An Insight into miR-1290: An Oncogenic miRNA with Diagnostic Potential
Abstract
1. Introduction
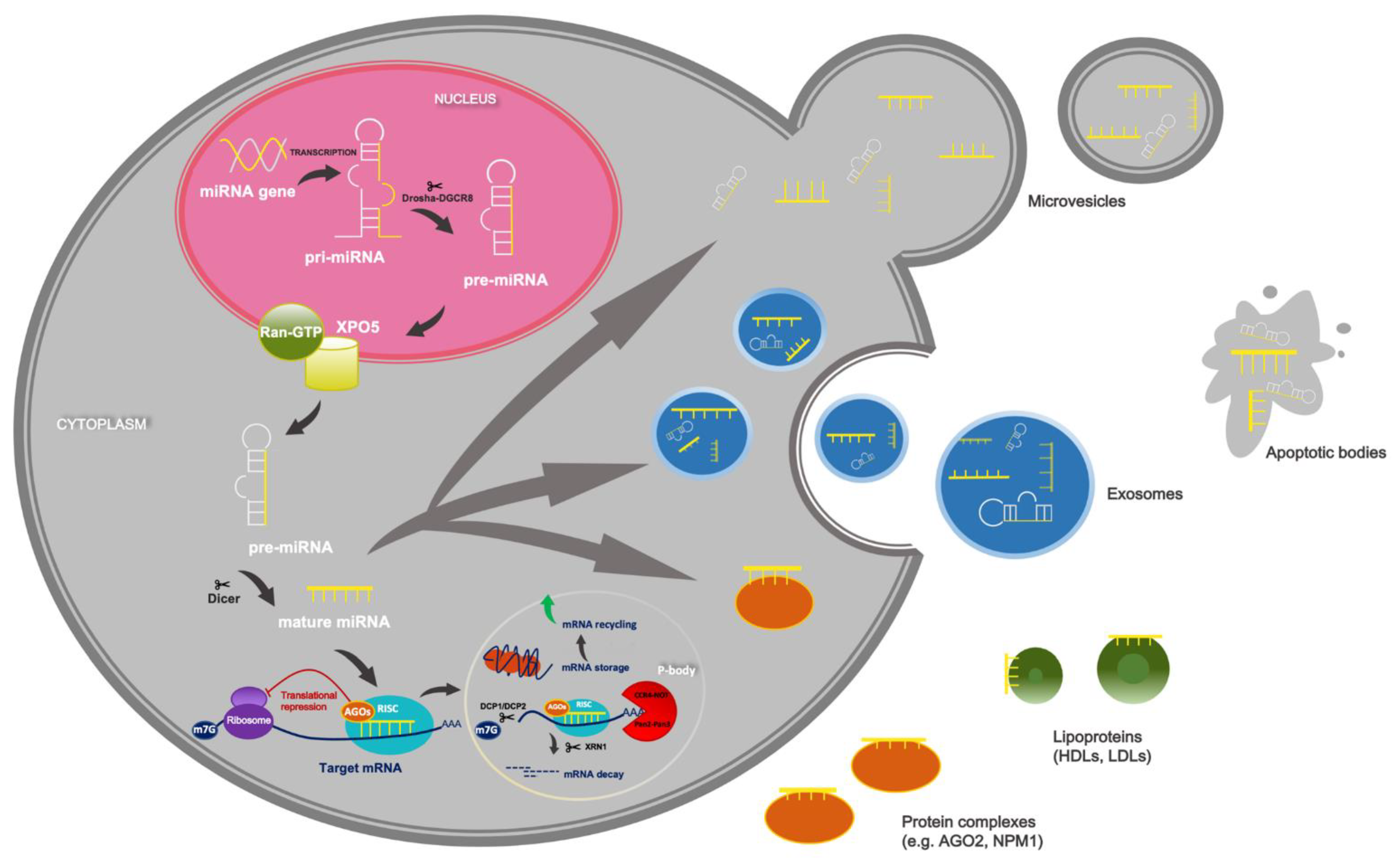
2. Overview of miRNAs’ Biogenesis and Function
3. Circulating miRNAs as Promising Non-Invasive Markers
4. Bioinformatics Analysis of miR-1290 Target Genes and Their Functional Annotations
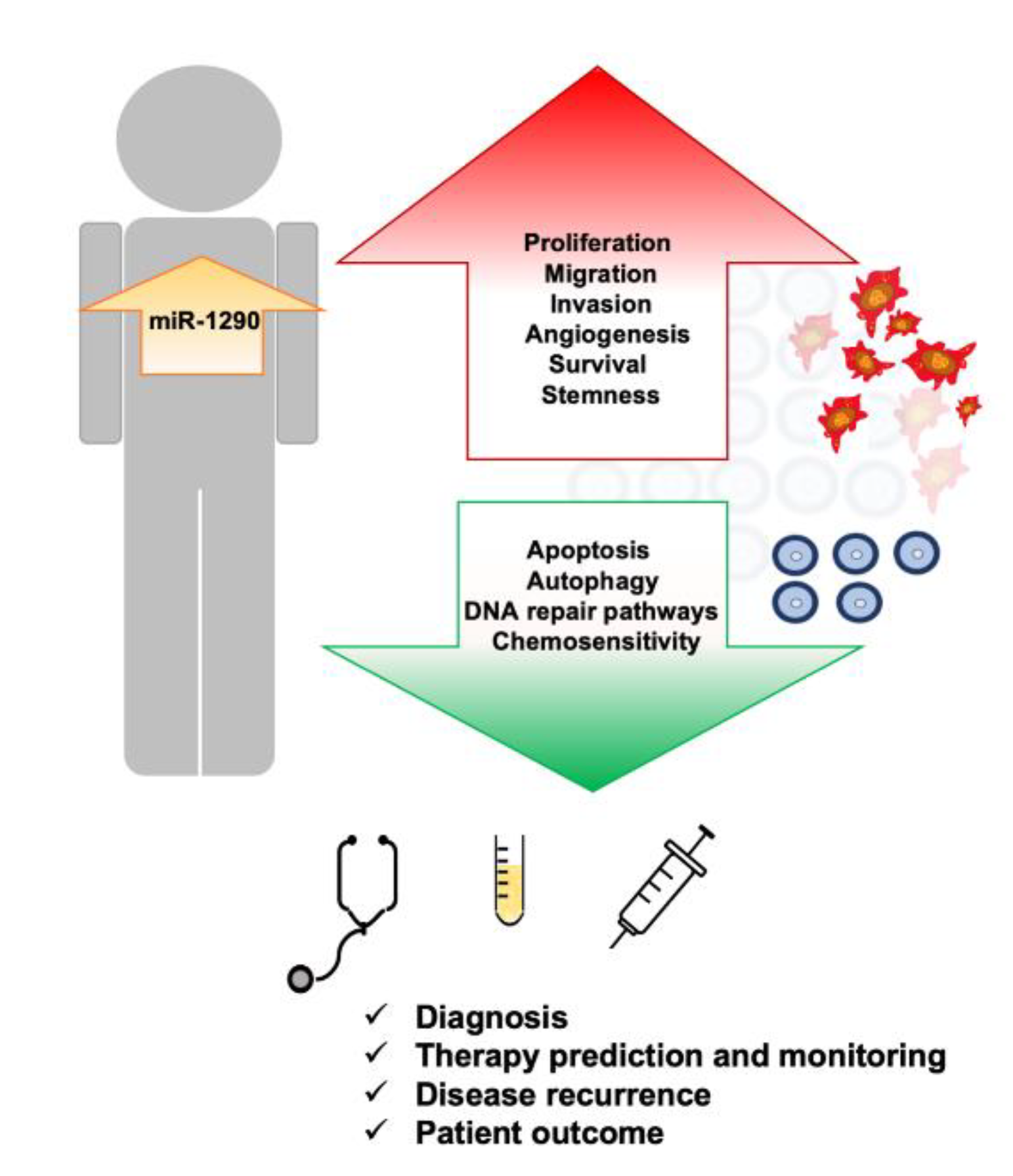
5. Physiological Role of miR-1290
6. The Role of miR-1290 in Non-Neoplastic Diseases
7. Upregulation of miR-1290 Is Associated with Different Types of Cancers
7.1. Gastrointestinal Cancers
7.1.1. Colorectal Cancer
7.1.2. Pancreatic Cancer
7.1.3. Gastric Cancer
7.1.4. Liver Cancer
7.1.5. Esophageal Cancer
7.2. Lung Cancer
7.3. Female Cancers
7.3.1. Breast Cancer
7.3.2. Cervical Cancer
7.3.3. Ovarian Cancer
7.4. Prostate Cancer
7.5. Head and Neck Cancers
7.5.1. Oral Cancer
7.5.2. Laryngeal Cancer
7.6. Cutaneous Cancer
7.7. Brain Cancer
7.8. Leukemia
7.9. Conjunctival Melanoma
8. Conclusions and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: An organizing principle for cancer medicine. In DeVita, Hellman, and Rosenberg’s Cancer: Principles & Practice of Oncology, 11th ed.; Wolters Kluwer: Philadelphia, PA, USA, 2018; pp. 44–66. ISBN 9781496394644. [Google Scholar]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Darwiche, N. Epigenetic mechanisms and the hallmarks of cancer: An intimate affair. Am. J. Cancer Res. 2020, 10, 1954–1978. [Google Scholar]
- Senga, S.S.; Grose, R.P. Hallmarks of cancer—The new testament. Open Biol. 2021, 11, 200358. [Google Scholar] [CrossRef]
- Roberti, A.; Valdes, A.F.; Torrecillas, R.; Fraga, M.F.; Fernandez, A.F. Epigenetics in cancer therapy and nanomedicine. Clin. Epigenetics 2019, 11, 81. [Google Scholar] [CrossRef]
- Ralston, A. Gene Expression Regulates Cell Differentiation. Nat. Educ. 2008, 1, 127–131. [Google Scholar]
- Kagohara, L.T.; Stein-O’Brien, G.L.; Kelley, D.; Flam, E.; Wick, H.C.; Danilova, L.V.; Easwaran, H.; Favorov, A.V.; Qian, J.; Gaykalova, D.A.; et al. Epigenetic regulation of gene expression in cancer: Techniques, resources and analysis. Brief. Funct. Genom. 2018, 17, 49–63. [Google Scholar] [CrossRef]
- Cheng, Y.; He, C.; Wang, M.; Ma, X.; Mo, F.; Yang, S.; Han, J.; Wei, X. Targeting epigenetic regulators for cancer therapy: Mechanisms and advances in clinical trials. Signal Transduct. Target. Ther. 2019, 4, 62. [Google Scholar] [CrossRef]
- Lao, T.D.; Huyen Le, T.A. MicroRNAs: Biogenesis, functions and potential biomarkers for early screening, prognosis and therapeutic molecular monitoring of nasopharyngeal carcinoma. Processes 2020, 8, 966. [Google Scholar] [CrossRef]
- Zhao, C.; Sun, X.; Li, L. Biogenesis and function of extracellular miRNAs. ExRNA 2019, 1, 387. [Google Scholar] [CrossRef]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Khuu, C.; Utheim, T.P.; Sehic, A. The Three Paralogous MicroRNA Clusters in Development and Disease, miR-17-92, miR-106a-363, and miR-106b-25. Scientifica 2016, 2016, 1379643. [Google Scholar] [CrossRef] [PubMed]
- Naruse, K.; Matsuura-Suzuki, E.; Watanabe, M.; Iwasaki, S.; Tomari, Y. In vitro reconstitution of chaperone-mediated human RISC assembly. RNA 2018, 24, 6–11. [Google Scholar] [CrossRef]
- Stroynowska-Czerwinska, A.; Fiszer, A.; Krzyzosiak, W.J. The panorama of miRNA-mediated mechanisms in mammalian cells. Cell. Mol. Life Sci. 2014, 71, 2253–2270. [Google Scholar] [CrossRef] [PubMed]
- Biasini, A.; Abdulkarim, B.; Pretis, S.; Tan, J.Y.; Arora, R.; Wischnewski, H.; Dreos, R.; Pelizzola, M.; Ciaudo, C.; Marques, A.C. Translation is required for miRNA-dependent decay of endogenous transcripts. EMBO J. 2021, 40, e104569. [Google Scholar] [CrossRef]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef]
- Wilczynska, A.; Bushell, M. The complexity of miRNA-mediated repression. Cell Death Differ. 2015, 22, 22–33. [Google Scholar] [CrossRef]
- Chen, C.Y.A.; Zheng, D.; Xia, Z.; Shyu, A. Bin Ago-TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat. Struct. Mol. Biol. 2009, 16, 1160–1166. [Google Scholar] [CrossRef]
- Rouya, C.; Siddiqui, N.; Morita, M.; Duchaine, T.F.; Fabian, M.R.; Sonenberg, N. Human DDX6 effects miRNA-mediated gene silencing via direct binding to CNOT1. RNA 2014, 20, 1398–1409. [Google Scholar] [CrossRef] [PubMed]
- Tenekeci, U.; Poppe, M.; Beuerlein, K.; Buro, C.; Müller, H.; Weiser, H.; Kettner-Buhrow, D.; Porada, K.; Newel, D.; Xu, M.; et al. K63-Ubiquitylation and TRAF6 Pathways Regulate Mammalian P-Body Formation and mRNA Decapping. Mol. Cell 2016, 62, 943–957. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.Y.; Ferrajoli, A.; Sood, A.K.; Lopez-Berestein, G.; Calin, G.A. microRNA Therapeutics in Cancer—An Emerging Concept. EBioMedicine 2016, 12, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Liu, M.; Cao, Y. New insight into microRNA functions in cancer: Oncogene-microRNA-tumor suppressor gene network. Front. Mol. Biosci. 2017, 4, 46. [Google Scholar] [CrossRef] [PubMed]
- Svoronos, A.A.; Engelman, D.M.; Slack, F.J. OncomiR or tumor suppressor? The duplicity of MicroRNAs in cancer. Cancer Res. 2016, 76, 3666–3670. [Google Scholar] [CrossRef] [PubMed]
- Anfossi, S.; Fu, X.; Nagvekar, R.; Calin, G.A. MicroRNAs, regulatory messengers inside and outside cancer cells. In Advances in Experimental Medicine and Biology Series: Exosomes, Stem Cells and MicroRNA; Springer: Cham, Switzerland, 2018; Volume 1056, pp. 87–108. [Google Scholar] [CrossRef]
- Lin, S.; Gregory, R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 2015, 15, 321–333. [Google Scholar] [CrossRef]
- Syeda, Z.A.; Langden, S.S.S.; Munkhzul, C.; Lee, M.; Song, S.J. Regulatory mechanism of microrna expression in cancer. Int. J. Mol. Sci. 2020, 21, 1723. [Google Scholar] [CrossRef]
- Stavast, C.J.; Erkeland, S.J. The Non-Canonical Aspects of MicroRNAs: Many Roads to Gene Regulation. Cells 2019, 8, 1465. [Google Scholar] [CrossRef]
- Ruby, J.G.; Jan, C.H.; Bartel, D.P. Intronic microRNA precursors that bypass Drosha processing. Nature 2007, 448, 83–86. [Google Scholar] [CrossRef]
- Xie, M.; Li, M.; Vilborg, A.; Lee, N.; Shu, M.-D.; Yartseva, V.; Šestan, N.; Steitz, J.A. Mammalian 5′-capped microRNA precursors that generate a single microRNA. Cell 2013, 155, 1568–1580. [Google Scholar] [CrossRef]
- Havens, M.A.; Reich, A.A.; Duelli, D.M.; Hastings, M.L. Biogenesis of mammalian microRNAs by a non-canonical processing pathway. Nucleic Acids Res. 2012, 40, 4626–4640. [Google Scholar] [CrossRef]
- Ahmad, J.; Hasnain, S.E.; Siddiqui, M.A.; Ahamed, M.; Musarrat, J.; Al-Khedhairy, A.A. MicroRNA in carcinogenesis & cancer diagnostics: A new paradigm. Indian J. Med. Res. 2013, 137, 680–694. [Google Scholar] [PubMed]
- Yang, J.S.; Maurin, T.; Lai, E.C. Functional parameters of Dicer-independent microRNA biogenesis. RNA 2012, 18, 945–957. [Google Scholar] [CrossRef] [PubMed]
- Yi, T.; Arthanari, H.; Akabayov, B.; Song, H.; Papadopoulos, E.; Qi, H.H.; Jedrychowski, M.; Güttler, T.; Guo, C.; Luna, R.E.; et al. EIF1A augments Ago2-mediated Dicer-independent miRNA biogenesis and RNA interference. Nat. Commun. 2015, 6, 7194. [Google Scholar] [CrossRef]
- Macias, S.; Cordiner, R.A.; Gautier, P.; Plass, M.; Cáceres, J.F. DGCR8 Acts as an Adaptor for the Exosome Complex to Degrade Double-Stranded Structured RNAs. Mol. Cell 2015, 60, 873–885. [Google Scholar] [CrossRef] [PubMed]
- Rnas, D.; Babiarz, J.E.; Ruby, J.G.; Wang, Y.; Bartel, D.P.; Blelloch, R. Mouse ES cells express endogenous shRNAs, siRNAs, and other. Genes Dev. 2008, 22, 2773–2785. [Google Scholar]
- Zhang, Y.; Deng, Q.; Tu, L.; Lv, D.; Liu, D. TRNA-derived small RNAs: A novel class of small RNAs in human hypertrophic scar fibroblasts. Int. J. Mol. Med. 2020, 45, 115–130. [Google Scholar] [CrossRef]
- Li, S.; Xu, Z.; Sheng, J. tRNA-derived small RNA: A novel regulatory small non-coding RNA. Genes 2018, 9, 246. [Google Scholar] [CrossRef]
- Anderson, P.; Ivanov, P. TRNA fragments in human health and disease. FEBS Lett. 2014, 588, 4297–4304. [Google Scholar] [CrossRef]
- Rubio, M.; Bustamante, M.; Hernandez-Ferrer, C.; Fernandez-Orth, D.; Pantano, L.; Sarria, Y.; Piqué-Borras, M.; Vellve, K.; Agramunt, S.; Carreras, R.; et al. Circulating miRNAs, isomiRs and small RNA clusters in human plasma and breast milk. PLoS ONE 2018, 13, e0193527. [Google Scholar] [CrossRef]
- Cui, M.; Wang, H.; Yao, X.; Zhang, D.; Xie, Y.; Cui, R.; Zhang, X. Circulating MicroRNAs in Cancer: Potential and Challenge. Front. Genet. 2019, 10, 626. [Google Scholar] [CrossRef] [PubMed]
- Piao, X.M.; Cha, E.J.; Yun, S.J.; Kim, W.J. Role of exosomal miRNA in bladder cancer: A promising liquid biopsy biomarker. Int. J. Mol. Sci. 2021, 22, 1713. [Google Scholar] [CrossRef] [PubMed]
- Sohel, M.H. Extracellular/Circulating MicroRNAs: Release Mechanisms, Functions and Challenges. Achiev. Life Sci. 2016, 10, 175–186. [Google Scholar] [CrossRef]
- Desgagné, V.; Bouchard, L.; Guérin, R. MicroRNAs in lipoprotein and lipid metabolism: From biological function to clinical application. Clin. Chem. Lab. Med. 2017, 55, 667–686. [Google Scholar] [CrossRef]
- Felekkis, K.; Papaneophytou, C. Challenges in using circulating micro-rnas as biomarkers for cardiovascular diseases. Int. J. Mol. Sci. 2020, 21, 561. [Google Scholar] [CrossRef] [PubMed]
- Kamal, N.N.S.B.N.M.; Shahidan, W.N.S. Non-exosomal and exosomal circulatory MicroRNAs: Which are more valid as biomarkers? Front. Pharmacol. 2020, 10, 1500. [Google Scholar] [CrossRef]
- Cheung, A.H.K.; Chow, C.; To, K.F. Latest development of liquid biopsy. J. Thorac. Dis. 2018, 10, S1645–S1651. [Google Scholar] [CrossRef]
- Janiszewska, M. The microcosmos of intratumor heterogeneity: The space-time of cancer evolution. Oncogene 2020, 39, 2031–2039. [Google Scholar] [CrossRef]
- Wang, H.; Peng, R.; Wang, J.; Qin, Z.; Xue, L. Circulating microRNAs as potential cancer biomarkers: The advantage and disadvantage. Clin. Epigenetics 2018, 10, 59. [Google Scholar] [CrossRef]
- Fan, T.; Mao, Y.; Sun, Q.; Liu, F.; Lin, J.S.; Liu, Y.; Cui, J.; Jiang, Y. Branched rolling circle amplification method for measuring serum circulating microRNA levels for early breast cancer detection. Cancer Sci. 2018, 109, 2897–2906. [Google Scholar] [CrossRef]
- Usuba, W.; Urabe, F.; Yamamoto, Y.; Matsuzaki, J.; Sasaki, H.; Ichikawa, M.; Takizawa, S.; Aoki, Y.; Niida, S.; Kato, K.; et al. Circulating miRNA panels for specific and early detection in bladder cancer. Cancer Sci. 2019, 110, 408–419. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Ding, Y.; Ma, Q.; Zhao, L.; Guo, X.; Shao, Y.; Niu, C.; He, Y.; Zhang, F.; Zheng, D.; et al. Identification of novel circulating miRNA biomarkers for the diagnosis of esophageal squamous cell carcinoma and squamous dysplasia. Cancer Epidemiol. Biomarkers Prev. 2019, 28, 1212–1220. [Google Scholar] [CrossRef] [PubMed]
- Yersal, O.; Barutca, S. Biological subtypes of breast cancer: Prognostic and therapeutic implications. World J. Clin. Oncol. 2014, 5, 412. [Google Scholar] [CrossRef] [PubMed]
- Eichelser, C.; Stückrath, I.; Müller, V.; Milde-Langosch, K.; Wikman, H.; Pantel, K.; Schwarzenbach, H. Increased serum levels of circulating exosomal microRNA-373 in receptor-negative breast cancer patients. Oncotarget 2014, 5, 9650–9663. [Google Scholar] [CrossRef] [PubMed]
- Bonneau, E.; Neveu, B.; Kostantin, E.; Tsongalis, G.J.; De Guire, V. How close are miRNAs from clinical practice? A perspective on the diagnostic and therapeutic market. Electron. J. Int. Fed. Clin. Chem. Lab. Med. 2019, 30, 114–127. [Google Scholar]
- Tiberio, P.; Callari, M.; Angeloni, V.; Daidone, M.G.; Appierto, V. Challenges in using circulating miRNAs as cancer biomarkers. Biomed Res. Int. 2015, 2015, 731479. [Google Scholar] [CrossRef]
- Xu, W.; Zhou, Y.; Xu, G.; Geng, B.; Cui, Q. Transcriptome analysis reveals non-identical microRNA profiles between arterial and venous plasma. Oncotarget 2017, 8, 28471–28480. [Google Scholar] [CrossRef]
- Jin, L.; Li, M.; Wang, H.; Yin, Z.; Chen, L.; Zhou, Y.; Han, Y.; Cui, Q.; Zhou, Y.; Xue, L. Transcriptome analysis of arterial and venous circulating miRNAs during hypertension. Sci. Rep. 2021, 11, 3469. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef]
- McDonald, J.S.; Milosevic, D.; Reddi, H.V.; Grebe, S.K.; Algeciras-Schimnich, A. Analysis of circulating microRNA: Preanalytical and analytical challenges. Clin. Chem. 2011, 57, 833–840. [Google Scholar] [CrossRef]
- Wang, K.; Yuan, Y.; Cho, J.H.; McClarty, S.; Baxter, D.; Galas, D.J. Comparing the MicroRNA spectrum between serum and plasma. PLoS ONE 2012, 7, e41561. [Google Scholar] [CrossRef] [PubMed]
- Mompeón, A.; Ortega-Paz, L.; Vidal-Gómez, X.; Costa, T.J.; Pérez-Cremades, D.; Garcia-Blas, S.; Brugaletta, S.; Sanchis, J.; Sabate, M.; Novella, S.; et al. Disparate miRNA expression in serum and plasma of patients with acute myocardial infarction: A systematic and paired comparative analysis. Sci. Rep. 2020, 10, 5373. [Google Scholar] [CrossRef] [PubMed]
- Mussbacher, M.; Krammer, T.L.; Heber, S.; Schrottmaier, W.C.; Zeibig, S.; Holthoff, H.P.; Pereyra, D.; Starlinger, P.; Hackl, M.; Assinger, A. Impact of Anticoagulation and Sample Processing on the Quantification of Human Blood-Derived microRNA Signatures. Cells 2020, 9, 1915. [Google Scholar] [CrossRef] [PubMed]
- Blondal, T.; Jensby Nielsen, S.; Baker, A.; Andreasen, D.; Mouritzen, P.; Wrang Teilum, M.; Dahlsveen, I.K. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods 2013, 59, S1–S6. [Google Scholar] [CrossRef]
- Faraldi, M.; Gomarasca, M.; Sansoni, V.; Perego, S.; Banfi, G.; Lombardi, G. Normalization strategies differently affect circulating miRNA profile associated with the training status. Sci. Rep. 2019, 9, 1584. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D. Single nucleotide alterations in MicroRNAs and human cancer-A not fully explored field. Non-Coding RNA Res. 2020, 5, 27–31. [Google Scholar] [CrossRef]
- Noren Hooten, N.; Fitzpatrick, M.; Wood, W.H.; De, S.; Ejiogu, N.; Zhang, Y.; Mattison, J.A.; Becker, K.G.; Zonderman, A.B.; Evans, M.K. Age-related changes in microRNA levels in serum. Aging 2013, 5, 725–740. [Google Scholar] [CrossRef]
- Duttagupta, R.; Jiang, R.; Gollub, J.; Getts, R.C.; Jones, K.W. Impact of cellular miRNAs on circulating miRNA biomarker signatures. PLoS ONE 2011, 6, e20769. [Google Scholar] [CrossRef] [PubMed]
- Chang, X.; Li, S.; Li, J.; Yin, L.; Zhou, T.; Zhang, C.; Chen, X.; Sun, K. Ethnic differences in microRNA-375 expression level and DNA methylation status in type 2 diabetes of Han and Kazak populations. J. Diabetes Res. 2014, 2014, 761938. [Google Scholar] [CrossRef]
- Dluzen, D.F.; Noren Hooten, N.; Zhang, Y.; Kim, Y.; Glover, F.E.; Tajuddin, S.M.; Jacob, K.D.; Zonderman, A.B.; Evans, M.K. Racial differences in microRNA and gene expression in hypertensive women. Sci. Rep. 2016, 6, 35815. [Google Scholar] [CrossRef]
- Searles, C.D.; Weber, M.; Baker, M.B.; Patel, R.S.; Quyyumi, A.A.; Bao, G. MicroRNA expression profile in CAD patients and the impact of ACEI/ARB. Cardiol. Res. Pract. 2011, 2011, 532915. [Google Scholar] [CrossRef]
- Dong, Z.; Jiang, H.; Jian, X.; Zhang, W. Change of miRNA expression profiles in patients with knee osteoarthritis before and after celecoxib treatment. J. Clin. Lab. Anal. 2019, 33, e22648. [Google Scholar] [CrossRef] [PubMed]
- Ferrero, G.; Carpi, S.; Polini, B.; Pardini, B.; Nieri, P.; Impeduglia, A.; Grioni, S.; Tarallo, S.; Naccarati, A. Intake of natural compounds and circulating microrna expression levels: Their relationship investigated in healthy subjects with different dietary habits. Front. Pharmacol. 2021, 11, 2214. [Google Scholar] [CrossRef] [PubMed]
- Cadieux, Z.; Lewis, H.; Esquela-Kerscher, A. Role of Nutrition, the Epigenome, and MicroRNAs in Cancer Pathogenesis. In RSC Drug Discovery Series: MicroRNAs in Diseases and Disorders: Emerging Therapeutic Targets; Royal Society of Chemistry: London, UK, 2019; pp. 1–35. [Google Scholar] [CrossRef]
- Macdonald-Ramos, K.; Martínez-Ibarra, A.; Monroy, A.; Miranda-Ríos, J.; Cerbón, M. Effect of dietary fatty acids on microrna expression related to metabolic disorders and inflammation in human and animal trials. Nutrients 2021, 13, 1830. [Google Scholar] [CrossRef]
- Li, F.; Bai, M.; Xu, J.; Zhu, L.; Liu, C.; Duan, R. Long-Term Exercise Alters the Profiles of Circulating Micro-RNAs in the Plasma of Young Women. Front. Physiol. 2020, 11, 372. [Google Scholar] [CrossRef]
- Kuji, T.; Sugasawa, T.; Fujita, S.; Ono, S.; Kawakami, Y.; Takekoshi, K. A Pilot Study of miRNA Expression Profile as a Liquid Biopsy for Full-Marathon Participants. Sports 2021, 9, 134. [Google Scholar] [CrossRef]
- Liu, H.; Xu, W.; Feng, J.; Ma, H.; Zhang, J.; Xie, X.; Zhuang, D.; Shen, W.; Liu, H.; Zhou, W. Increased Expression of Plasma miRNA-320a and let-7b-5p in Heroin-Dependent Patients and Its Clinical Significance. Front. Psychiatry 2021, 12, 679206. [Google Scholar] [CrossRef]
- Ignacio, C.; Hicks, S.D.; Burke, P.; Lewis, L.; Szombathyne-Meszaros, Z.; Middleton, F.A. Alterations in serum microRNA in humans with alcohol use disorders impact cell proliferation and cell death pathways and predict structural and functional changes in brain. BMC Neurosci. 2015, 16, 55. [Google Scholar] [CrossRef]
- Ibáñez, F.; Ureña-Peralta, J.R.; Costa-Alba, P.; Torres, J.L.; Laso, F.J.; Marcos, M.; Guerri, C.; Pascual, M. Circulating micrornas in extracellular vesicles as potential biomarkers of alcohol-induced neuroinflammation in adolescence: Gender differences. Int. J. Mol. Sci. 2020, 21, 6730. [Google Scholar] [CrossRef]
- Huang, J.; Wu, J.; Li, Y.; Li, X.; Yang, T.; Yang, Q.; Jiang, Y. Deregulation of serum MicroRNA expression is associated with cigarette smoking and lung cancer. Biomed Res. Int. 2014, 2014, 344316. [Google Scholar] [CrossRef]
- Takahashi, K.; Yokota, S.-I.; Tatsumi, N.; Fukami, T.; Yokoi, T.; Nakajima, M. Cigarette smoking substantially alters plasma microRNA profiles in healthy subjects. Toxicol. Appl. Pharmacol. 2013, 272, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Morin, R.D.; O’Connor, M.D.; Griffith, M.; Kuchenbauer, F.; Delaney, A.; Prabhu, A.L.; Zhao, Y.; McDonald, H.; Zeng, T.; Hirst, M.; et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008, 18, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Sticht, C.; De La Torre, C.; Parveen, A.; Gretz, N. miRWalk: An online resource for prediction of microRNA binding sites. PLoS ONE 2018, 13, e0206239. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019, 20, 18. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Kim, S.; Park, M.; Kim, J.Y.; Kim, T.; Hwang, J.Y.; Ha, K.S.; Won, M.H.; Ryoo, S.; Kwon, Y.G.; Kim, Y.M. Circulating miRNAs Associated with Dysregulated Vascular and Trophoblast Function as Target-Based Diagnostic Biomarkers for Preeclampsia. Cells 2020, 9, 2003. [Google Scholar] [CrossRef]
- Ponsuksili, S.; Tesfaye, D.; Schellander, K.; Hoelker, M.; Hadlich, F.; Schwerin, M.; Wimmers, K. Differential expression of miRNAs and their target mRNAs in endometria prior to maternal recognition of pregnancy associates with endometrial receptivity for in vivo- and in vitro-produced bovine embryos. Biol. Reprod. 2014, 91, 135. [Google Scholar] [CrossRef]
- Shi, S.; Tan, Q.; Liang, J.; Cao, D.; Wang, S.; Liang, J.; Chen, K.; Wang, Z. Placental trophoblast cell-derived exosomal microRNA-1290 promotes the interaction between endometrium and embryo by targeting LHX6. Mol. Ther. Nucleic Acids 2021, 26, 760–772. [Google Scholar] [CrossRef]
- Rahman, M.L.; Liang, L.; Valeri, L.; Su, L.; Zhu, Z.; Gao, S.; Mostofa, G.; Qamruzzaman, Q.; Hauser, R.; Baccarelli, A.; et al. Regulation of birthweight by placenta-derived miRNAs: Evidence from an arsenic-exposed birth cohort in Bangladesh. Epigenetics 2018, 13, 573–590. [Google Scholar] [CrossRef] [PubMed]
- Moore, D.; Meays, B.M.; Madduri, L.S.V.; Shahjin, F.; Chand, S.; Niu, M.; Albahrani, A.; Guda, C.; Pendyala, G.; Fox, H.S.; et al. Downregulation of an Evolutionary Young miR-1290 in an iPSC-Derived Neural Stem Cell Model of Autism Spectrum Disorder. Stem Cells Int. 2019, 2019, 8710180. [Google Scholar] [CrossRef] [PubMed]
- Yelamanchili, S.V.; Morsey, B.; Harrison, E.B.; Rennard, D.A.; Emanuel, K.; Thapa, I.; Bastola, D.R.; Fox, H.S. The evolutionary young miR-1290 favors mitotic exit and differentiation of human neural progenitors through altering the cell cycle proteins. Cell Death Dis. 2014, 5, e982. [Google Scholar] [CrossRef] [PubMed]
- Zbucka-Kretowska, M.; Niemira, M.; Paczkowska-Abdulsalam, M.; Bielska, A.; Szalkowska, A.; Parfieniuk, E.; Ciborowski, M.; Wolczynski, S.; Kretowski, A. Prenatal circulating microRNA signatures of foetal Down syndrome. Sci. Rep. 2019, 9, 2394. [Google Scholar] [CrossRef] [PubMed]
- Che, J.; Xu, C.; Wu, Y.; Jia, P.; Han, Q.; Ma, Y.; Wang, X.; Zheng, Y. MiR-1290 promotes myoblast differentiation and protects against myotube atrophy via Akt/p70/FoxO3 pathway regulation. Skelet. Muscle 2021, 11, 6. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Chen, L.; Feng, Z.; Chen, W.; Yan, S.; Yang, R.; Xiao, J.; Gao, J.; Zhang, D.; Ke, X. EPC-Derived Exosomal miR-1246 and miR-1290 Regulate Phenotypic Changes of Fibroblasts to Endothelial Cells to Exert Protective Effects on Myocardial Infarction by Targeting ELF5 and SP1. Front. Cell Dev. Biol. 2021, 9, 9356. [Google Scholar] [CrossRef]
- Yue, K.Y.; Zhang, P.R.; Zheng, M.H.; Cao, X.L.; Cao, Y.; Zhang, Y.Z.; Zhang, Y.F.; Wu, H.N.; Lu, Z.H.; Liang, L.; et al. Neurons can upregulate Cav-1 to increase intake of endothelial cells-derived extracellular vesicles that attenuate apoptosis via miR-1290. Cell Death Dis. 2019, 10, 869. [Google Scholar] [CrossRef]
- Sun, H.; Hu, S.; Zhang, Z.; Lun, J.; Liao, W.; Zhang, Z. Expression of exosomal microRNAs during chondrogenic differentiation of human bone mesenchymal stem cells. J. Cell. Biochem. 2019, 120, 171–181. [Google Scholar] [CrossRef]
- Cui, L.; Zhou, X.; Li, J.; Wang, L.; Wang, J.; Li, Q.; Chu, J.; Zheng, L.; Wu, Q.; Han, Z.; et al. Dynamic microRNA Profiles of Hepatic Differentiated Human Umbilical Cord Lining-Derived Mesenchymal Stem Cells. PLoS ONE 2012, 7, e44737. [Google Scholar] [CrossRef]
- Zhou, X.; Cui, L.; Zhou, X.; Yang, Q.; Wang, L.; Guo, G.; Hou, Y.; Cai, W.; Han, Z.; Shi, Y.; et al. Induction of hepatocyte-like cells from human umbilical cord-derived mesenchymal stem cells by defined microRNAs. J. Cell. Mol. Med. 2017, 21, 881–893. [Google Scholar] [CrossRef]
- Zhuang, H.; Zhang, R.; Zhang, S.; Shu, Q.; Zhang, D.; Xu, G. Altered expression of microRNAs in the neuronal differentiation of human Wharton’s Jelly mesenchymal stem cells. Neurosci. Lett. 2015, 600, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Sang, Q.; Yao, Z.; Wang, H.; Feng, R.; Wang, H.; Zhao, X.; Xing, Q.; Jin, L.; He, L.; Wu, L.; et al. Identification of microRNAs in human follicular fluid: Characterization of microRNAs that govern steroidogenesis in vitro and are associated with polycystic ovary syndrome in vivo. J. Clin. Endocrinol. Metab. 2013, 98, 3068–3079. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wen, Q.; Chen, H.; Wu, X.; Liu, B.; Li, H.; Su, L.; Tong, H. Exosomes Derived From Heat Stroke Cases Carry miRNAs Associated With Inflammation and Coagulation Cascade. Front. Immunol. 2021, 12, 2149. [Google Scholar] [CrossRef]
- Xu, H.; Cui, Y.; Liu, X.; Zheng, X.; Liu, J.; Hu, X.; Gao, F.; Hu, X.; Li, M.; Wei, X.; et al. miR-1290 promotes IL-8-mediated vascular endothelial cell adhesion by targeting GSK-3β. Mol. Biol. Rep. 2021. [Google Scholar] [CrossRef]
- Di Mauro, S.; Ragusa, M.; Urbano, F.; Filippello, A.; Di Pino, A.; Scamporrino, A.; Pulvirenti, A.; Ferro, A.; Rabuazzo, A.M.; Purrello, M.; et al. Intracellular and extracellular miRNome deregulation in cellular models of NAFLD or NASH: Clinical implications. Nutr. Metab. Cardiovasc. Dis. 2016, 26, 1129–1139. [Google Scholar] [CrossRef]
- Tan, Y.; Ge, G.; Pan, T.; Wen, D.; Gan, J. A pilot study of serum microRNAs panel as potential biomarkers for diagnosis of nonalcoholic fatty liver disease. PLoS ONE 2014, 9, e105192. [Google Scholar] [CrossRef]
- Ng, P.C.; Chan, K.Y.Y.; Leung, K.T.; Tam, Y.H.; Ma, T.P.Y.; Lam, H.S.; Cheung, H.M.; Lee, K.H.; To, K.F.; Li, K.; et al. Comparative MiRNA Expressional Profiles and Molecular Networks in Human Small Bowel Tissues of Necrotizing Enterocolitis and Spontaneous Intestinal Perforation. PLoS ONE 2015, 10, e0135737. [Google Scholar] [CrossRef]
- Ng, P.C.; Chan, K.Y.Y.; Yuen, T.P.; Sit, T.; Lam, H.S.; Leung, K.T.; Wong, R.P.O.; Chan, L.C.N.; Pang, Y.L.I.; Cheung, H.M.; et al. Plasma miR-1290 Is a Novel and Specific Biomarker for Early Diagnosis of Necrotizing Enterocolitis-Biomarker Discovery with Prospective Cohort Evaluation. J. Pediatr. 2019, 205, 83–90.e10. [Google Scholar] [CrossRef]
- Ma, Z.X.; Tan, X.; Shen, Y.; Ke, X.; Yang, Y.C.; He, X.B.; Wang, Z.H.; Dai, Y.B.; Hong, S.L.; Hu, G.H. MicroRNA expression profile of mature dendritic cell in chronic rhinosinusitis. Inflamm. Res. 2015, 64, 885–893. [Google Scholar] [CrossRef] [PubMed]
- Vaira, V.; Roncoroni, L.; Barisani, D.; Gaudioso, G.; Bosari, S.; Bulfamante, G.; Doneda, L.; Conte, D.; Tomba, C.; Bardella, M.T.; et al. microRNA profiles in coeliac patients distinguish different clinical phenotypes and are modulated by gliadin peptides in primary duodenal fibroblasts. Clin. Sci. 2014, 126, 417–423. [Google Scholar] [CrossRef]
- Guan, S.; Wu, Y.; Zhang, Q.; Zhou, J. TGF-β1 induces CREB1-mediated miR-1290 upregulation to antagonize lung fibrosis via Napsin A. Int. J. Mol. Med. 2020, 46, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Chickooree, D.; Zhu, K.; Ram, V.; Wu, H.J.; He, Z.J.; Zhang, S. A preliminary microarray assay of the miRNA expression signatures in buccal mucosa of oral submucous fibrosis patients. J. Oral Pathol. Med. 2016, 45, 691–697. [Google Scholar] [CrossRef]
- Adyshev, D.M.; Moldobaeva, N.; Mapes, B.; Elangovan, V.; Garcia, J.G.N. MicroRNA regulation of nonmuscle myosin light chain kinase expression in human lung endothelium. Am. J. Respir. Cell Mol. Biol. 2013, 49, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Ahmed, M.M.; Hasan, P.M.Z.; Sharma, A.; Bilgrami, A.L.; Manda, K.; Ishrat, R.; Syed, M.A. Identification and Validation of Potential miRNAs, as Biomarkers for Sepsis and Associated Lung Injury: A Network-Based Approach. Genes 2020, 11, 1327. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhou, W.; Li, Z.; Guan, F. Hsa_circ_0056558 regulates cyclin-dependent kinase 6 by sponging microRNA-1290 to suppress the proliferation and differentiation in ankylosing spondylitis. Autoimmunity 2021, 54, 114–128. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.Y.; Huang, C.H.; Chen, C.J.; Chen, T.W.; Lin, C.Y.; Lin, Y.-T.; Kuo, S.M.; Huang, C.G.; Lee, L.A.; Chen, Y.H.; et al. Novel Role for miR-1290 in Host Species Specificity of Influenza A Virus. Mol. Ther.-Nucleic Acids 2019, 17, 10–23. [Google Scholar] [CrossRef]
- Wang, P.; Qu, X.; Zhou, X.; Shen, Y.; Ji, H.; Fu, Z.; Deng, J.; Lu, P.; Yu, W.; Lu, H.; et al. Two cellular microRNAs, miR-196b and miR-1290, contribute to HIV-1 latency. Virology 2015, 486, 228–238. [Google Scholar] [CrossRef]
- Soares, C.T.; Trombone, A.P.F.; Fachin, L.R.V.; Rosa, P.S.; Ghidella, C.C.; Ramalho, R.F.; Pinilla, M.G.; Carvalho, A.F.; Carrara, D.N.; Soares, F.A.; et al. Differential Expression of MicroRNAs in Leprosy Skin Lesions. Front. Immunol. 2017, 8, 1053. [Google Scholar] [CrossRef]
- Sørensen, A.E.; Udesen, P.B.; Maciag, G.; Geiger, J.; Saliani, N.; Januszewski, A.S.; Jiang, G.; Ma, R.C.; Hardikar, A.A.; Wissing, M.L.M.; et al. Hyperandrogenism and Metabolic Syndrome Are Associated With Changes in Serum-Derived microRNAs in Women With Polycystic Ovary Syndrome. Front. Med. 2019, 6, 242. [Google Scholar] [CrossRef]
- Ye, L.; Jiang, T.; Shao, H.; Zhong, L.; Wang, Z.; Liu, Y.; Tang, H.; Qin, B.; Zhang, X.; Fan, J. miR-1290 Is a Biomarker in DNA-Mismatch-Repair-Deficient Colon Cancer and Promotes Resistance to 5-Fluorouracil by Directly Targeting hMSH2. Mol. Ther.-Nucleic Acids 2017, 7, 453–464. [Google Scholar] [CrossRef]
- Liu, X.; Xu, X.; Pan, B.; He, B.; Chen, X.; Zeng, K.; Xu, M.; Pan, Y.; Sun, H.; Xu, T.; et al. Circulating miR-1290 and miR-320d as novel diagnostic biomarkers of human colorectal cancer. J. Cancer 2019, 10, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Zhuang, Y.; Zhang, J.; Chen, M.; Wu, S. Four circulating exosomal miRNAs as novel potential biomarkers for the early diagnosis of human colorectal cancer. Tissue Cell 2021, 70, 101499. [Google Scholar] [CrossRef] [PubMed]
- Imaoka, H.; Toiyama, Y.; Fujikawa, H.; Hiro, J.; Saigusa, S.; Tanaka, K.; Inoue, Y.; Mohri, Y.; Mori, T.; Kato, T.; et al. Circulating microRNA-1290 as a novel diagnostic and prognostic biomarker in human colorectal cancer. Ann. Oncol. 2016, 27, 1879–1886. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Yu, J.; Kim, H.; Wolfgang, C.L.; Canto, M.I.; Hruban, R.H.; Goggins, M. MicroRNA array analysis finds elevated serum miR-1290 accurately distinguishes patients with low-stage pancreatic cancer from healthy and disease controls. Clin. Cancer Res. 2013, 19, 3600–3610. [Google Scholar] [CrossRef]
- Tavano, F.; Gioffreda, D.; Valvano, M.R.; Palmieri, O.; Tardio, M.; Latiano, T.P.; Piepoli, A.; Maiello, E.; Pirozzi, F.; Andriulli, A. Droplet digital PCR quantification of miR-1290 as a circulating biomarker for pancreatic cancer. Sci. Rep. 2018, 8, 16389. [Google Scholar] [CrossRef]
- Wei, J.; Yang, L.; Wu, Y.N.; Xu, J. Serum miR-1290 and miR-1246 as Potential Diagnostic Biomarkers of Human Pancreatic Cancer. J. Cancer 2020, 11, 1325–1333. [Google Scholar] [CrossRef]
- Xie, R.; Wu, S.N.; Gao, C.C.; Yang, X.Z.; Wang, H.G.; Zhang, J.L.; Yan, W.; Ma, T.H. Prognostic value of combined and individual expression of microRNA-1290 and its target gene nuclear factor I/X in human esophageal squamous cell carcinoma. Cancer Biomark. 2017, 20, 325–331. [Google Scholar] [CrossRef]
- Mo, D.; Gu, B.; Gong, X.; Wu, L.; Wang, H.; Jiang, Y.; Zhang, B.; Zhang, M.; Zhang, Y.; Xu, J.; et al. miR-1290 is a potential prognostic biomarker in non-small cell lung cancer. J. Thorac. Dis. 2015, 7, 1570–1579. [Google Scholar] [CrossRef]
- Wu, Y.; Wei, J.; Zhang, W.; Xie, M.; Wang, X.; Xu, J. Serum exosomal miR-1290 is a potential biomarker for lung adenocarcinoma. Onco. Targets. Ther. 2020, 13, 7809–7818. [Google Scholar] [CrossRef]
- Nagamitsu, Y.; Nishi, H.; Sasaki, T.; Takaesu, Y.; Terauchi, F.; Isaka, K. Profiling analysis of circulating microRNA expression in cervical cancer. Mol. Clin. Oncol. 2016, 5, 189–194. [Google Scholar] [CrossRef]
- Torres, A.; Torres, K.; Pesci, A.; Ceccaroni, M.; Paszkowski, T.; Cassandrini, P.; Zamboni, G.; MacIejewski, R. Diagnostic and prognostic significance of miRNA signatures in tissues and plasma of endometrioid endometrial carcinoma patients. Int. J. Cancer 2013, 132, 1633–1645. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Sawada, K.; Nakamura, K.; Yoshimura, A.; Miyamoto, M.; Shimizu, A.; Ishida, K.; Nakatsuka, E.; Kodama, M.; Hashimoto, K.; et al. Exosomal miR-1290 is a potential biomarker of high-grade serous ovarian carcinoma and can discriminate patients from those with malignancies of other histological types 11 Medical and Health Sciences 1112 Oncology and Carcinogenesis. J. Ovarian Res. 2018, 11, 85. [Google Scholar] [CrossRef] [PubMed]
- Záveský, L.; Jandáková, E.; Weinberger, V.; Minář, L.; Hanzíková, V.; Dušková, D.; Záveská Drábková, L.; Svobodová, I.; Hořínek, A. Ascites-Derived Extracellular microRNAs as Potential Biomarkers for Ovarian Cancer. Reprod. Sci. 2019, 26, 510–522. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.M.; Liu, Z.Z.; Lin, J.N. Prognostic value of plasma miR-1290 expression in patients with oral squamous cell carcinoma. Hua Xi Kou Qiang Yi Xue Za Zhi 2020, 38, 371–375. [Google Scholar] [CrossRef]
- Nakashima, H.; Yoshida, R.; Hirosue, A.; Kawahara, K.; Sakata, J.; Arita, H.; Yamamoto, T.; Toya, R.; Murakami, R.; Hiraki, A.; et al. Circulating miRNA-1290 as a potential biomarker for response to chemoradiotherapy and prognosis of patients with advanced oral squamous cell carcinoma: A single-center retrospective study. Tumor Biol. 2019, 41, 1010428319826853. [Google Scholar] [CrossRef]
- Mori, G.; Pasca, M.R. Gut microbial signatures in sporadic and hereditary colorectal cancer. Int. J. Mol. Sci. 2021, 22, 1312. [Google Scholar] [CrossRef]
- Sawicki, T.; Ruszkowska, M.; Danielewicz, A. A Review of Colorectal Cancer in Terms of Epidemiology, Risk Factors, Development, Symptoms and Diagnosis. Cancers 2021, 13, 2025. [Google Scholar] [CrossRef]
- Rawla, P.; Sunkara, T.; Barsouk, A. Epidemiology of colorectal cancer: Incidence, mortality, survival, and risk factors. Prz. Gastroenterol. 2019, 14, 89–103. [Google Scholar] [CrossRef]
- De’angelis, G.L.; Bottarelli, L.; Azzoni, C.; De’angelis, N.; Leandro, G.; Di Mario, F.; Gaiani, F.; Negri, F. Microsatellite instability in colorectal cancer. Acta Biomed. 2018, 17, 159. [Google Scholar]
- Vacante, M.; Ciuni, R.; Basile, F.; Biondi, A. Gut microbiota and colorectal cancer development: A closer look to the adenoma-carcinoma sequence. Biomedicines 2020, 8, 489. [Google Scholar] [CrossRef]
- Stigliano, V.; Sanchez-Mete, L.; Martayan, A.; Anti, M. Early-onset colorectal cancer: A sporadic or inherited disease? World J. Gastroenterol. 2014, 20, 12420. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Wang, Y.; Zhang, H.; Wang, F. miR-1290 contributes to colorectal cancer cell proliferation by targeting INPP4B. Oncol. Res. 2018, 26, 1167–1174. [Google Scholar] [CrossRef] [PubMed]
- Zhai, S.; Liu, Y.; Lu, X.; Qian, H.; Tang, X.; Cheng, X.; Wang, Y.; Shi, Y.; Deng, X. INPP4B as a prognostic and diagnostic marker regulates cell growth of pancreatic cancer via activating AKT. Onco. Targets. Ther. 2019, ume 12, 8287–8299. [Google Scholar] [CrossRef]
- Hodgson, M.C.; Shao, L.J.; Frolov, A.; Li, R.; Peterson, L.E.; Ayala, G.; Ittmann, M.M.; Weigel, N.L.; Agoulnik, I.U. Decreased expression and androgen regulation of the tumor suppressor gene INPP4B in prostate cancer. Cancer Res. 2011, 71, 572–582. [Google Scholar] [CrossRef] [PubMed]
- Vo, T.T.T.; Fruman, D.A. INPP4B Is a Tumor Suppressor in the Context of PTEN Deficiency. Cancer Discov. 2015, 5, 697–700. [Google Scholar] [CrossRef][Green Version]
- Hsu, I.; Yeh, C.R.; Slavin, S.; Miyamoto, H.; Netto, G.J.; Tsai, Y.C.; Muyan, M.; Wu, X.R.; Messing, E.M.; Guancial, E.A.; et al. Estrogen receptor alpha prevents bladder cancer development via INPP4B inhibited Akt pathway in vitro and in vivo. Oncotarget 2014, 5, 7917–7935. [Google Scholar] [CrossRef]
- Wu, J.; Ji, X.; Zhu, L.; Jiang, Q.; Wen, Z.; Xu, S.; Shao, W.; Cai, J.; Du, Q.; Zhu, Y.; et al. Up-regulation of microRNA-1290 impairs cytokinesis and affects the reprogramming of colon cancer cells. Cancer Lett. 2013, 329, 155–163. [Google Scholar] [CrossRef]
- Kheirelseid, E.A.H.; Miller, N.; Chang, K.H.; Curran, C.; Hennessey, E.; Sheehan, M.; Kerin, M.J. Mismatch repair protein expression in colorectal cancer. J. Gastrointest. Oncol. 2013, 4, 397–408. [Google Scholar] [CrossRef]
- Nojadeh, J.N.; Sharif, S.B.; Sakhinia, E. Microsatellite instability in colorectal cancer. EXCLI J. 2018, 17, 159. [Google Scholar]
- Wheeler, J.M.D.; Bodmer, W.F.; McC Mortensen, N.J. DNA mismatch repair genes and colorectal cancer. Gut 2000, 47, 148–153. [Google Scholar] [CrossRef]
- Pehserl, A.M.; Ress, A.L.; Stanzer, S.; Resel, M.; Karbiener, M.; Stadelmeyer, E.; Stiegelbauer, V.; Gerger, A.; Mayr, C.; Scheideler, M.; et al. Comprehensive analysis of miRNome alterations in response to sorafenib treatment in colorectal cancer cells. Int. J. Mol. Sci. 2016, 17, 2011. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.; Petrillo, A.; Smyth, E.C.; Shaida, N.; Khwaja, S.; Cheow, H.; Duckworth, A.; Heister, P.; Praseedom, R.; Jah, A.; et al. Colorectal liver metastases: Current management and future perspectives. World J. Clin. Oncol. 2020, 11, 761–808. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, H.; Sun, L.; Shen, S.; Zhou, Q.; Yuan, Y.; Xing, C. Epigenetic Alternations of MicroRNAs and DNA Methylation Contribute to Liver Metastasis of Colorectal Cancer. Dig. Dis. Sci. 2019, 64, 1523–1534. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Wei, Y.; Wei, X. AXL receptor tyrosine kinase as a promising anti-cancer approach: Functions, molecular mechanisms and clinical applications. Mol. Cancer 2019, 18, 153. [Google Scholar] [CrossRef]
- Li, M.; An, W.; Xu, L.; Lin, Y.; Su, L.; Liu, X. The arginine methyltransferase PRMT5 and PRMT1 distinctly regulate the degradation of anti-apoptotic protein CFLAR L in human lung cancer cells. J. Exp. Clin. Cancer Res. 2019, 38, 64. [Google Scholar] [CrossRef]
- Zhang, Z.; Zheng, F.; You, Y.; Ma, Y.; Lu, T.; Yue, W.; Zhang, D. Growth arrest specific gene 7 is associated with schizophrenia and regulates neuronal migration and morphogenesis. Mol. Brain 2016, 9, 54. [Google Scholar] [CrossRef]
- Guerra, C.; Molinari, M. Thioredoxin-Related Transmembrane Proteins: TMX1 and Little Brothers TMX2, TMX3, TMX4 and TMX5. Cells 2020, 9, 2000. [Google Scholar] [CrossRef]
- Pellino, G.; Gallo, G.; Pallante, P.; Capasso, R.; De Stefano, A.; Maretto, I.; Malapelle, U.; Qiu, S.; Nikolaou, S.; Barina, A.; et al. Noninvasive biomarkers of colorectal cancer: Role in diagnosis and personalised treatment perspectives. Gastroenterol. Res. Pract. 2018, 2018, 2397863. [Google Scholar] [CrossRef]
- Capasso, M.; Franceschi, M.; Rodriguez-Castro, K.I.; Crafa, P.; Cambiè, G.; Miraglia, C.; Barchi, A.; Nouvenne, A.; Leandro, G.; Meschi, T.; et al. Epidemiology and risk factors of pancreatic cancer. Acta Biomed. 2018, 89, 141–146. [Google Scholar]
- Ying, H.; Dey, P.; Yao, W.; Kimmelman, A.C.; Draetta, G.F.; Maitra, A.; Depinho, R.A. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2016, 30, 355–385. [Google Scholar] [CrossRef]
- McGuigan, A.; Kelly, P.; Turkington, R.C.; Jones, C.; Coleman, H.G.; McCain, R.S. Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes. World J. Gastroenterol. 2018, 24, 4846–4861. [Google Scholar] [CrossRef] [PubMed]
- Sikdar, N.; Saha, G.; Dutta, A.; Ghosh, S.; Shrikhande, S.V.; Banerjee, S. Genetic Alterations of Periampullary and Pancreatic Ductal Adenocarcinoma: An Overview. Curr. Genom. 2018, 19, 444–463. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Roberts, N.J.; Klein, A.P. Inherited pancreatic cancer. Chin. Clin. Oncol. 2017, 6, 58. [Google Scholar] [CrossRef]
- Ta, N.; Huang, X.; Zheng, K.; Zhang, Y.; Gao, Y.; Deng, L.; Zhang, B.; Jiang, H.; Zheng, J. MiRNA-1290 promotes aggressiveness in pancreatic ductal adenocarcinoma by targeting IKK1. Cell. Physiol. Biochem. 2018, 51, 711–728. [Google Scholar] [CrossRef] [PubMed]
- Kabacaoglu, D.; Ruess, D.A.; Ai, J.; Algül, H. NF-kB/rel transcription factors in pancreatic cancer: Focusing on relA, c-rel, and relB. Cancers 2019, 11, 937. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Chen, Z.; Zhu, F.; Hu, Y. IκB kinase alpha and cancer. J. Interf. Cytokine Res. 2012, 34, 152–158. [Google Scholar] [CrossRef]
- Zhao, F.; Wei, C.; Cui, M.Y.; Xia, Q.Q.; Wang, S.-B.; Zhang, Y. Prognostic value of microRNAs in pancreatic cancer: A meta-analysis. Aging 2020, 12, 9380–9404. [Google Scholar] [CrossRef]
- Karasek, P.; Gablo, N.; Hlavsa, J.; Kiss, I.; Vychytilova-Faltejskova, P.; Hermanova, M.; Kala, Z.; Slaby, O.; Prochazka, V. Pre-operative plasma miR-21-5p is a sensitive biomarker and independent prognostic factor in patients with pancreatic ductal adenocarcinoma undergoing surgical resection. Cancer Genom. Proteom. 2018, 15, 321–327. [Google Scholar] [CrossRef]
- Masamune, A.; Yoshida, N.; Hamada, S.; Takikawa, T.; Nabeshima, T.; Shimosegawa, T. Exosomes derived from pancreatic cancer cells induce activation and profibrogenic activities in pancreatic stellate cells. Biochem. Biophys. Res. Commun. 2018, 495, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Sitarz, R.; Skierucha, M.; Mielko, J.; Offerhaus, G.J.A.; Maciejewski, R.; Polkowski, W.P. Gastric cancer: Epidemiology, prevention, classification, and treatment. Cancer Manag. Res. 2018, ume 10, 239–248. [Google Scholar] [CrossRef]
- Cheng, X.J.; Lin, J.C.; Tu, S.P. Etiology and Prevention of Gastric Cancer. Gastrointest. Tumors 2016, 3, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Katona, B.W.; Rustgi, A.K. Gastric Cancer Genomics: Advances and Future Directions. Cell. Mol. Gastroenterol. Hepatol. 2017, 3, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Laurén, P. The two histological main types of gastric carcinoma: Diffuse and so-called intestinal-type carcinoma. Acta Pathol. Microbiol. Scand. 1965, 64, 31–49. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; El Hajj, N.; Sittler, S.; Lammert, N.; Barnes, R.; Meloni-Ehrig, A. Gastric cancer: Classification, histology and application of molecular pathology. J. Gastrointest. Oncol. 2012, 3, 251–261. [Google Scholar] [PubMed]
- Cisło, M.; Filip, A.A.; Offerhaus, G.J.A.; Ciseł, B.; Rawicz-Pruszyński, K.; Skierucha, M.; Polkowski, W.P. Distinct molecular subtypes of gastric cancer: From Laurén to molecular pathology. Oncotarget 2018, 9, 19427–19442. [Google Scholar] [CrossRef] [PubMed]
- Ansari, S.; Gantuya, B.; Tuan, V.P.; Yamaoka, Y. Diffuse gastric cancer: A summary of analogous contributing factors for its molecular pathogenicity. Int. J. Mol. Sci. 2018, 19, 2424. [Google Scholar] [CrossRef]
- Quadri, H.S.; Smaglo, B.G.; Morales, S.J.; Phillips, A.C.; Martin, A.D.; Chalhoub, W.M.; Haddad, N.G.; Unger, K.R.; Levy, A.D.; Al-Refaie, W.B. Gastric Adenocarcinoma: A Multimodal Approach. Front. Surg. 2017, 4, 42. [Google Scholar] [CrossRef]
- Lott, P.C.; Carvajal-Carmona, L.G. Resolving gastric cancer aetiology: An update in genetic predisposition. Lancet Gastroenterol. Hepatol. 2018, 3, 874–883. [Google Scholar] [CrossRef]
- Lin, M.; Shi, C.; Lin, X.; Pan, J.; Shen, S.; Xu, Z.; Chen, Q. SMicroRNA-1290 inhibits cells proliferation and migration by targeting FOXA1 in gastric cancer cells. Gene 2016, 582, 137–142. [Google Scholar] [CrossRef]
- Ren, H.; Zhang, P.; Tang, Y.; Wu, M.; Zhang, W. Forkhead box protein A1 is a prognostic predictor and promotes tumor growth of gastric cancer. Onco. Targets. Ther. 2015, 8, 3029–3039. [Google Scholar] [CrossRef]
- Lin, M.; Pan, J.; Chen, Q.; Xu, Z.; Lin, X.; Shi, C. Overexpression of FOXA1 inhibits cell proliferation and EMT of human gastric cancer AGS cells. Gene 2018, 642, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Shen, M.; Yan, M.; Cui, Y.; Gao, Z.; Meng, X. Exosome-mediated transfer of MIR-1290 promotes cell proliferation and invasion in gastric cancer via NKD1. Acta Biochim. Biophys. Sin. 2019, 51, 900–907. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Zheng, Q.; Wu, H.; Wang, C.; Liu, T.; Zhou, W. miR-532 promoted gastric cancer migration and invasion by targeting NKD1. Life Sci. 2017, 177, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Liu, Y.; Zhang, Q.; Zhang, H.; Du, J. Tumor-derived extracellular vesicles containing microRNA-1290 promote immune escape of cancer cells through the Grhl2/ZEB1/PD-L1 axis in gastric cancer. Transl. Res. 2021, 231, 102–112. [Google Scholar] [CrossRef]
- Barsouk, A.; Thandra, K.C.; Saginala, K.; Rawla, P.; Barsouk, A. Chemical Risk Factors of Primary Liver Cancer: An Update. Hepatic Med. Evid. Res. 2021, 12, 179–188. [Google Scholar] [CrossRef]
- Ghouri, Y.A.; Mian, I.; Rowe, J.H. Review of hepatocellular carcinoma: Epidemiology, etiology, and carcinogenesis. J. Carcinog. 2017, 16, 1–8. [Google Scholar] [CrossRef]
- Gelband, H.; Jha, P.; Sankaranarayanan, R.; Horton, S. Cancer: Disease Control Priorities, 3rd ed.; The International Bank for Reconstruction and Development/The World Bank: Washington, DC, USA, 2015; Volume 3. [Google Scholar] [CrossRef]
- Savitha, G.; Vishnupriya, V.; Krishnamohan, S. Hepatocellular carcinoma—A review. J. Pharm. Sci. Res. 2017, 9, 1276. [Google Scholar]
- Rao, C.V.; Asch, A.S.; Yamada, H.Y. Frequently mutated genes/pathways and genomic instability as prevention targets in liver cancer. Carcinogenesis 2017, 38, 2–11. [Google Scholar] [CrossRef]
- Feng, M.; Pan, Y.; Kong, R.; Shu, S. Therapy of Primary Liver Cancer. Innovation 2020, 1, 100032. [Google Scholar] [CrossRef]
- Liu, L.J.; Xie, S.X.; Chen, Y.T.; Xue, J.L.; Zhang, C.J.; Zhu, F. Aberrant regulation of WNT signaling in hepatocellular carcinoma. World J. Gastroenterol. 2016, 22, 7486–7499. [Google Scholar] [CrossRef]
- Zhang, Q.; Wu, J.; Bai, X.; Liang, T. Evaluation of Intra-Tumoral Vascularization in Hepatocellular Carcinomas. Front. Med. 2020, 7, 584250. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wang, G.; Niu, L.; Zhao, S.; Li, J.; Zhang, Z.; Jiang, H.; Zhang, Q.; Wang, H.; Sun, P.; et al. Exosomal MiR-1290 Promotes Angiogenesis of Hepatocellular Carcinoma via Targeting SMEK1. J. Oncol. 2021, 2021, 6617700. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.R.; Park, S.; Han, S.; Ahn, J.H.; Kim, S.; Sinn, D.H.; Jeong, W.K.; Ko, J.S.; Gwak, M.S.; Kim, G.S. Sarcopenia as a predictor of post-transplant tumor recurrence after living donor liver transplantation for hepatocellular carcinoma beyond the Milan criteria. Sci. Rep. 2018, 8, 7157. [Google Scholar] [CrossRef] [PubMed]
- Abreu, P.; Gorgen, A.; Oldani, G.; Hibi, T.; Sapisochin, G. Recent advances in liver transplantation for cancer: The future of transplant oncology. JHEP Rep. 2019, 1, 377–391. [Google Scholar] [CrossRef] [PubMed]
- Ng, K.T.P.; Lo, C.M.; Wong, N.; Li, C.X.; Qi, X.; Liu, X.B.; Geng, W.; Yeung, O.W.H.; Ma, Y.Y.; Chan, S.C.; et al. Early-phase circulating miRNAs predict tumor recurrence and survival of hepatocellular carcinoma patients after liver transplantation. Oncotarget 2016, 7, 19824–19839. [Google Scholar] [CrossRef]
- Liu, Y.; Guo, J.; Shen, K.; Wang, R.; Chen, C.; Liao, Z.; Zhou, J. Paclitaxel suppresses hepatocellular carcinoma tumorigenesis through regulating circ-birc6/ mir-877-5p/ywhaz axis. OncoTargets Ther. 2020, 13, 9377–9388. [Google Scholar] [CrossRef]
- Yan, H.; Wang, S.; Yu, H.; Zhu, J.; Chen, C. Molecular pathways and functional analysis of miRNA expression associated with paclitaxel-induced apoptosis in hepatocellular carcinoma cells. Pharmacology 2013, 92, 167–174. [Google Scholar] [CrossRef]
- Watanabe, M. Risk factors and molecular mechanisms of esophageal cancer: Differences between the histologic subtype. J. Cancer Metastasis Treat. 2015, 1, 1–7. [Google Scholar] [CrossRef]
- Lu, P.; Gu, J.; Zhang, N.; Sun, Y.; Wang, J. Risk factors for precancerous lesions of esophageal squamous cell carcinoma in high-risk areas of rural China: A population-based screening study. Medicine 2020, 99, e21426. [Google Scholar] [CrossRef]
- Liang, H.; Fan, J.H.; Qiao, Y.L. Epidemiology, etiology, and prevention of esophageal squamous cell carcinoma in China. Cancer Biol. Med. 2017, 14, 33. [Google Scholar]
- Tarazi, M.; Chidambaram, S.; Markar, S.R. Risk factors of esophageal squamous cell carcinoma beyond alcohol and smoking. Cancers 2021, 14, 1009. [Google Scholar] [CrossRef] [PubMed]
- Businello, G.; Parente, P.; Mastracci, L.; Pennelli, G.; Traverso, G.; Milione, M.; Bellan, E.; Michelotto, M.; Kotsafti, A.; Grillo, F.; et al. The pathologic and molecular landscape of esophageal squamous cell carcinogenesis. Cancers 2020, 12, 2160. [Google Scholar] [CrossRef] [PubMed]
- Codipilly, D.C.; Qin, Y.; Dawsey, S.M.; Kisiel, J.; Topazian, M.; Ahlquist, D.; Iyer, P.G. Screening for esophageal squamous cell carcinoma: Recent advances. Gastrointest. Endosc. 2018, 88, 413–426. [Google Scholar] [CrossRef]
- Marabotto, E.; Pellegatta, G.; Sheijani, A.D.; Ziola, S.; Zentilin, P.; De Marzo, M.G.; Giannini, E.G.; Ghisa, M.; Barberio, B.; Scarpa, M.; et al. Prevention Strategies for Esophageal Cancer—An Expert Review. Cancers 2021, 13, 2183. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, D.; Peng, H.; Chen, X.; Han, X.; Yu, J.; Wang, W.; Liang, L.; Liu, Z.; Zheng, Y.; et al. Epigenetically upregulated oncoprotein PLCE1 drives esophageal carcinoma angiogenesis and proliferation via activating the PI-PLCϵ-NF-κB signaling pathway and VEGF-C/ Bcl-2 expression. Mol. Cancer 2019, 18, 1. [Google Scholar] [CrossRef] [PubMed]
- Abnet, C.C.; Arnold, M.; Wei, W.Q. Epidemiology of Esophageal Squamous Cell Carcinoma. Gastroenterology 2018, 154, 360–373. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Su, M.; Ou, W.; Wang, H.; Tian, B.; Ma, J.; Tang, J.; Wu, J.; Wu, Z.; Wang, W.; et al. Involvement of noncoding RNAs in epigenetic modifications of esophageal cancer. Biomed. Pharmacother. 2019, 117, 109192. [Google Scholar] [CrossRef]
- Mao, Y.; Liu, J.; Zhang, D.; Li, B. MiR-1290 promotes cancer progression by targeting nuclear factor I/X(NFIX) in esophageal squamous cell carcinoma (ESCC). Biomed. Pharmacother. 2015, 76, 82–93. [Google Scholar] [CrossRef]
- Li, M.; He, X.Y.; Zhang, Z.M.; Li, S.; Ren, L.H.; Cao, R.S.; Feng, Y.D.; Ji, Y.L.; Zhao, Y.; Shi, R.H. MicroRNA-1290 promotes esophageal squamous cell carcinoma cell proliferation and metastasis. World J. Gastroenterol. 2015, 21, 3245–3255. [Google Scholar] [CrossRef]
- Sun, H.; Wang, L.; Zhao, Q.; Dai, J. Diagnostic and prognostic value of serum miRNA-1290 in human esophageal squamous cell carcinoma. Cancer Biomark. 2019, 25, 381–387. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Travis, W.D.; Brambilla, E.; Nicholson, A.G.; Yatabe, Y.; Austin, J.H.M.; Beasley, M.B.; Chirieac, L.R.; Dacic, S.; Duhig, E.; Flieder, D.B.; et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances since the 2004 Classification. J. Thorac. Oncol. 2015, 10, 1243–1260. [Google Scholar] [CrossRef] [PubMed]
- Wadowska, K.; Bil-Lula, I.; Trembecki, Ł.; Śliwińska-Mossoń, M. Genetic markers in lung cancer diagnosis: A review. Int. J. Mol. Sci. 2020, 21, 4569. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ma, C.; Luo, H.; Zhang, J.; Wang, J.; Guo, H. Identification of the differential expression of genes and upstream microRNAs in small cell lung cancer compared with normal lung based on bioinformatics analysis. Medicine 2020, 99, e19086. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Sun, C.; Liang, X.; Xie, S.; Huang, J.; Li, D. Integrative analysis of microRNA and mRNA expression profiles in non-small-cell lung cancer. Cancer Gene Ther. 2016, 23, 90–97. [Google Scholar] [CrossRef]
- Zhang, W.C.; Chin, T.M.; Yang, H.; Nga, M.E.; Lunny, D.P.; Lim, E.K.H.; Sun, L.L.; Pang, Y.H.; Leow, Y.N.; Malusay, S.R.Y.; et al. Tumour-initiating cell-specific MIR-1246 and MIR-1290 expression converge to promote non-small cell lung cancer progression. Nat. Commun. 2016, 7, 11702. [Google Scholar] [CrossRef]
- Kim, G.; An, H.J.; Lee, M.J.; Song, J.Y.; Jeong, J.Y.; Lee, J.H.; Jeong, H.C. Hsa-miR-1246 and hsa-miR-1290 are associated with stemness and invasiveness of non-small cell lung cancer. Lung Cancer 2016, 91, 15–22. [Google Scholar] [CrossRef]
- Sun, B.; Yang, N.; Jiang, Y.; Zhang, H.; Hou, C.; Ji, C.; Liu, Y.; Zuo, P. Antagomir-1290 suppresses CD133+ cells in non-small cell lung cancer by targeting fyn-related Src family tyrosine kinase. Tumor Biol. 2015, 36, 6223–6230. [Google Scholar] [CrossRef]
- Jin, J.-J.; Liu, Y.-H.; Si, J.-M.; Ni, R.; Wang, J. Overexpression of miR-1290 contributes to cell proliferation and invasion of non small cell lung cancer by targeting interferon regulatory factor 2. Int. J. Biochem. Cell Biol. 2018, 95, 113–120. [Google Scholar] [CrossRef]
- Xiao, X.; Yang, D.; Gong, X.; Mo, D.; Pan, S.; Xu, J. miR-1290 promotes lung adenocarcinoma cell proliferation and invasion by targeting SOCS4. Oncotarget 2018, 9, 11977–11988. [Google Scholar] [CrossRef]
- Wu, L.; Liu, T.; Xiao, Y.; Li, X.; Zhu, Y.; Zhao, Y.; Bao, J.; Wu, C. Polygonatum odoratum lectin induces apoptosis and autophagy by regulation of microRNA-1290 and microRNA-15a-3p in human lung adenocarcinoma A549 cells. Int. J. Biol. Macromol. 2016, 85, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.B.; Kim, K.; Bae, S.; Choi, Y.; Cha, H.J.; Kim, S.Y.; Lee, J.H.; Jeon, S.H.; Jung, H.J.; Ahn, K.J.; et al. MicroRNA-1290 promotes asiatic acid-induced apoptosis by decreasing BCL2 protein level in A549 non-small cell lung carcinoma cells. Oncol. Rep. 2014, 32, 1029–1036. [Google Scholar] [CrossRef] [PubMed]
- Seijo, L.M.; Peled, N.; Ajona, D.; Boeri, M.; Field, J.K.; Sozzi, G.; Pio, R.; Zulueta, J.J.; Spira, A.; Massion, P.P.; et al. Biomarkers in Lung Cancer Screening: Achievements, Promises, and Challenges. J. Thorac. Oncol. 2019, 14, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Jella, K.K.; Jaafar, L.; Moreno, C.S.; Dynan, W.S. Characterization of exosome release and extracellular vesicle-associated miRNAs for human bronchial epithelial cells irradiated with high charge and energy ions. Life Sci. Sp. Res. 2021, 28, 11–17. [Google Scholar] [CrossRef]
- Szejniuk, W.M.; Robles, A.I.; McCulloch, T.; Falkmer, U.G.I.; Røe, O.D. Epigenetic predictive biomarkers for response or outcome to platinum-based chemotherapy in non-small cell lung cancer, current state-of-art. Pharm. J. 2019, 19, 5–14. [Google Scholar] [CrossRef]
- Saito, M.; Shiraishi, K.; Matsumoto, K.; Schetter, A.J.; Ogata-Kawata, H.; Tsuchiya, N.; Kunitoh, H.; Nokihara, H.; Watanabe, S.I.; Tsuta, K.; et al. A Three-microRNA signature predicts responses to platinum-based doublet chemotherapy in patients with lung adenocarcinoma. Clin. Cancer Res. 2014, 20, 4784–4793. [Google Scholar] [CrossRef]
- Provenzano, E.; Ulaner, G.A.; Chin, S.F. Molecular Classification of Breast Cancer. PET Clin. 2018, 13, 325–338. [Google Scholar] [CrossRef]
- Tsang, J.Y.S.; Tse, G.M. Molecular Classification of Breast Cancer. Adv. Anat. Pathol. 2020, 27, 27–35. [Google Scholar] [CrossRef]
- Richard, V.; Davey, M.G.; Annuk, H.; Miller, N.; Dwyer, R.M.; Lowery, A.; Kerin, M.J. MicroRNAs in Molecular Classification and Pathogenesis of Breast Tumors. Cancers 2021, 13, 5332. [Google Scholar] [CrossRef]
- Li, S.; Zhang, M.; Xu, F.; Wang, Y.; Leng, D. Detection significance of miR-3662, miR-146a, and miR-1290 in serum exosomes of breast cancer patients. J. Cancer Res. Ther. 2021, 17, 749–755. [Google Scholar] [CrossRef]
- Hamam, R.; Ali, A.M.; Alsaleh, K.A.; Kassem, M.; Alfayez, M.; Aldahmash, A.; Alajez, N.M. microRNA expression profiling on individual breast cancer patients identifies novel panel of circulating microRNA for early detection. Sci. Rep. 2016, 6, 25997. [Google Scholar] [CrossRef] [PubMed]
- Endo, Y.; Toyama, T.; Takahashi, S.; Yoshimoto, N.; Iwasa, M.; Asano, T.; Fujii, Y.; Yamashita, H. MiR-1290 and its potential targets are associated with characteristics of estrogen receptor α-positive breast cancer. Endocr. Relat. Cancer 2013, 20, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Endo, Y.; Yamashita, H.; Takahashi, S.; Sato, S.; Yoshimoto, N.; Asano, T.; Hato, Y.; Dong, Y.; Fujii, Y.; Toyama, T. Immunohistochemical determination of the miR-1290 target arylamine N-acetyltransferase 1 (NAT1) as a prognostic biomarker in breast cancer. BMC Cancer 2014, 14, 990. [Google Scholar] [CrossRef] [PubMed]
- Zaka, M.; Sutton, C.W.; Peng, Y.; Konur, S. Model-Based Integration Analysis Revealed Presence of Novel Prognostic miRNA Targets and Important Cancer Driver Genes in Triple-Negative Breast Cancers. Cancers 2020, 12, 632. [Google Scholar] [CrossRef]
- Okunade, K.S. Human papillomavirus and cervical cancer. J. Obstet. Gynaecol. 2020, 40, 602–608. [Google Scholar] [CrossRef]
- Balasubramaniam, S.D.; Balakrishnan, V.; Oon, C.E.; Kaur, G. Key molecular events in cervical cancer development. Medicina 2019, 55, 384. [Google Scholar] [CrossRef]
- Tornesello, M.L.; Faraonio, R.; Buonaguro, L.; Annunziata, C.; Starita, N.; Cerasuolo, A.; Pezzuto, F.; Tornesello, A.L.; Buonaguro, F.M. The Role of microRNAs, Long Non-coding RNAs, and Circular RNAs in Cervical Cancer. Front. Oncol. 2020, 10, 150. [Google Scholar] [CrossRef]
- Sadri Nahand, J.; Moghoofei, M.; Salmaninejad, A.; Bahmanpour, Z.; Karimzadeh, M.; Nasiri, M.; Mirzaei, H.R.; Pourhanifeh, M.H.; Bokharaei-Salim, F.; Mirzaei, H.; et al. Pathogenic role of exosomes and microRNAs in HPV-mediated inflammation and cervical cancer: A review. Int. J. Cancer 2020, 146, 305–320. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Chen, J.; Ren, Z.; Chen, Y.; Li, J.; Miao, X.; Song, Y.; Zhao, T.; Li, Y.; Shi, Y.; et al. A specific miRNA signature promotes radioresistance of human cervical cancer cells. Cancer Cell Int. 2013, 13, 118. [Google Scholar] [CrossRef]
- Fu, J.; Wang, W.; Wang, Y.; Liu, C.; Wang, P. The role of squamous cell carcinoma antigen (SCC Ag) in outcome prediction after concurrent chemoradiotherapy and treatment decisions for patients with cervical cancer. Radiat. Oncol. 2019, 14, 146. [Google Scholar] [CrossRef]
- Yao, T.; Rao, Q.; Liu, L.; Zheng, C.; Xie, Q.; Liang, J.; Lin, Z. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in cervical cancer. Virol. J. 2013, 10, 175–177. [Google Scholar] [CrossRef] [PubMed]
- Lheureux, S.; Braunstein, M.; Oza, A.M. Epithelial ovarian cancer: Evolution of management in the era of precision medicine. CA Cancer J. Clin. 2019, 69, 280–304. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, I.; Matsuura, T. Screening and prevention for high-grade serous carcinoma of the ovary based on carcinogenesis—Fallopian tube- And ovarian-derived tumors and incessant retrograde bleeding. Diagnostics 2020, 10, 120. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Hu, H.; Wang, X.; Zhu, Y.; Jiang, M. WFDC Protein: A Promising Diagnosis Biomarker of Ovarian Cancer. J. Cancer 2021, 12, 5404–5412. [Google Scholar] [CrossRef]
- Dochez, V.; Caillon, H.; Vaucel, E.; Dimet, J.; Winer, N.; Ducarme, G. Biomarkers and algorithms for diagnosis of ovarian cancer: CA125, HE4, RMI and ROMA, a review. J. Ovarian Res. 2019, 12, 28. [Google Scholar] [CrossRef]
- Žilovič, D.; Čiurlienė, R.; Sabaliauskaitė, R.; Jarmalaitė, S. Future Screening Prospects for Ovarian Cancer. Cancers 2021, 13, 3840. [Google Scholar] [CrossRef]
- Chong, G.O.; Jeon, H.S.; Han, H.S.; Son, J.W.; Lee, Y.H.; Hong, D.G.; Lee, Y.S.; Cho, Y.L. Differential microRNA expression profiles in primary and recurrent epithelial ovarian cancer. Anticancer Res. 2015, 35, 2611–2617. [Google Scholar]
- Li, Y.; Yao, L.; Liu, F.; Hong, J.; Chen, L.; Zhang, B.; Zhang, W. Characterization of microRNA expression in serous ovarian carcinoma. Int. J. Mol. Med. 2014, 34, 491–498. [Google Scholar] [CrossRef]
- Lai, X.J.; Cheng, H.F. LncRNA colon cancer-associated transcript 1 (CCAT1) promotes proliferation and metastasis of ovarian cancer via miR-1290. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 322–328. [Google Scholar] [CrossRef]
- Shapira, I.; Oswald, M.; Lovecchio, J.; Khalili, H.; Menzin, A.; Whyte, J.; Dos Santos, L.; Liang, S.; Bhuiya, T.; Keogh, M.; et al. Circulating biomarkers for detection of ovarian cancer and predicting cancer outcomes. Br. J. Cancer 2014, 110, 976–983. [Google Scholar] [CrossRef]
- Teo, M.Y.; Rathkopf, D.E.; Kantoff, P. Treatment of advanced prostate cancer. Annu. Rev. Med. 2019, 70, 479–499. [Google Scholar] [CrossRef] [PubMed]
- Scaravilli, M.; Porkka, K.P.; Brofeldt, A.; Annala, M.; Tammela, T.L.J.; Jenster, G.W.; Nykter, M.; Visakorpi, T. MiR-1247-5p is overexpressed in castration resistant prostate cancer and targets MYCBP2. Prostate 2015, 75, 798–805. [Google Scholar] [CrossRef] [PubMed]
- Packer, J.R.; Maitland, N.J. The molecular and cellular origin of human prostate cancer. Biochim. Biophys. Acta Mol. Cell Res. 2016, 1863, 1238–1260. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.P.; Lai, H.M.; Guo, Z. Prostate cancer early diagnosis: Circulating microRNA pairs potentially beyond single microRNAs upon 1231 serum samples. Brief. Bioinform. 2021, 22, bbaa111. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Yuan, T.; Liang, M.; Du, M.; Xia, S.; Dittmar, R.; Wang, D.; See, W.; Costello, B.A.; Quevedo, F.; et al. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur. Urol. 2015, 67, 33–41. [Google Scholar] [CrossRef]
- Xu, Y.; Qin, S.; An, T.; Tang, Y.; Huang, Y.; Zheng, L. MiR-145 detection in urinary extracellular vesicles increase diagnostic efficiency of prostate cancer based on hydrostatic filtration dialysis method. Prostate 2017, 77, 1167–1175. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.J.; Wang, W.P.; Wang, X.B.; Zhang, X.T.; Du, J.D. MiR-1290 targets CCNG2 to promote the metastasis of oral squamous cell carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 10332–10342. [Google Scholar] [CrossRef]
- Rivera, C. Essentials of oral cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 11884. [Google Scholar] [PubMed]
- Jiang, L.; Wang, Z.; Liu, C.; Gong, Z.; Yang, Y.; Kang, H.; Li, Y.; Hu, G. TrkB promotes laryngeal cancer metastasis via activation PI3K/ AKT pathway. Oncotarget 2017, 8, 108726–108737. [Google Scholar] [CrossRef]
- SUN, X.; SONG, Y.; TAI, X.; LIU, B.; JI, W. MicroRNA expression and its detection in human supraglottic laryngeal squamous cell carcinoma. Biomed. Rep. 2013, 1, 743–746. [Google Scholar] [CrossRef][Green Version]
- Ciolofan, M.S.; Vlăescu, A.N.; Mogoantă, C.-A.; Ioniță, E.; Ioniță, I.; Căpitănescu, A.-N.; Mitroi, M.-R.; Anghelina, F. Clinical, Histological and Immunohistochemical Evaluation of Larynx Cancer. Curr. Health Sci. J. 2017, 43, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Xu, Z.; Wang, K.; Sun, L.; Liu, G.; Han, B. MicroRNA and gene networks in human laryngeal cancer. Exp. Ther. Med. 2015, 10, 2245–2252. [Google Scholar] [CrossRef] [PubMed]
- Janiszewska, J.; Bodnar, M.; Paczkowska, J.; Ustaszewski, A.; Smialek, M.J.; Szylberg, L.; Marszalek, A.; Kiwerska, K.; Grenman, R.; Szyfter, K.; et al. Loss of the maf transcription factor in laryngeal squamous cell carcinoma. Biomolecules 2021, 11, 1035. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Yang, J.; Zheng, J.; Hsueh, C.; Guo, Y.; Zhou, L. Characterization of selective exosomal microRNA expression profile derived from laryngeal squamous cell carcinoma detected by next generation sequencing. Oncol. Rep. 2018, 40, 2584–2594. [Google Scholar] [CrossRef]
- Lopez, A.T.; Carvajal, R.D.; Geskin, L. Secondary Prevention Strategies for Nonmelanoma Skin Cancer. Oncology 2018, 32, 195–200. [Google Scholar]
- Cives, M.; Mannavola, F.; Lospalluti, L.; Sergi, M.C.; Cazzato, G.; Filoni, E.; Cavallo, F.; Giudice, G.; Stucci, L.S.; Porta, C.; et al. Non-melanoma skin cancers: Biological and clinical features. Int. J. Mol. Sci. 2020, 21, 5394. [Google Scholar] [CrossRef]
- Geusau, A.; Borik-Heil, L.; Skalicky, S.; Mildner, M.; Grillari, J.; Hackl, M.; Sunder-Plassmann, R. Dysregulation of tissue and serum microRNAs in organ transplant recipients with cutaneous squamous cell carcinomas. Health Sci. Rep. 2020, 3, e205. [Google Scholar] [CrossRef]
- Yan, L.; Cai, K.; Sun, K.; Gui, J.; Liang, J. MiR-1290 promotes proliferation, migration, and invasion of glioma cells by targeting LHX6. J. Cell. Physiol. 2018, 233, 6621–6629. [Google Scholar] [CrossRef]
- Reifenberger, G.; Wirsching, H.G.; Knobbe-Thomsen, C.B.; Weller, M. Advances in the molecular genetics of gliomas-implications for classification and therapy. Nat. Rev. Clin. Oncol. 2017, 14, 434–452. [Google Scholar] [CrossRef]
- Khalighfard, S.; Kalhori, M.R.; Haddad, P.; Khori, V.; Alizadeh, A.M. Enhancement of resistance to chemo-radiation by hsa-miR-1290 expression in glioblastoma cells. Eur. J. Pharmacol. 2020, 880, 173144. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, Z.; Liu, X.; Yang, C.; Xie, D. Gain of circBRAF Represses Glioma Progression by Regulating miR-1290/FBXW7 Axis. Neurochem. Res. 2021, 46, 1203–1213. [Google Scholar] [CrossRef]
- Kang, X.; Li, H.; Zhang, Z. Sevoflurane blocks glioma malignant development by upregulating circRELN through circRELN-mediated miR-1290/RORA axis. BMC Anesthesiol. 2021, 21, 213. [Google Scholar] [CrossRef]
- Kling, T.; Wenger, A.; Lunavat, T.R.; Jang, S.C.; Rydenhag, B.; Lötvall, J.; Pollard, S.M.; Danielsson, A.; Carén, H. Pediatric brain tumor cells release exosomes with a miRNA repertoire that differs from exosomes secreted by normal cells. Oncotarget 2017, 8, 90164–90175. [Google Scholar] [CrossRef]
- Zhou, W.; Shunqing, W.; Yi, Y.; Zhou, R.; Mao, P. MiR-196b/miR-1290 participate in the antitumor effect of resveratrol via regulation of IGFBP3 expression in acute lymphoblastic leukemia. Oncol. Rep. 2017, 37, 1075–1083. [Google Scholar] [CrossRef]
- Moriyama, T.; Liu, S.; Li, J.; Meyer, J.; Zhao, X.; Yang, W.; Shao, Y.; Heath, R.; Hnízda, A.; Carroll, W.L.; et al. Mechanisms of NT5C2-mediated thiopurine resistance in acute lymphoblastic leukemia. Mol. Cancer Ther. 2019, 18, 1887–1895. [Google Scholar] [CrossRef]
- Avigad, S.; Verly, I.R.; Lebel, A.; Kordi, O.; Shichrur, K.; Ohali, A.; Hameiri-Grossman, M.; Kaspers, G.J.; Cloos, J.; Fronkova, E.; et al. miR expression profiling at diagnosis predicts relapse in pediatric precursor B-cell acute lymphoblastic leukemia. Genes Chromosom. Cancer 2016, 55, 328–339. [Google Scholar] [CrossRef]
- Mikkelsen, L.H.; Andersen, M.K.; Andreasen, S.; Larsen, A.C.; Tan, Q.; Toft, P.B.; Wadt, K.; Heegaard, S. Global microRNA profiling of metastatic conjunctival melanoma. Melanoma Res. 2019, 29, 465–473. [Google Scholar] [CrossRef]
| Type of Cancer | Source of miR-1290 | Adjuvant Therapy | Clinical Significance of miR-1290 | References |
|---|---|---|---|---|
| Colorectal cancer | Stage II and III CRC tissues (n = 291) resected before adjuvant therapy | 5-FU-based chemotherapy after radical resection | miR-1290 is an independent prognostic factor for OS (HR = 1.48; 95% CI: 0.85–2.90) and DFS (HR = 1.59; 95% CI: 1.32–2.95). | [123] |
| Plasma from stage I-IV CRC patients (n = 80) collected before adjuvant therapy, colorectal adenoma (n = 50), and HCs (n = 30) | Not indicated | miR-1290 can differentiate colorectal adenoma from HCs with an AUC = 0.78 (95% CI: 0.69–0.88), 75.53% SE and 87.41% SP, and CRC patients from HCs with an AUC = 0.88 (95% CI: 0.82–0.95), 76.65% SE and 90.23% SP. | [124] | |
| Serum exosomes from stage I-IV CRC patients (n = 100) collected before adjuvant therapy and HCs (n = 35) | Not indicated | miR-1290 can discriminate stage I CRC patients from HCs with an AUC = 0.89 (95% CI: 0.81–0.97), 83.33% SE and 85.71% SP, and CRC patients at different stages from HCs with an AUC = 0.92 (95% CI: 0.87–0.97), 85% SE and 88.57% SP. | [125] | |
| Stage I-IV CRC tissues (n = 179) resected before adjuvant therapy. Preoperative sera from stage I-IV CRC patients (n = 211), colorectal adenoma (n = 56), and HCs (n = 57) | 5-FU-based chemotherapy for patients with stage III/IV CRC, no adjuvant therapy to stage I/II CRC patients | Tissue miR-1290 is not an independent prognostic factor for OS. Serum miR-1290 can distinguish colorectal adenoma from HCs with an AUC = 0.72, 46.4% SE and 91.2% SP, and CRC patients from HCs with an AUC = 0.83, 70.1% SE and 91.2% SP. Serum miR-1290 is an independent prognostic marker for OS (HR = 4.51; 95% CI: 1.23–23.69) and an independent predictor for tumor recurrence after curative surgery (HR = 3.92; 95% CI: 1.11–25.14). | [126] | |
| Pancreatic cancer | Preoperative sera from patients with stage I-III PanC (n = 41), stage I-III pancreatic neuroendocrine tumors (n = 18), CP (n = 35), and HCs (n = 19). Preoperative sera from patients with PanC (n = 56). | Pancreatic resection followed by 5-FU-based and palliative therapies (including gemcitabine) | miR-1290 can discriminate subjects with PanC relative to HCs, CP, and pancreatic neuroendocrine tumors with an AUC = 0.96 (95% CI: 0.91–1.00), 0.81 (0.71–0.91), and 0.80 (0.67–0.93), respectively. miR-1290 is an independent prognostic biomarker for OS (HR = 2.24; 95% CI: 1.16–4.33). | [127] |
| Plasma from stage I-IV PanC patients (n = 167, collected before surgery or before chemotherapy for patients with advanced disease) and HCs (n = 267) | Adjuvant chemotherapy administered to 61% of patients | miR-1290 can discriminate patients with PanC from HCs with an AUC = 0.73 (95% CI: 0.68–0.79), 56.3% SE and 89.5% SP. miR-1290 is not an independent prognostic factor for OS and DFS. | [128] | |
| Sera from stage I-IV PanC patients (n = 120, obtained before any therapeutic procedures), benign pancreatic disease controls (n = 40), and HCs (n = 40) | Not indicated | miR-1290 can discriminate PanC from HCs and benign controls with an AUC = 0.93 (95% CI: 0.89–0.97), 75.0% SE and 97.5% SP, and an AUC = 0.89 (95% CI: 0.84–0.94), 88.3% SE and 72.5% SP, respectively. miR-1290 is an independent risk factor for PanC (OR = 12.35). | [129] | |
| Esophageal cancer | ESCC tissues (n = 100) resected before adjuvant therapy | Not indicated | miR-1290 is an independent prognostic factor for OS (HR = 1.97; 95% CI: 1.00–4.19) and DFS (HR = 1.81; 1.00–4.06). | [130] |
| Lung cancer | Sera of stage I-IV NSCLC patients (n = 66) collected before any therapeutic procedures | Not indicated | miR-1290 is an independent prognostic factor for OS (HR = 1.79; 95% CI: 1.17–2.98). | [131] |
| Serum exosomes from stage I-IV LADC patients (n = 60) collected before any antitumor therapy | Not indicated | miR-1290 can discriminate LADC from HCs with an AUC = 0.94 (95% CI: 0.89–0.99), 80.0% SE and 96.7% SP. miR-1290 is an independent predictor of PFS (HR = 7.80, 95% CI: 1.44–42.41). | [132] | |
| Cervical cancer | Sera of stage I-IV CC patients (n = 45, collected before adjuvant therapy), 55 CIN, and 31 HCs | Not indicated | miR-1290 can differentiate subjects with CC from HCs with an AUC of 0.80 (95% CI: 0.69–0.90), 90.3% SE and 62.2% SP. | [133] |
| Endometrioid endometrial carcinoma | Plasma of stage I-IV EEC patients (n = 34) and HCs (n = 14) | Radiotherapy and/or chemotherapy | miR-1290 can discriminate subjects with EEC from HCs with an AUC = 0.77 (95% CI: 0.63–0.88), 76% SE and 86% SP. | [134] |
| Ovarian cancer | Sera from stage I-IV OC patients (n = 70; including HGSOC, n = 30) and HCs (n = 13) | Not indicated | miR-1290 can distinguish OC patients from HCs with an AUC = 0.48, 51% SE and 57% SP, and HGSOC patients from HCs with an AUC = 0.71, 63% SE and 85% SP. | [135] |
| Ascitic fluid or peritoneal lavages from stage I-IV OC patients (n = 23) and plasma from HCs (n = 34) | Not indicated | miR-1290 can discriminate OC patients from HCs with an AUC = 1.00. | [136] | |
| Oral cancer | Plasma from OSCC patients (n = 70) collected before adjuvant therapy and HCs (n = 40) | Not indicated | miR-1290 can distinguish OSCC patients from HCs with an AUC = 0.90 (95% CI: 0.84–0.96), 89.2% SE and 85.0% SP. Low miR-1290 level is an independent risk factor for the poor prognosis (HR = 2.74, 95% CI: 2.15–6.12). | [137] |
| Preoperative plasma from stage II-IV OSCC patients (n = 55) | Preoperative 5-FU-based chemoradiotherapy | Low miR-1290 level is an independent predictor of OS (HR = 3.35, 95% CI: 1.08–12.0) and DFS (HR = 3.31, 95% CI: 1.08–11.6). | [138] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guz, M.; Jeleniewicz, W.; Cybulski, M. An Insight into miR-1290: An Oncogenic miRNA with Diagnostic Potential. Int. J. Mol. Sci. 2022, 23, 1234. https://doi.org/10.3390/ijms23031234
Guz M, Jeleniewicz W, Cybulski M. An Insight into miR-1290: An Oncogenic miRNA with Diagnostic Potential. International Journal of Molecular Sciences. 2022; 23(3):1234. https://doi.org/10.3390/ijms23031234
Chicago/Turabian StyleGuz, Małgorzata, Witold Jeleniewicz, and Marek Cybulski. 2022. "An Insight into miR-1290: An Oncogenic miRNA with Diagnostic Potential" International Journal of Molecular Sciences 23, no. 3: 1234. https://doi.org/10.3390/ijms23031234
APA StyleGuz, M., Jeleniewicz, W., & Cybulski, M. (2022). An Insight into miR-1290: An Oncogenic miRNA with Diagnostic Potential. International Journal of Molecular Sciences, 23(3), 1234. https://doi.org/10.3390/ijms23031234






