 15, 59. [Google Scholar] [CrossRef] [PubMed]
- Sauter, M.; Schommer, S.; Kremmer, E.; Remberger, K.; Dölken, G.; Lemm, I.; Buck, M.; Best, B.; Neumann-Haefelin, D.; Mueller-Lantzsch, N. Human endogenous retrovirus K10: Expression of Gag protein and detection of antibodies in patients with seminomas. J. Virol. 1995, 69, 414–421. [Google Scholar] [CrossRef]
- Bronson, D.L.; Fraley, E.E.; Fogh, J.; Kalter, S.S. Induction of retrovirus particles in human testicular tumor (Tera-1) cell cultures: An electron microscopic study. J. Natl. Cancer Inst. 1979, 63, 337–339. [Google Scholar] [CrossRef]
- Sauter, M.; Roemer, K.; Best, B.; Afting, M.; Schommer, S.; Seitz, G.; Hartmann, M.; Mueller-Lantzsch, N. Specificity of antibodies directed against Env protein of human endogenous retroviruses in patients with germ cell tumors. Cancer Res. 1996, 56, 4362–4365. [Google Scholar]
- Löwer, R.; Tönjes, R.R.; Korbmacher, C.; Kurth, R.; Löwer, J. Identification of a Rev-related protein by analysis of spliced transcripts of the human endogenous retroviruses HTDV/HERV-K. J. Virol. 1995, 69, 141–149. [Google Scholar] [CrossRef]
- Boller, K.; Janssen, O.; Schuldes, H.; Tönjes, R.R.; Kurth, R. Characterization of the antibody response specific for the human endogenous retrovirus HTDV/HERV-K. J. Virol. 1997, 71, 4581–4588. [Google Scholar] [CrossRef]
- Goedert, J.J.; Sauter, M.E.; Jacobson, L.P.; Vessella, R.L.; Hilgartner, M.W.; Leitman, S.F.; Fraser, M.C.; Mueller-Lantzsch, N.G. High prevalence of antibodies against HERV-K10 in patients with testicular cancer but not with AIDS. Cancer Epidemiol. Biomark. Prev. 1999, 8, 293–296. [Google Scholar]
- Götzinger, N.; Sauter, M.; Roemer, K.; Mueller-Lantzsch, N.; Gotzinger, N.; Sauter, M.; Roemer, K.; Mueller-Lantzsch, N. Regulation of human endogenous retrovirus-K Gag expression in teratocarcinoma cell lines and human tumours. J. Gen. Virol. 1996, 77, 2983–2990. [Google Scholar] [CrossRef] [PubMed]
- Boese, A.; Sauter, M.; Galli, U.; Best, B.; Herbst, H.; Mayer, J.; Kremmer, E.; Roemer, K.; Mueller-Lantzsch, N. Human endogenous retrovirus protein cORF supports cell transformation and associates with the promyelocytic leukemia zinc finger protein. Oncogene 2000, 19, 4328–4336. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.M.; Sapir, T.; Park, S.S.; Rual, J.F.; Contreras-Galindo, R.; Reiner, O.; Markovitz, D.M. The HERV-K accessory protein Np9 controls viability and migration of teratocarcinoma cells. PLoS ONE 2019, 14, e0212970. [Google Scholar] [CrossRef]
- Ruprecht, K.; Ferreira, H.; Flockerzi, A.; Wahl, S.; Sauter, M.; Mayer, J.; Mueller-Lantzsch, N. Human Endogenous Retrovirus Family HERV-K(HML-2) RNA Transcripts Are Selectively Packaged into Retroviral Particles Produced by the Human Germ Cell Tumor Line Tera-1 and Originate Mainly from a Provirus on Chromosome 22q11.21. J. Virol. 2008, 82, 10008–10016. [Google Scholar] [CrossRef] [PubMed]
- Bieda, K.; Hoffman, A.; Boller, K. Phenotypic heterogeneity of human endogenous retrovirus particles produced by teratocarcinoma cell lines. J. Gen. Virol. 2001, 82, 591–596. [Google Scholar] [CrossRef] [PubMed]
- Kleiman, A.; Senyuta, N.; Tryakin, A.; Sauter, M.; Karseladze, A.; Tjulandin, S.; Gurtsevitch, V.; Mueller-Lantzsch, N. HERV-K(HML-2) GAG/ENV antibodies as indicator for therapy effect in patients with germ cell tumors. Int. J. Cancer 2004, 110, 459–461. [Google Scholar] [CrossRef] [PubMed]
- Rakoff-Nahoum, S.J.; Kuebler, P.; Heymann, J.J.; Sheehy, M.E.; Ortiz, G.M.; Ogg, G.S.; Barbour, J.D.; Lenz, J.; Steinfeld, A.D.; Nixon, D.F. Detection of T Lymphocytes Specific for Human Endogenous Retrovirus K (HERV-K) in Patients with Seminoma. AIDS Res. Hum. Retroviruses 2006, 22, 52–56. [Google Scholar] [CrossRef]
- Doucet-O’Hare, T.T.; DiSanza, B.L.; DeMarino, C.; Atkinson, A.L.; Rosenblum, J.S.; Henderson, L.J.; Johnson, K.R.; Kowalak, J.; Garcia-Montojo, M.; Allen, S.J.; et al. SMARCB1 deletion in atypical teratoid rhabdoid tumors results in human endogenous retrovirus K (HML-2) expression. Sci. Rep. 2021, 11, 12893. [Google Scholar] [CrossRef]
- Maze, E.A.; Agit, B.; Reeves, S.; Hilton, D.A.; Parkinson, D.B.; Laraba, L.; Ercolano, E.; Kurian, K.M.; Hanemann, C.O.; Belshaw, R.D.; et al. Human endogenous retrovirus type K promotes proliferation and confers sensitivity to anti-retroviral drugs in Merlin-negative schwannoma and meningioma. Cancer Res. 2021, 82, 235–247. [Google Scholar] [CrossRef]
- Muster, T.; Waltenberger, A.; Grassauer, A.; Hirschl, S.; Caucig, P.; Romirer, I.; Födinger, D.; Seppele, H.; Schanab, O.; Magin-Lachmann, C.; et al. An Endogenous Retrovirus Derived from Human Melanoma Cells. Cancer Res. 2003, 63, 8735–8741. [Google Scholar]
- Büscher, K.; Trefzer, U.; Hofmann, M.; Sterry, W.; Kurth, R.; Denner, J. Expression of human endogenous retrovirus K in melanomas and melanoma cell lines. Cancer Res. 2005, 65, 4172–4180. [Google Scholar] [CrossRef]
- Büscher, K.; Hahn, S.; Hofmann, M.; Trefzer, U.; Ozel, M.; Sterry, W.; Löwer, J.; Löwer, R.; Kurth, R.; Denner, J. Expression of the human endogenous retrovirus-K transmembrane envelope, Rec and Np9 proteins in melanomas and melanoma cell lines. Melanoma Res. 2006, 16, 223–234. [Google Scholar] [CrossRef] [PubMed]
- Serafino, A.; Balestrieri, E.; Pierimarchi, P.; Matteucci, C.; Moroni, G.; Oricchio, E.; Rasi, G.; Mastino, A.; Spadafora, C.; Garaci, E.; et al. The activation of human endogenous retrovirus K (HERV-K) is implicated in melanoma cell malignant transformation. Exp. Cell Res. 2009, 315, 849–862. [Google Scholar] [CrossRef] [PubMed]
- Oricchio, E.; Sciamanna, I.; Beraldi, R.; Tolstonog, G.V.; Schumann, G.G.; Spadafora, C. Distinct roles for LINE-1 and HERV-K retroelements in cell proliferation, differentiation and tumor progression. Oncogene 2007, 26, 4226–4233. [Google Scholar] [CrossRef] [PubMed]
- Humer, J.; Waltenberger, A.; Grassauer, A.; Kurz, M.; Valencak, J.; Rapberger, R.; Hahn, S.; Löwer, R.; Wolff, K.; Bergmann, M.; et al. Identification of a melanoma marker derived from melanoma-associated endogenous retroviruses. Cancer Res. 2006, 66, 1658–1663. [Google Scholar] [CrossRef] [PubMed]
- Hahn, S.; Ugurel, S.; Hanschmann, K.M.; Strobel, H.; Tondera, C.; Schadendorf, D.; Löwer, J.; Löwer, R. Serological response to human endogenous retrovirus K in melanoma patients correlates with survival probability. AIDS Res. Hum. Retrovir. 2008, 24, 717–723. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, J.; Rabinovich, B.A.; Mi, T.; Switzer, K.C.; Olivares, S.; Maiti, S.N.; Plummer, J.B.; Singh, H.; Kumaresan, P.R.; Huls, H.M.; et al. Genetic Engineering of T Cells to Target HERV-K, an Ancient Retrovirus on Melanoma. Clin. Cancer Res. 2015, 21, 3241–3251. [Google Scholar] [CrossRef]
- Sciamanna, I.; Landriscina, M.; Pittoggi, C.; Quirino, M.; Mearelli, C.; Beraldi, R.; Mattei, E.; Serafino, A.; Cassano, A.; Sinibaldi-Vallebona, P.; et al. Inhibition of endogenous reverse transcriptase antagonizes human tumor growth. Oncogene 2005, 24, 3923–3931. [Google Scholar] [CrossRef]
- Maliniemi, P.; Vincendeau, M.; Mayer, J.; Frank, O.; Hahtola, S.; Karenko, L.; Carlsson, E.; Mallet, F.; Seifarth, W.; Leib-Mösch, C.; et al. Expression of Human Endogenous Retrovirus-W Including Syncytin-1 in Cutaneous T-Cell Lymphoma. PLoS ONE 2013, 8, 76281. [Google Scholar] [CrossRef]
- Laukkanen, K.; Saarinen, M.; Mallet, F.; Aatonen, M.; Hau, A.; Ranki, A. Cutaneous T-Cell Lymphoma (CTCL) Cell Line-Derived Extracellular Vesicles Contain HERV-W-Encoded Fusogenic Syncytin-1. J. Investig. Dermatol. 2020, 140, 1466–1469.e4. [Google Scholar] [CrossRef]
- Kudo-Saito, C.; Yura, M.; Yamamoto, R.; Kawakami, Y. Induction of immunoregulatory CD271+ cells by metastatic tumor cells that express human endogenous retrovirus H. Cancer Res. 2014, 74, 1361–1370. [Google Scholar] [CrossRef] [PubMed]
- Schiavetti, F.; Thonnard, J.; Colau, D.; Boon, T.; Coulie, P.G. A human endogenous retroviral sequence encoding an antigen recognized on melanoma by cytolytic T lymphocytes. Cancer Res. 2002, 62, 5510–5516. [Google Scholar] [PubMed]
- Saini, S.K.; Ørskov, A.D.; Bjerregaard, A.-M.M.; Unnikrishnan, A.; Holmberg-Thydén, S.; Borch, A.; Jensen, K.V.; Anande, G.; Bentzen, A.K.; Marquard, A.M.; et al. Human endogenous retroviruses form a reservoir of T cell targets in hematological cancers. Nat. Commun. 2020, 11, 5660. [Google Scholar] [CrossRef]
- Contreras-Galindo, R.; Kaplan, M.H.; Leissner, P.; Verjat, T.; Ferlenghi, I.; Bagnoli, F.; Giusti, F.; Dosik, M.H.; Hayes, D.F.; Gitlin, S.D.; et al. Human Endogenous Retrovirus K (HML-2) Elements in the Plasma of People with Lymphoma and Breast Cancer. J. Virol. 2008, 82, 9329–9336. [Google Scholar] [CrossRef] [PubMed]
- Tatkiewicz, W.; Dickie, J.; Bedford, F.; Jones, A.; Atkin, M.; Kiernan, M.; Maze, E.A.; Agit, B.; Farnham, G.; Kanapin, A.; et al. Characterising a human endogenous retrovirus(HERV)-derived tumour-associated antigen: Enriched RNA-Seq analysis of HERV-K(HML-2) in mantle cell lymphoma cell lines. Mob. DNA 2020, 11, 9. [Google Scholar] [CrossRef]
- Alqahtani, S.; Promtong, P.; Oliver, A.W.; He, X.T.; Walker, T.D.; Povey, A.; Hampson, L.; Hampson, I.N. Silver nanoparticles exhibit size-dependent differential toxicity and induce expression of syncytin-1 in FA-AML1 and MOLT-4 leukaemia cell lines. Mutagenesis 2016, 31, 695–702. [Google Scholar] [CrossRef][Green Version]
- Sun, Y.; Ouyang, D.-Y.; Pang, W.; Tu, Y.-Q.; Li, Y.-Y.; Shen, X.-M.; Tam, S.-C.; Yang, H.-Y.; Zheng, Y.-T. Expression of syncytin in leukemia and lymphoma cells. Leuk. Res. 2010, 34, 1195–1202. [Google Scholar] [CrossRef]
- Rezaei, S.D.; Hayward, J.A.; Norden, S.; Pedersen, J.; Mills, J.; Hearps, A.C.; Tachedjian, G. Herv-k gag rna and protein levels are elevated in malignant regions of the prostate in males with prostate cancer. Viruses 2021, 13, 449. [Google Scholar] [CrossRef]
- Patnala, R.; Lee, S.-H.; Dahlstrom, J.E.; Ohms, S.; Chen, L.; Dheen, S.T.; Rangasamy, D. Inhibition of LINE-1 retrotransposon-encoded reverse transcriptase modulates the expression of cell differentiation genes in breast cancer cells. Breast Cancer Res. Treat. 2014, 143, 239–253. [Google Scholar] [CrossRef]
- Ibba, G.; Piu, C.; Uleri, E.; Serra, C.; Dolei, A. Disruption by SaCas9 endonuclease of HERV-Kenv, a retroviral gene with oncogenic and neuropathogenic potential, inhibits molecules involved in cancer and amyotrophic lateral sclerosis. Viruses 2018, 10, 412. [Google Scholar] [CrossRef]
- Reis, B.S.; Jungbluth, A.A.; Frosina, D.; Holz, M.; Ritter, E.; Nakayama, E.; Ishida, T.; Obata, Y.; Carver, B.; Scher, H.; et al. Prostate cancer progression correlates with increased humoral immune response to a human endogenous retrovirus GAG protein. Clin. Cancer Res. 2013, 19, 6112–6125. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, A.; Ali, A.; Tan, S.H.; Banerjee, S.; Chen, Y.; Cullen, J.; Xavier, C.P.; Mohamed, A.A.; Ravindranath, L.; Srivastav, J.; et al. Autoantibodies against oncogenic ERG protein in prostate cancer: Potential use in diagnosis and prognosis in a panel with C-MYC, AMACR and HERV-K gag. Genes Cancer 2016, 7, 394–413. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Goering, W.; Schmitt, K.; Dostert, M.; Schaal, H.; Deenen, R.; Mayer, J.; Schulz, W.A. Human endogenous retrovirus HERV-K(HML-2) activity in prostate cancer is dominated by a few loci. Prostate 2015, 75, 1958–1971. [Google Scholar] [CrossRef] [PubMed]
- Wang-Johanning, F.; Radvanyi, L.; Rycaj, K.; Plummer, J.B.; Yan, P.; Sastry, K.J.; Piyathilake, C.J.; Hunt, K.K.; Johanning, G.L. Human Endogenous Retrovirus K Triggers an Antigen-Specific Immune Response in Breast Cancer Patients. Cancer Res. 2008, 68, 5869–5877. [Google Scholar] [CrossRef]
- Wang-Johanning, F.; Rycaj, K.; Plummer, J.B.; Li, M.; Yin, B.; Frerich, K.; Garza, J.G.; Shen, J.; Lin, K.; Yan, P.; et al. Immunotherapeutic potential of anti-human endogenous retrovirus-K envelope protein antibodies in targeting breast tumors. J. Natl. Cancer Inst. 2012, 104, 189–210. [Google Scholar] [CrossRef]
- Wang-Johanning, F.; Li, M.; Esteva, F.J.; Hess, K.R.; Yin, B.; Rycaj, K.; Plummer, J.B.; Garza, J.G.; Ambs, S.; Johanning, G.L. Human endogenous retrovirus type K antibodies and mRNA as serum biomarkers of early-stage breast cancer. Int. J. Cancer 2014, 134, 587–595. [Google Scholar] [CrossRef]
- Zhao, J.; Rycaj, K.; Geng, S.; Li, M.; Plummer, J.B.; Yin, B.; Liu, H.; Xu, X.; Zhang, Y.; Yan, Y.; et al. Expression of Human Endogenous Retrovirus Type K Envelope Protein is a Novel Candidate Prognostic Marker for Human Breast Cancer. Genes Cancer 2011, 2, 914. [Google Scholar] [CrossRef]
- Zhou, F.; Li, M.; Wei, Y.; Lin, K.; Lu, Y.; Shen, J.; Johanning, G.L.; Wang-Johanning, F. Activation of HERV-K Env protein is essential for tumorigenesis and metastasis of breast cancer cells. Oncotarget 2016, 7, 84093–84117. [Google Scholar] [CrossRef]
- Rycaj, K.; Plummer, J.B.; Yin, B.; Li, M.; Garza, J.; Radvanyi, L.; Ramondetta, L.M.; Lin, K.; Johanning, G.L.; Tang, D.G.; et al. Cytotoxicity of human endogenous retrovirus K-specific T cells toward autologous ovarian cancer cells. Clin. Cancer Res. 2015, 21, 471–483. [Google Scholar] [CrossRef]
- Wang-Johanning, F.; Liu, J.; Rycaj, K.; Huang, M.; Tsai, K.; Rosen, D.G.; Chen, D.T.; Lu, D.W.; Barnhart, K.F.; Johanning, G.L. Expression of multiple human endogenous retrovirus surface envelope proteins in ovarian cancer. Int. J. Cancer 2007, 120, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Radvanyi, L.; Yin, B.; Rycaj, K.; Li, J.; Chivukula, R.; Lin, K.; Lu, Y.; Shen, J.; Chang, D.Z.; et al. Downregulation of Human Endogenous Retrovirus Type K (HERV-K) Viral env RNA in Pancreatic Cancer Cells Decreases Cell Proliferation and Tumor Growth. Clin. Cancer Res. 2017, 23, 5892–5911. [Google Scholar] [CrossRef] [PubMed]
- Patience, C.; Simpson, G.R.; Colletta, A.A.; Welch, H.M.; Weiss, R.A.; Boyd, M.T. Human endogenous retrovirus expression and reverse transcriptase activity in the T47D mammary carcinoma cell line. J. Virol. 1996, 70, 2654–2657. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, L.; Cafora, M.; Rota, F.; Hoxha, M.; Iodice, S.; Tarantini, L.; Dolci, M.; Delbue, S.; Pistocchi, A.; Bollati, V. Extracellular vesicles released by colorectal cancer cell lines modulate innate immune response in zebrafish model: The possible role of human endogenous retroviruses. Int. J. Mol. Sci. 2019, 20, 3669. [Google Scholar] [CrossRef] [PubMed]
- Mullins, C.S.; Hühns, M.; Krohn, M.; Peters, S.; Cheynet, V.; Oriol, G.; Guillotte, M.; Ducrot, S.; Mallet, F.; Linnebacher, M. Generation, characterization and application of antibodies directed against HERV-H Gag protein in colorectal samples. PLoS ONE 2016, 11, e0153349. [Google Scholar] [CrossRef]
- Larsen, J.M.; Christensen, I.J.; Nielsen, H.J.; Hansen, U.; Bjerregaard, B.; Talts, J.F.; Larsson, L.-I. Syncytin immunoreactivity in colorectal cancer: Potential prognostic impact. Cancer Lett. 2009, 280, 44–49. [Google Scholar] [CrossRef]
- Díaz-Carballo, D.; Saka, S.; Acikelli, A.H.; Homp, E.; Erwes, J.; Demmig, R.; Klein, J.; Schröer, K.; Malak, S.; D’Souza, F.; et al. Enhanced antitumoral activity of TLR7 agonists via activation of human endogenous retroviruses by HDAC inhibitors. Commun. Biol. 2021, 4, 276. [Google Scholar] [CrossRef]
- Strick, R.; Ackermann, S.; Langbein, M.; Swiatek, J.; Schubert, S.W.; Hashemolhosseini, S.; Koscheck, T.; Fasching, P.A.; Schild, R.L.; Beckmann, M.W.; et al. Proliferation and cell–cell fusion of endometrial carcinoma are induced by the human endogenous retroviral Syncytin-1 and regulated by TGF-β. J. Mol. Med. 2006, 85, 23–38. [Google Scholar] [CrossRef]
- Muir, A.; Lever, A.M.L.; Moffett, A. Human endogenous retrovirus-W envelope (syncytin) is expressed in both villous and extravillous trophoblast populations. J. Gen. Virol. 2006, 87, 2067–2071. [Google Scholar] [CrossRef]
- Heidmann, O.; Béguin, A.; Paternina, J.; Berthier, R.; Deloger, M.; Bawa, O.; Heidmann, T. HEMO, an ancestral endogenous retroviral envelope protein shed in the blood of pregnant women and expressed in pluripotent stem cells and tumors. Proc. Natl. Acad. Sci. USA 2017, 114, E6642–E6651. [Google Scholar] [CrossRef]
- Hummel, J.; Kämmerer, U.; Müller, N.; Avota, E.; Schneider-Schaulies, S. Human endogenous retrovirus envelope proteins target dendritic cells to suppress T-cell activation. Eur. J. Immunol. 2015, 45, 1748–1759. [Google Scholar] [CrossRef]
- Schulte, A.M.; Malerczyk, C.; Cabal-Manzano, R.; Gajarsa, J.J.; List, H.-J.; Riegel, A.T.; Wellstein, A. Influence of the human endogenous retrovirus-like element HERV-E.PTN on the expression of growth factor pleiotrophin: A critical role of a retroviral Sp1-binding site. Oncogene 2000, 19, 3988–3998. [Google Scholar] [CrossRef] [PubMed]
- Strissel, P.L.; Ruebner, M.; Thiel, F.; Wachter, D.; Ekici, A.B.; Wolf, F.; Thieme, F.; Ruprecht, K.; Beckmann, M.W.; Strick, R. Reactivation of codogenic endogenous retroviral (ERV) envelope genes in human endometrial carcinoma and prestages: Emergence of new molecular targets. Oncotarget 2012, 3, 1204–1219. [Google Scholar] [CrossRef] [PubMed]
- Jeon, K.-Y.Y.; Ko, E.-J.J.; Oh, Y.L.; Kim, H.; Eo, W.K.; Kim, A.; Sun, H.G.; Ock, M.S.; Kim, K.H.; Cha, H.-J.J. Analysis of KAP1 expression patterns and human endogenous retrovirus Env proteins in ovarian cancer. Genes Genom. 2020, 42, 1145–1150. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Liu, T.; Zhao, Z.; Chen, Y.; Zeng, J.; Liu, S.; Zhu, F. Mutations in 3′-long terminal repeat of HERV-W family in chromosome 7 upregulate syncytin-1 expression in urothelial cell carcinoma of the bladder through interacting with c-Myb. Oncogene 2014, 33, 3947–3958. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Harashima, N.; Kajigaya, S.; Yokoyama, H.; Cherkasova, E.; McCoy, J.P.; Hanada, K.I.; Mena, O.; Kurlander, R.; Abdul, T.; et al. Regression of human kidney cancer following allogeneic stem cell transplantation is associated with recognition of an HERV-E antigen by T cells. J. Clin. Investig. 2008, 118, 1099–1109. [Google Scholar] [CrossRef] [PubMed]
- Cherkasova, E.; Malinzak, E.; Rao, S.; Takahashi, Y.; Senchenko, V.N.; Kudryavtseva, A.V.; Nickerson, M.L.; Merino, M.; Hong, J.A.; Schrump, D.S.; et al. Inactivation of the von Hippel-Lindau tumor suppressor leads to selective expression of a human endogenous retrovirus in kidney cancer. Oncogene 2011, 30, 4697–4706. [Google Scholar] [CrossRef]
- Cherkasova, E.; Scrivani, C.; Doh, S.; Weisman, Q.; Takahashi, Y.; Harashima, N.; Yokoyama, H.; Srinivasan, R.; Linehan, W.M.; Lerman, M.I.; et al. Detection of an immunogenic HERV-E envelope with selective expression in clear cell kidney cancer. Cancer Res. 2016, 76, 2177–2185. [Google Scholar] [CrossRef]
- Childs, R.W. HERV-E TCR Transduced Autologous T Cells in People with Metastatic Clear Cell Renal Cell Carcinoma. Available online: https://clinicaltrials.gov/ct2/show/NCT03354390 (accessed on 2 December 2021).
- Weyerer, V.; Strissel, P.L.; Stöhr, C.; Eckstein, M.; Wach, S.; Taubert, H.; Brandl, L.; Geppert, C.I.; Wullich, B.; Cynis, H.; et al. Endogenous Retroviral-K Envelope Is a Novel Tumor Antigen and Prognostic Indicator of Renal Cell Carcinoma. Front. Oncol. 2021, 11, 657187. [Google Scholar] [CrossRef]
- Buslei, R.; Strissel, P.L.; Henke, C.; Schey, R.; Lang, N.; Ruebner, M.; Stolt, C.C.; Fabry, B.; Buchfelder, M.; Strick, R. Activation and regulation of endogenous retroviral genes in the human pituitary gland and related endocrine tumours. Neuropathol. Appl. Neurobiol. 2015, 41, 180–200. [Google Scholar] [CrossRef]
- Dai, L.; Del Valle, L.; Miley, W.; Whitby, D.; Ochoa, A.C.; Flemington, E.K.; Qin, Z. Transactivation of human endogenous retrovirus K (HERV-K) by KSHV promotes Kaposi’s sarcoma development. Oncogene 2018, 37, 4534–4545. [Google Scholar] [CrossRef]
- Gröger, V.; Wieland, L.; Naumann, M.; Meinecke, A.-C.; Meinhardt, B.; Rossner, S.; Ihling, C.; Emmer, A.; Staege, M.S.; Cynis, H. Formation of HERV-K and HERV-Fc1 Envelope Family Members is Suppressed on Transcriptional and Translational Level. Int. J. Mol. Sci. 2020, 21, 7855. [Google Scholar] [CrossRef] [PubMed]
- Tan, I.B.; Drexhage, H.A.; Mullink, R.; Hensen-Logmans, S.; De Haan-Meulman, M.; Snow, G.B.; Balm, A.J.M. Immunohistochemical Detection of Retroviral—P15E—Related Material in Carcinomas of the Head and Neck. Otolaryngol. Neck Surg. 1987, 96, 251–255. [Google Scholar] [CrossRef] [PubMed]
- Lang, M.S.; Oostendorp, R.A.J.; Simons, P.J.; Boersma, W.; Knegt, P.; van Ewijk, W. New monoclonal antibodies against the putative immunosuppressive site of retroviral p15E. Cancer Res. 1994, 54, 1831–1836. [Google Scholar] [PubMed]
- Kolbe, A.R.; Bendall, M.L.; Pearson, A.T.; Paul, D.; Nixon, D.F.; Pérez-Losada, M.; Crandall, K.A. Human Endogenous Retrovirus Expression Is Associated with Head and Neck Cancer and Differential Survival. Viruses 2020, 12, 956. [Google Scholar] [CrossRef]
- Ma, W.; Hong, Z.; Liu, H.; Chen, X.; Ding, L.; Liu, Z.; Zhou, F.; Yuan, Y. Human Endogenous Retroviruses-K (HML-2) Expression Is Correlated with Prognosis and Progress of Hepatocellular Carcinoma. BioMed Res. Int. 2016, 2016, 8201642. [Google Scholar] [CrossRef]
- Hu, L.; Uzhameckis, D.; Hedborg, F.; Blomberg, J. Dynamic and selective HERV RNA expression in neuroblastoma cells subjected to variation in oxygen tension and demethylation. APMIS 2016, 124, 140–149. [Google Scholar] [CrossRef]
- Wieland, L.; Engel, K.; Volkmer, I.; Krüger, A.; Posern, G.; Kornhuber, M.E.; Staege, M.S.; Emmer, A. Overexpression of Endogenous Retroviruses and Malignancy Markers in Neuroblastoma Cell Lines by Medium-Induced Microenvironmental Changes. Front. Oncol. 2021, 11, 637522. [Google Scholar] [CrossRef]
- Lemaître, C.; Tsang, J.; Bireau, C.; Heidmann, T.; Dewannieux, M. A human endogenous retrovirus-derived gene that can contribute to oncogenesis by activating the ERK pathway and inducing migration and invasion. PLoS Pathog. 2017, 13, e1006451. [Google Scholar] [CrossRef]
- Ammoun, S.; Maze, E.A.; Agit, B.; Belshaw, R.; Hanemann, C.O. Abstract 1164: Human Endogenous Retrovirus Type K Promotes Proliferation of Merlin Negative Schwannoma and Meningioma Which Can Be Inhibited by Anti-retroviral and Anti-TEAD Drugs. In Proceedings of the Experimental and Molecular Therapeutics, Philadelphia, PA, USA, 10–15 April 2021; p. 1164. [Google Scholar]
- Bahrami, S.; Gryz, E.A.; Graversen, J.H.; Troldborg, A.; Stengaard Pedersen, K.; Laska, M.J. Immunomodulating peptides derived from different human endogenous retroviruses (HERVs) show dissimilar impact on pathogenesis of a multiple sclerosis animal disease model. Clin. Immunol. 2018, 191, 37–43. [Google Scholar] [CrossRef]
- Mangeney, M.; Renard, M.; Schlecht-Louf, G.; Bouallaga, I.; Heidmann, O.; Letzelter, C.; Richaud, A.; Ducos, B.; Heidmann, T. Placental syncytins: Genetic disjunction between the fusogenic and immunosuppressive activity of retroviral envelope proteins. Proc. Natl. Acad. Sci. USA 2007, 104, 20534–20539. [Google Scholar] [CrossRef]
- Morozov, V.A.; Dao Thi, V.L.; Denner, J. The transmembrane protein of the human endogenous retrovirus—K (HERV-K) modulates cytokine release and gene expression. PLoS ONE 2013, 8, e70399. [Google Scholar] [CrossRef] [PubMed]
- Sacha, J.B.; Kim, I.-J.; Chen, L.; Ullah, J.H.; Goodwin, D.A.; Simmons, H.A.; Schenkman, D.I.; von Pelchrzim, F.; Gifford, R.J.; Nimityongskul, F.A.; et al. Vaccination with Cancer- and HIV Infection-Associated Endogenous Retrotransposable Elements Is Safe and Immunogenic. J. Immunol. 2012, 189, 1467–1479. [Google Scholar] [CrossRef] [PubMed]
- Rooney, M.S.; Shukla, S.A.; Wu, C.J.; Getz, G.; Hacohen, N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015, 160, 48–61. [Google Scholar] [CrossRef] [PubMed]
- Panda, A.; de Cubas, A.A.; Stein, M.; Riedlinger, G.; Kra, J.; Mayer, T.; Smith, C.C.; Vincent, B.G.; Serody, J.S.; Beckermann, K.E.; et al. Endogenous retrovirus expression is associated with response to immune checkpoint blockade in clear cell renal cell carcinoma. JCI Insight 2018, 3, e121522. [Google Scholar] [CrossRef]
- Parikh, A.R.; Szabolcs, A.; Allen, J.N.; Clark, J.W.; Wo, J.Y.; Raabe, M.; Thel, H.; Hoyos, D.; Mehta, A.; Arshad, S.; et al. Radiation therapy enhances immunotherapy response in microsatellite stable colorectal and pancreatic adenocarcinoma in a phase II trial. Nat. Cancer 2021, 2, 1124–1135. [Google Scholar] [CrossRef]
- EnaraBio. Available online: https://enarabio.com/ (accessed on 14 December 2021).
- Jones, P.A.; Ohtani, H.; Chakravarthy, A.; De Carvalho, D.D. Epigenetic therapy in immune-oncology. Nat. Rev. Cancer 2019, 19, 151–161. [Google Scholar] [CrossRef]
- Lee, A.K.; Pan, D.; Bao, X.; Hu, M.; Li, F.; Li, C.Y. Endogenous Retrovirus Activation as a Key Mechanism of Anti-Tumor Immune Response in Radiotherapy. Radiat. Res. 2020, 193, 305–317. [Google Scholar] [CrossRef]
- Garcia-Montojo, M.; Fathi, S.; Norato, G.; Smith, B.R.; Rowe, D.B.; Kiernan, M.C.; Vucic, S.; Mathers, S.; van Eijk, R.P.A.; Santamaria, U.; et al. Inhibition of HERV-K (HML-2) in amyotrophic lateral sclerosis patients on antiretroviral therapy. J. Neurol. Sci. 2021, 423, 117358. [Google Scholar] [CrossRef]
- Solovyov, A.; Vabret, N.; Arora, K.S.; Snyder, A.; Funt, S.A.; Bajorin, D.F.; Rosenberg, J.E.; Bhardwaj, N.; Ting, D.T.; Greenbaum, B.D. Global Cancer Transcriptome Quantifies Repeat Element Polarization between Immunotherapy Responsive and T Cell Suppressive Classes. Cell Rep. 2018, 23, 512–521. [Google Scholar] [CrossRef]
| Indication | HERV | Tissue | Cell Line | Controls | Case/Control | Conclusion | Reference |
|---|---|---|---|---|---|---|---|
| Glioblastoma | HERV-K | - | U87 | - | - | Expression | [43] |
| Unspecified brain tumor | HERV-K | Serum | - | - | 128/0 | Seroreactivity | [55] |
| Schwannoma and meningioma | HERV-K | Tumor | - | Healthy controls | 10/10 | Expression Tumor function Therapeutic | [69] |
| Atypical teratoid rhabdoid tumors | HERV-K | Tumor | CHLA 02, CHLA 04, CHLA 05, and CHLA 06 | Healthy controls | 37/3 | Expression Tumor function | [68] |
| Indication | HERV | Tissue | Cell Line | Controls | Case/Control | Conclusion | Reference |
|---|---|---|---|---|---|---|---|
| NSCLC | HERV-K | - | A549 | - | - | Expression | [43] |
| SCLC | HERV-K | - | H69 | - | - | Expression Tumor function Therapeutic | [78] |
| Indication | HERV | Tissue | Cell Line | Controls | Case/Control | Conclusion | Reference |
|---|---|---|---|---|---|---|---|
| Pancreatic adenocarcinoma | HERV-H | - | Panc-1 | - | - | Expression Tumor function | [81] |
| Pancreatic adenocarcinoma | HERV-K | Serum Tumor | Panc-1, Panc-2, HPDE-E6E7 | Adjacent Healthy controls | 106/40 1/1 | Expression Tumor function Seroreactivity | [101] |
| Pancreatic adenocarcinoma | HERV-K | - | Panc-1 | - | - | Expression Tumor function | [43] |
| Indication | HERV | Tissue | Cell Line | Controls | Case/Control | Conclusion | Reference |
|---|---|---|---|---|---|---|---|
| Pituitary adenoma | HERV-W | Tumor | - | From sellar exploration where no adenoma was found | 117/15 | Expression Tumor function | [120] |
| Indication | HERV | Tissue | Cell Line | Controls | Case/Control | Conclusion | Reference |
|---|---|---|---|---|---|---|---|
| Kaposi’s sarcoma | HERV-K | Blood | BCBL-1, BC-1, BC-3 BCP-1 | Blood from KSHV-neg HIV patients | 11/10 | Expression Tumor function | [121] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Müller, M.D.; Holst, P.J.; Nielsen, K.N. A Systematic Review of Expression and Immunogenicity of Human Endogenous Retroviral Proteins in Cancer and Discussion of Therapeutic Approaches. Int. J. Mol. Sci. 2022, 23, 1330. https://doi.org/10.3390/ijms23031330
Müller MD, Holst PJ, Nielsen KN. A Systematic Review of Expression and Immunogenicity of Human Endogenous Retroviral Proteins in Cancer and Discussion of Therapeutic Approaches. International Journal of Molecular Sciences. 2022; 23(3):1330. https://doi.org/10.3390/ijms23031330
Chicago/Turabian StyleMüller, Mikkel Dons, Peter Johannes Holst, and Karen Nørgaard Nielsen. 2022. "A Systematic Review of Expression and Immunogenicity of Human Endogenous Retroviral Proteins in Cancer and Discussion of Therapeutic Approaches" International Journal of Molecular Sciences 23, no. 3: 1330. https://doi.org/10.3390/ijms23031330
APA StyleMüller, M. D., Holst, P. J., & Nielsen, K. N. (2022). A Systematic Review of Expression and Immunogenicity of Human Endogenous Retroviral Proteins in Cancer and Discussion of Therapeutic Approaches. International Journal of Molecular Sciences, 23(3), 1330. https://doi.org/10.3390/ijms23031330






