Transcription Factor RrANT1 of Rosa rugosa Positively Regulates Flower Organ Size in Petunia hybrida
Abstract
:1. Introduction
2. Results
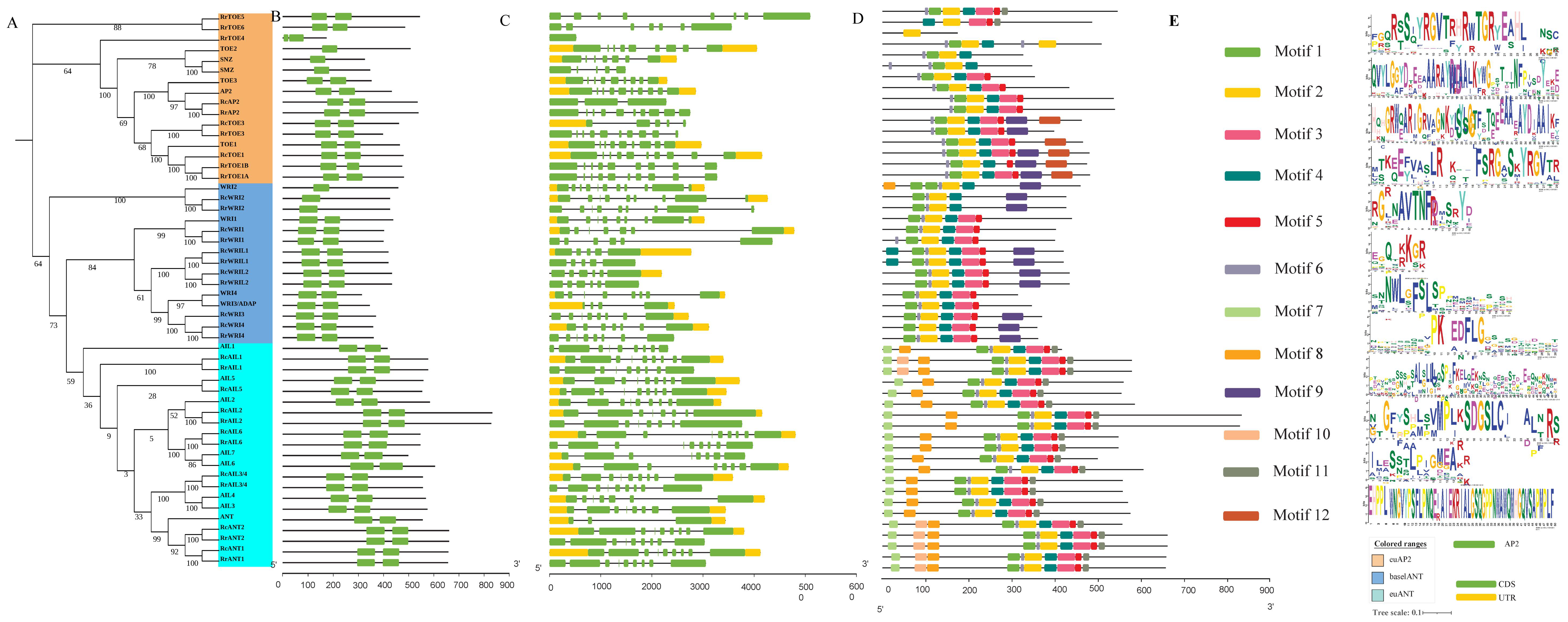
2.1. Identification and Phylogenetic Analysis of RrAP2 Family
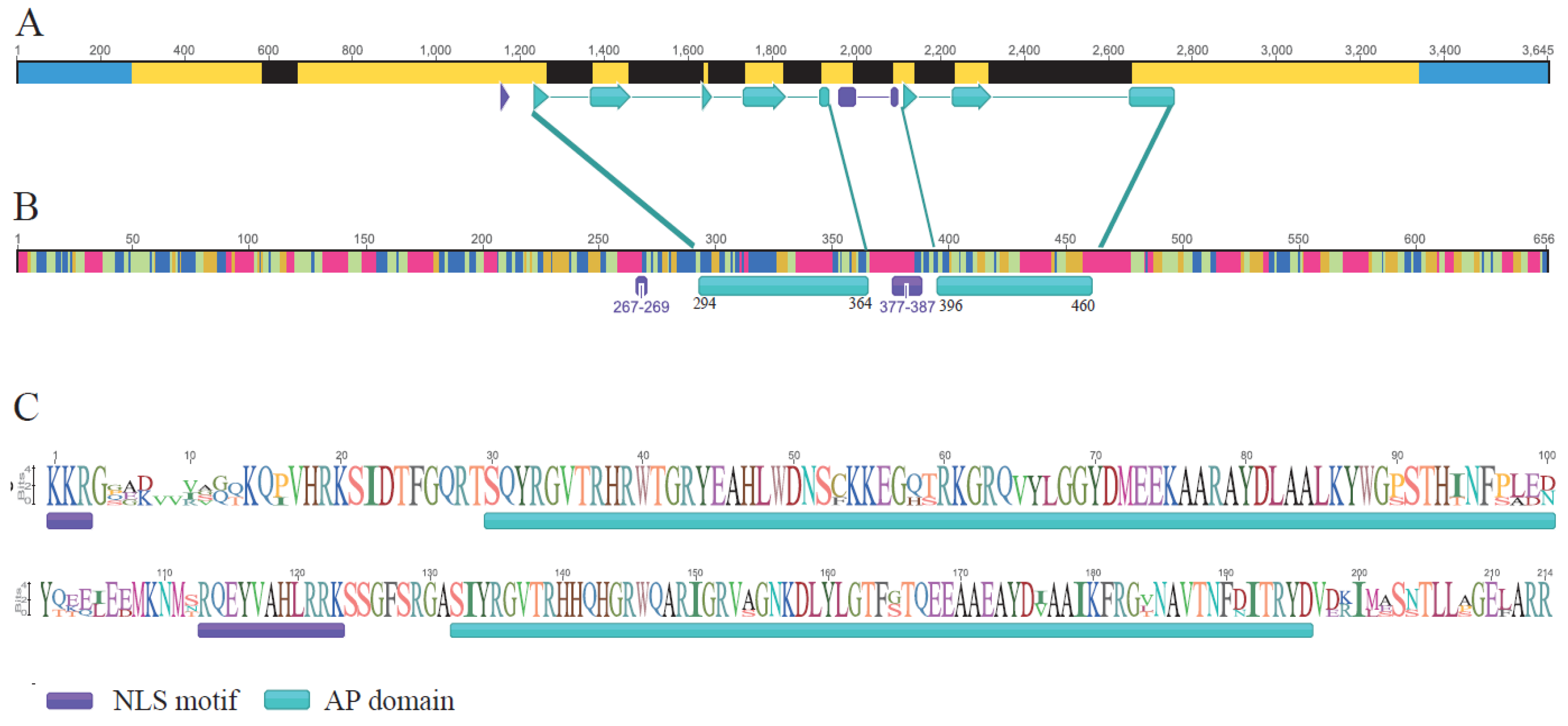
2.2. RrANT1 Identification and Characterization
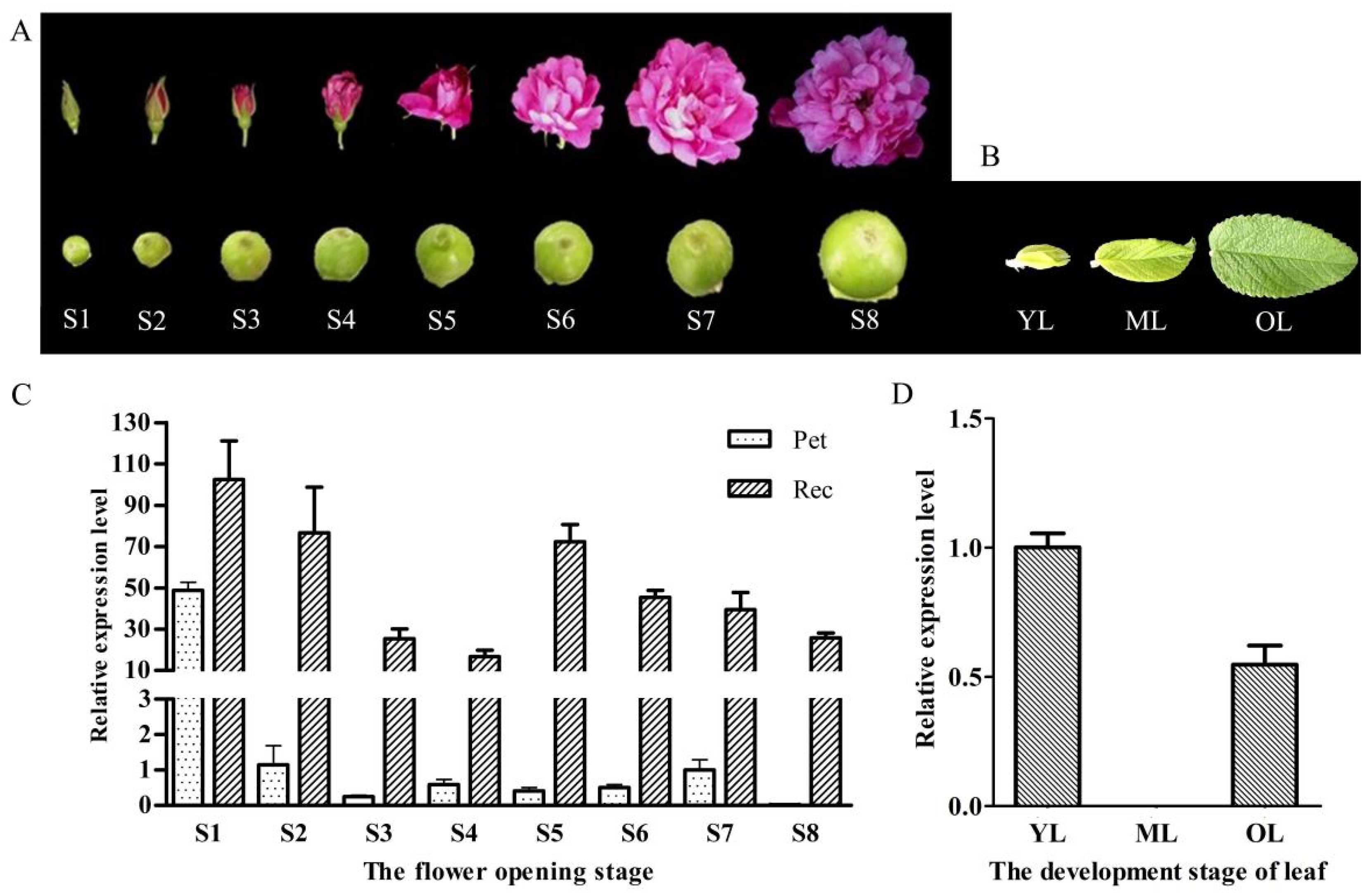
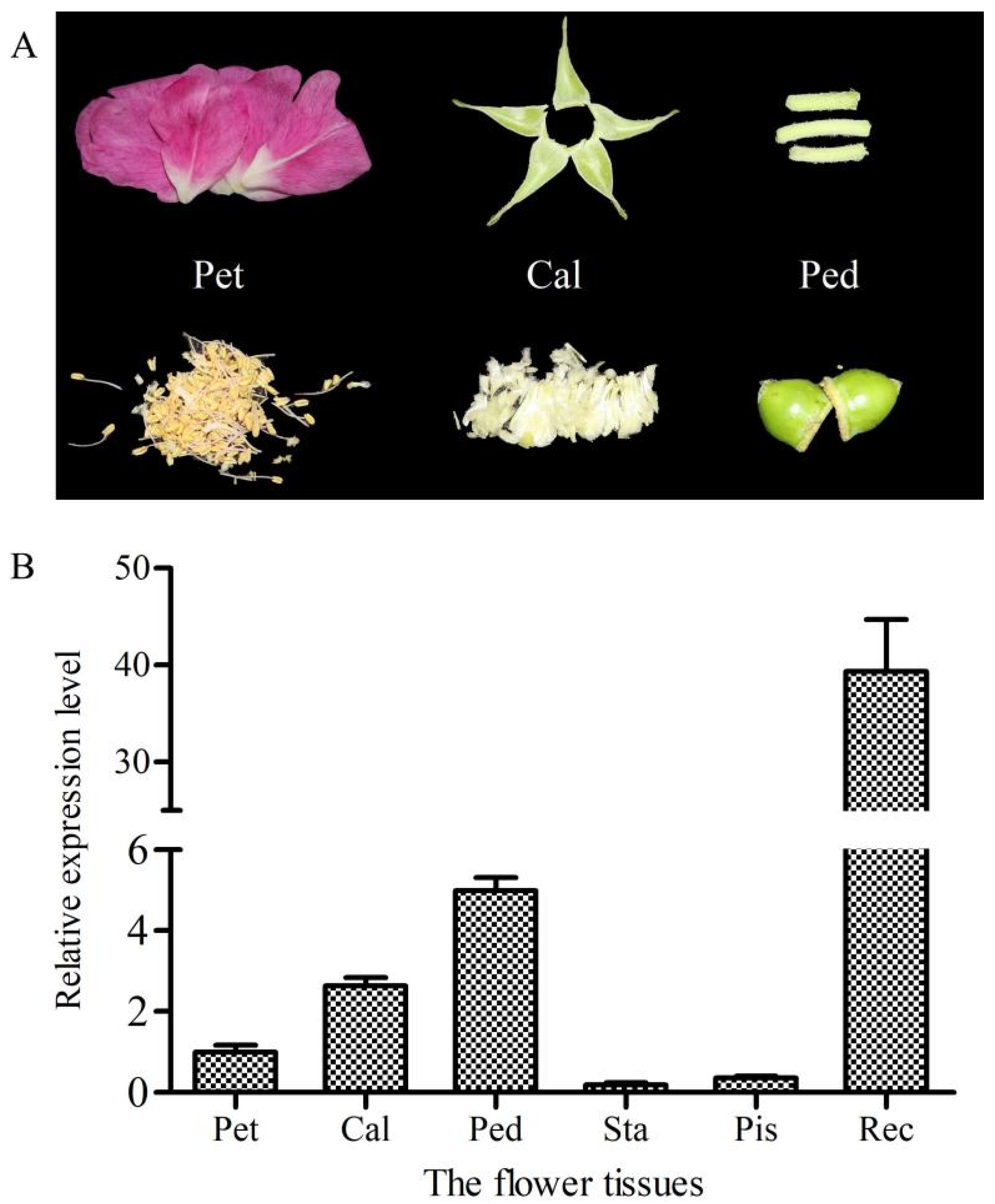
2.3. Relative Expression Level of RrANT1 in the Flower Tissues and Leaves of R. rugosa
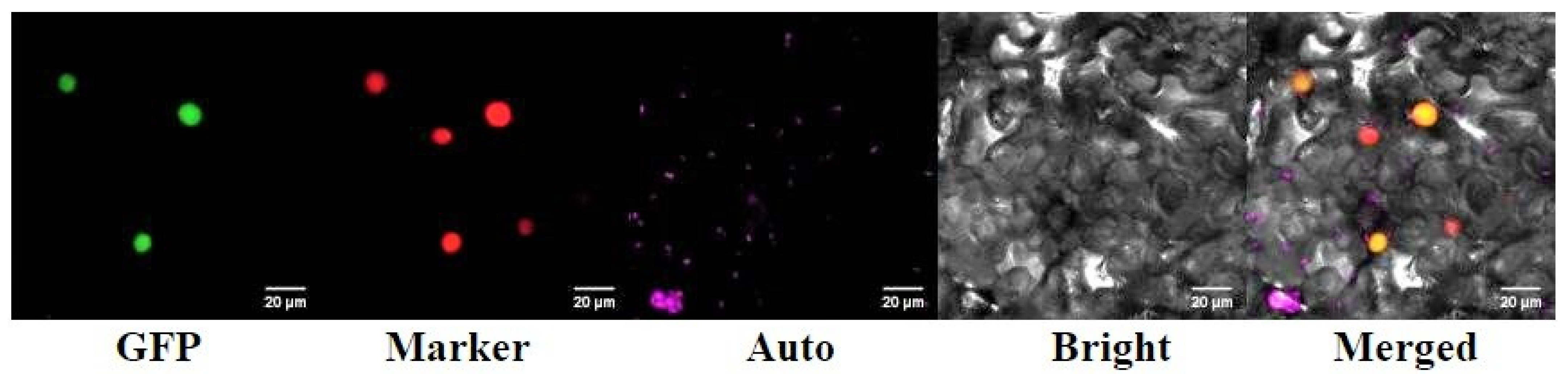
2.4. Subcellular Localization of RrANT1
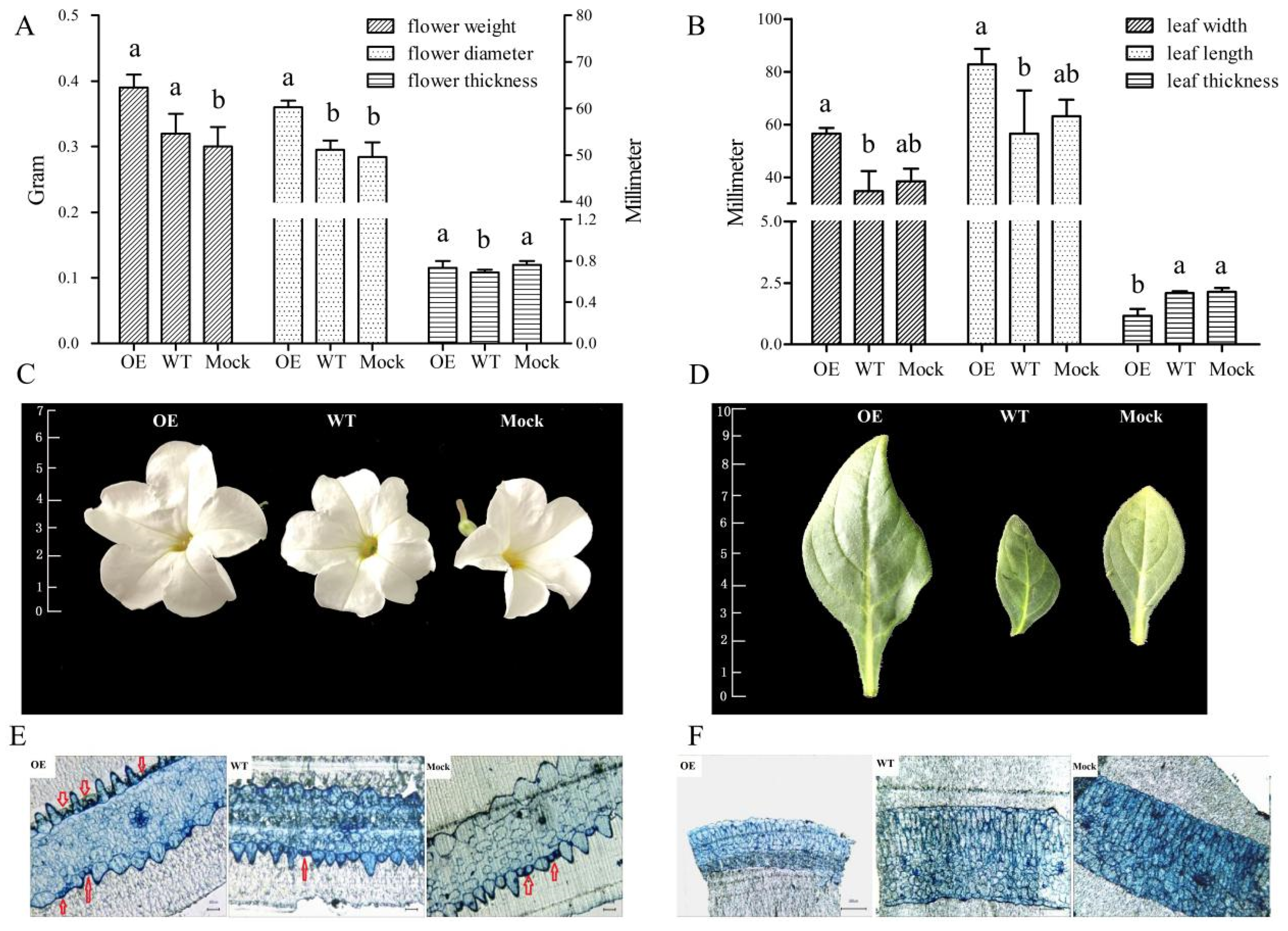
2.5. Phenotypic and Microstructure Observation of Leaves and Flowers of Transgenic Petunia
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Identification, Phylogenetic, Gene Structure, and Motif Composition Analysis of AP2 Family
4.3. The Extraction of Nucleic Acid
4.4. Cloning of Full-Length cDNA of RrANT1
4.5. Reverse Transcription Reaction and RrANT1 Expression Analysis
4.6. Vector Construction and Subcellular Localization of RrANT1
4.7. Overexpression (OE) of RrANT1 in P. hybrida and Microstructural Observation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- O’Maoileidigh, D.S.; Graciet, E.; Wellmer, F. Gene networks controlling Arabidopsis thaliana flower development. N. Phytol. 2014, 201, 16–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Powell, A.E.; Lenhard, M. Control of Organ Size in Plants. Curr. Biol. 2012, 22, R360–R367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anastasiou, E.; Kenz, S.; Gerstung, M.; MacLean, D.; Timmer, J.; Fleck, C.; Lenhard, M. Control of plant organ size by KLUH/CYP78A5-dependent intercellular signaling. Dev. Cell 2007, 13, 843–856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, B.H.; Ko, J.-H.; Lee, S.; Lee, Y.; Pak, J.-H.; Kim, J.H. The Arabidopsis GRF-INTERACTING FACTOR Gene Family Performs an Overlapping Function in Determining Organ Size as Well as Multiple Developmental Properties. Plant Physiol. 2009, 151, 655–668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.H.; Kende, H. A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 13374–13379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, D.; Song, Y.; Chen, Z.; Yu, D. Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiol. Plant. 2009, 136, 223–236. [Google Scholar] [CrossRef]
- Disch, S.; Anastasiou, E.; Sharma, V.K.; Laux, T.; Fletcher, J.C.; Lenhard, M. The E3 ubiquitin ligase BIG BROTHER controls Arabidopsis organ size in a dosage-dependent manner. Curr. Biol. 2006, 16, 272–279. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zheng, L.; Corke, F.; Smith, C.; Bevan, M.W. Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev. 2008, 22, 1331–1336. [Google Scholar] [CrossRef] [Green Version]
- Xu, R.; Li, Y. The Mediator complex subunit 8 regulates organ size in Arabidopsis thaliana. Plant Signal. Behav. 2012, 7, 182–183. [Google Scholar] [CrossRef] [Green Version]
- Szecsi, J.; Joly, C.; Bordji, K.; Varaud, E.; Cock, J.M.; Dumas, C.; Bendahmane, M. BIGPETALp, a bHLH transcription factor is involved in the control of Arabidopsis petal size. Embo J. 2006, 25, 3912–3920. [Google Scholar] [CrossRef] [Green Version]
- D’Ario, M.; Sablowski, R. Cell Size Control in Plants. Annu. Rev. Genet. 2019, 53, 45–65. [Google Scholar] [CrossRef] [PubMed]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta-Gene Regul. Mech. 2012, 1819, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Soltis, P.S.; Wall, K.; Soltis, D.E. Phylogeny and domain evolution in the APETALA2-like gene family. Mol. Biol. Evol. 2006, 23, 107–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shigyo, M.; Ito, M. Analysis of gymnosperm two-AP2-domain-containing genes. Dev. Genes Evol. 2004, 214, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Krizek, B.A.; Eaddy, M. AINTEGUMENTA-LIKE6 regulates cellular differentiation in flowers. Plant Mol. Biol. 2012, 78, 199–209. [Google Scholar] [CrossRef]
- Krizek, B.A. Aintegumenta and Aintegumenta-Like6 regulate auxin-mediated flower development in Arabidopsis. BMC Res. Notes 2011, 4, 176. [Google Scholar] [CrossRef] [Green Version]
- Horstman, A.; Willemsen, V.; Boutilier, K.; Heidstra, R. AINTEGUMENTA-LIKE proteins: Hubs in a plethora of networks. Trends Plant Sci. 2014, 19, 146–157. [Google Scholar] [CrossRef]
- Rashid, M.; He, G.; Yang, G.; Hussain, J.; Yan, X. AP2/ERF Transcription Factor in Rice: Genome-Wide Canvas and Syntenic Relationships between Monocots and Eudicots. Evol. Bioinform. 2012, 8, 321–355. [Google Scholar] [CrossRef]
- Wei, H.; Liu, Y.; Dou, M.; Yang, S.; Feng, X. Molecular Evolution of AP2/ERF Gene Family in Glycine max. Plant Physiol. J. 2015, 51, 1706–1718. (In Chinese) [Google Scholar]
- Meng, L.-S.; Wang, Z.-B.; Yao, S.-Q.; Liu, A. The ARF2-ANT-COR15A gene cascade regulates ABA-signaling-mediated resistance of large seeds to drought in Arabidopsis. J. Cell Sci. 2015, 128, 3922–3932. [Google Scholar]
- Krizek, B.A. AINTEGUMENTA and AINTEGUMENTA-LIKE6 Act Redundantly to Regulate Arabidopsis Floral Growth and Patterning. Plant Physiol. 2009, 150, 1916–1929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krizek, B.A.; Anderson, J.T. Control of flower size. J. Exp. Bot. 2013, 64, 1427–1437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krizek, B.A.; Bantle, A.T.; Heflin, J.M.; Han, H.; Freese, N.H.; Loraine, A.E. AINTEGUMENTA and AINTEGUMENTA-LIKE6 directly regulate floral homeotic, growth, and vascular development genes in young Arabidopsis flowers. J. Exp. Bot. 2021, 72, 5478–5493. [Google Scholar] [CrossRef] [PubMed]
- Klucher, K.M.; Chow, H.; Reiser, L.; Fischer, R.L. The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 1996, 8, 137–153. [Google Scholar]
- Nole-Wilson, S.; Krizek, B.A. AINTEGUMENTA contributes to organ polarity and regulates growth of lateral organs in combination with YABBY genes. Plant Physiol. 2006, 141, 977–987. [Google Scholar] [CrossRef] [Green Version]
- Manchado-Rojo, M.; Weiss, J.; Egea-Cortines, M. Validation of Aintegumenta as a gene to modify floral size in ornamental plants. Plant Biotechnol. J. 2014, 12, 1053–1065. [Google Scholar] [CrossRef] [Green Version]
- Krizek, B.A. Ectopic expression AINTEGUMENTA in Arabidopsis plants results in increased growth of floral organs. Dev. Genet. 1999, 25, 224–236. [Google Scholar] [CrossRef]
- Kuluev, B.; Avalbaev, A.; Nurgaleeva, E.; Knyazev, A.; Nikonorov, Y.; Chemeris, A. Role of AINTEGUMENTA-like gene NtANTL in the regulation of tobacco organ growth. J. Plant Physiol. 2015, 189, 11–23. [Google Scholar] [CrossRef]
- Wen, L.; Sun, X.; Fan, H.; Guo, Y.; Yu, Y.; Ren, H.; WANG, W.; ZHENG, C. Cloning and Functional Verification of AINTEGUMENTA Gene in Chrysanthemum. Sci. Agric. Sin. 2018, 51, 1771–1782. (In Chinese) [Google Scholar]
- Miao, L.; Li, S.Z.; Shi, A.K.; Li, Y.S.; He, C.X.; Yan, Y.; Wang, J.; Sun, M.T.; Yu, X.C. Genome-wide analysis of the AINTEGUMENTA-like (AIL) transcription factor gene family in pumpkin (Cucurbita moschata Duch.) and CmoANT1.2 response in graft union healing. Plant Physiol Biochem 2021, 162, 706–715. [Google Scholar] [CrossRef]
- Wei, T.; Zhu, M.; Zhou, J.; Chen, L.; Feng, L. Cytological evidence for the pathway of synthesis, accumulation, and secretion of rose essential oil. J. Essent. Oil-Bear. Plants JEOP 2019, 22, 301–310. [Google Scholar] [CrossRef]
- Nole-Wilson, S.; Tranby, T.L.; Krizek, B.A. AINTEGUMENTA-like (AIL) genes are expressed in young tissues and may specify meristematic or division-competent states. Plant Mol. Biol. 2005, 57, 613–628. [Google Scholar] [CrossRef] [PubMed]
- Nole-Wilson, S.; Krizek, B.A. DNA binding properties of the Arabidopsis floral development protein AINTEGUMENTA. Nucleic Acids Res. 2000, 28, 4076–4082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizukami, Y.; Fischer, R.L. Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Plant Biol. 2000, 2000, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juarez-Corona, A.G.; de Folter, S. ANT and AIL6: Masters of the master regulators during flower development. J. Exp. Bot. 2021, 72, 5263–5266. [Google Scholar] [CrossRef]
- Elliott, R.C.; Betzner, A.S.; Huttner, E.; Oakes, M.P.; Tucker, W.Q.J.; Gerentes, D.; Perez, P.; Smyth, D.R. AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 1996, 8, 155–168. [Google Scholar]
- Yamaguchi, N.; Jeong, C.W.; Nole-Wilson, S.; Krizek, B.A.; Wagner, D. AINTEGUMENTA and AINTEGUMENTA-LIKE6/PLETHORA3 Induce LEAFY Expression in Response to Auxin to Promote the Onset of Flower Formation in Arabidopsis. Plant Physiol. 2016, 170, 283–293. [Google Scholar] [CrossRef] [Green Version]
- Krizek, B.A.; Blakley, I.C.; Ho, Y.Y.; Freese, N.; Loraine, A.E. The Arabidopsis transcription factor AINTEGUMENTA orchestrates patterning genes and auxin signaling in the establishment of floral growth and form. Plant J. 2020, 103, 752–768. [Google Scholar] [CrossRef]
- Liu, W.-Y.; Lin, H.-H.; Yu, C.-P.; Chang, C.-K.; Chen, H.-J.; Lin, J.-J.; Lu, M.-Y.J.; Tu, S.-L.; Shiu, S.-H.; Wu, S.-H.; et al. Maize ANT1 modulates vascular development, chloroplast development, photosynthesis, and plant growth. Proc. Natl. Acad. Sci. USA 2020, 117, 21747–21756. [Google Scholar] [CrossRef]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Finn, R.D.; Mistry, J.; Tate, J.; Coggill, P.; Heger, A.; Pollington, J.E.; Gavin, O.L.; Gunasekaran, P.; Ceric, G.; Forslund, K.; et al. The Pfam protein families database. Nucleic Acids Res. 2010, 38, D211–D222. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- De Moraes Russo, C.A.; Selvatti, A.P. Bootstrap and Rogue Identification Tests for Phylogenetic Analyses. Mol. Biol. Evol. 2018, 35, 2327–2333. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Xing, Y.; Wei, T.; Wang, P.; Liang, Y.; Xu, M.; Ding, H.; Wang, J.; Feng, L. Transcription Factor RrANT1 of Rosa rugosa Positively Regulates Flower Organ Size in Petunia hybrida. Int. J. Mol. Sci. 2022, 23, 1236. https://doi.org/10.3390/ijms23031236
Xu Y, Xing Y, Wei T, Wang P, Liang Y, Xu M, Ding H, Wang J, Feng L. Transcription Factor RrANT1 of Rosa rugosa Positively Regulates Flower Organ Size in Petunia hybrida. International Journal of Molecular Sciences. 2022; 23(3):1236. https://doi.org/10.3390/ijms23031236
Chicago/Turabian StyleXu, Yong, Yongxiang Xing, Tiantian Wei, Pengqing Wang, Yue Liang, Mengmeng Xu, Haiquan Ding, Jianwen Wang, and Liguo Feng. 2022. "Transcription Factor RrANT1 of Rosa rugosa Positively Regulates Flower Organ Size in Petunia hybrida" International Journal of Molecular Sciences 23, no. 3: 1236. https://doi.org/10.3390/ijms23031236
APA StyleXu, Y., Xing, Y., Wei, T., Wang, P., Liang, Y., Xu, M., Ding, H., Wang, J., & Feng, L. (2022). Transcription Factor RrANT1 of Rosa rugosa Positively Regulates Flower Organ Size in Petunia hybrida. International Journal of Molecular Sciences, 23(3), 1236. https://doi.org/10.3390/ijms23031236





