Structural Insights into the Ligand-Binding and -Releasing Mechanism of Helicoverpa armigera Pheromone-Binding Protein PBP1
Abstract
1. Introduction
2. Results
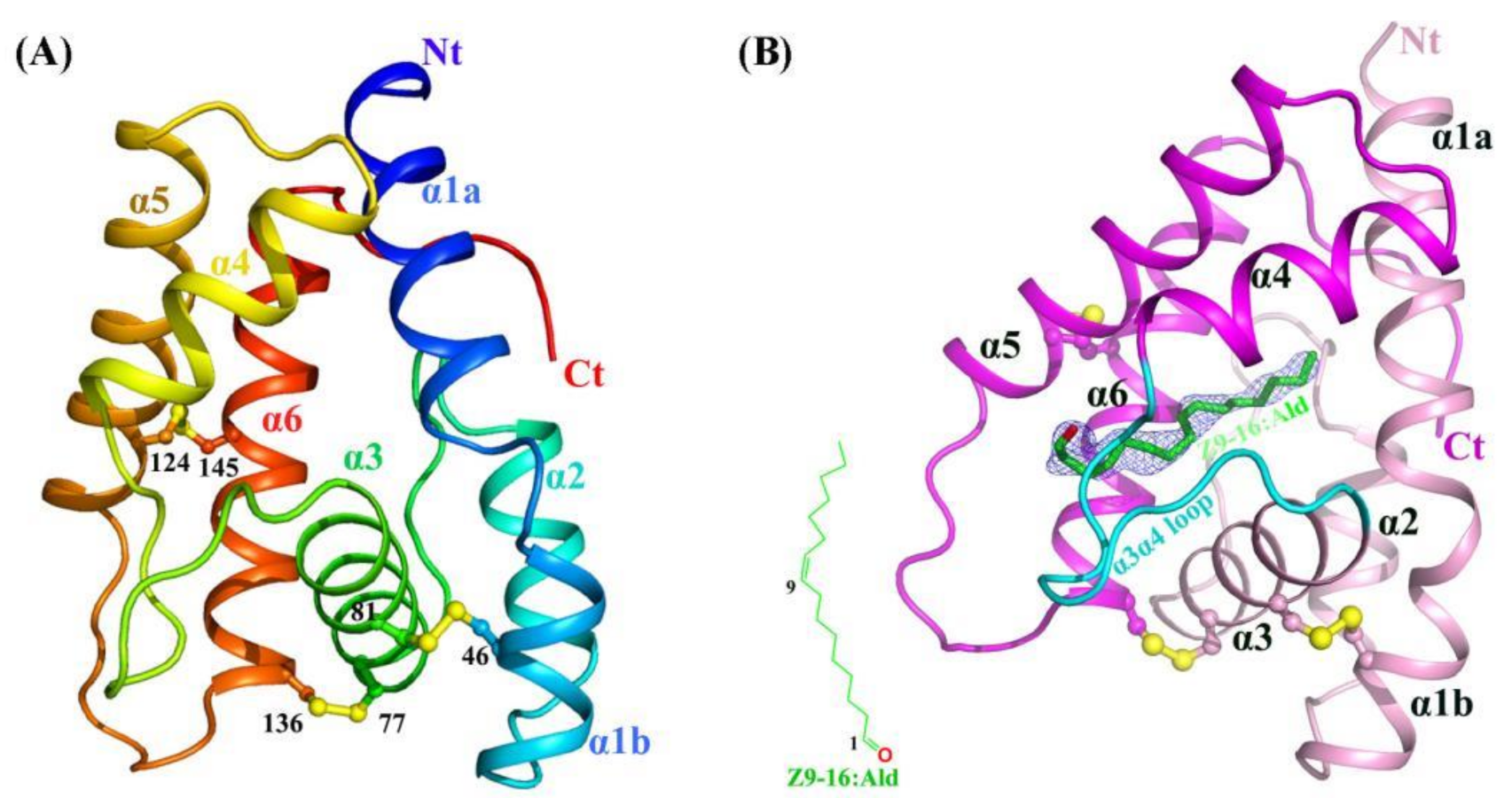
2.1. Crystal Structures of Apo-HarmPBP1 and HarmPBP1/Z9-16:Ald Complex
2.2. Structural Comparison Revealed a Specific Conformation of Apo-HarmPBP1
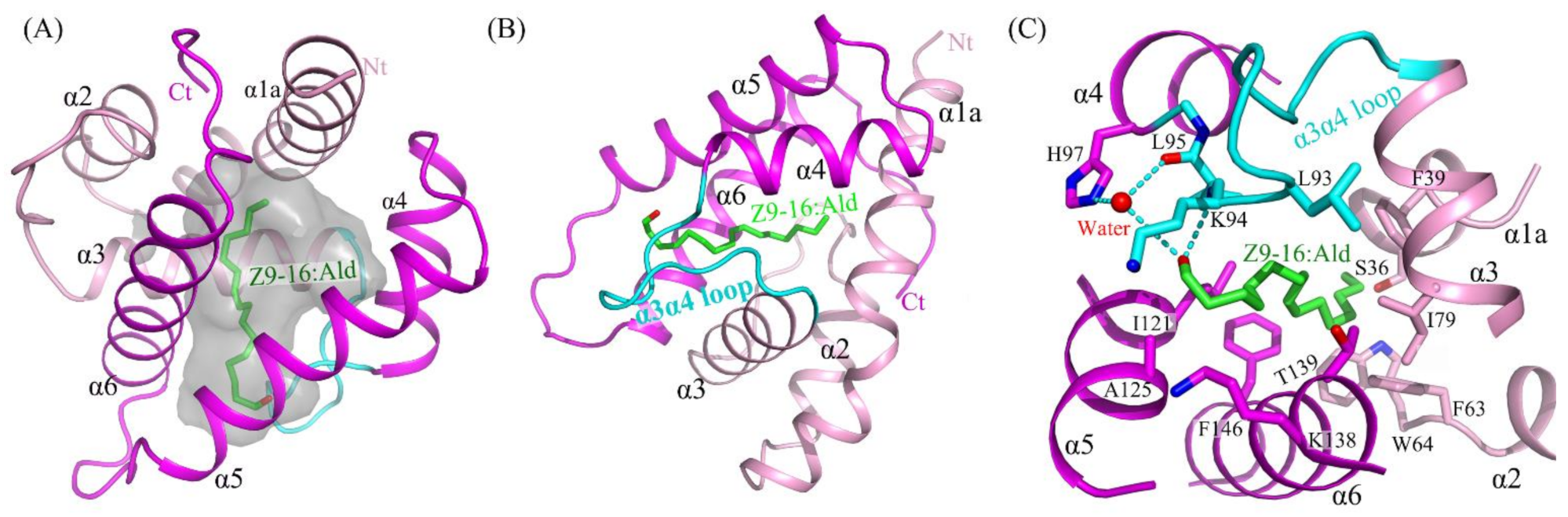
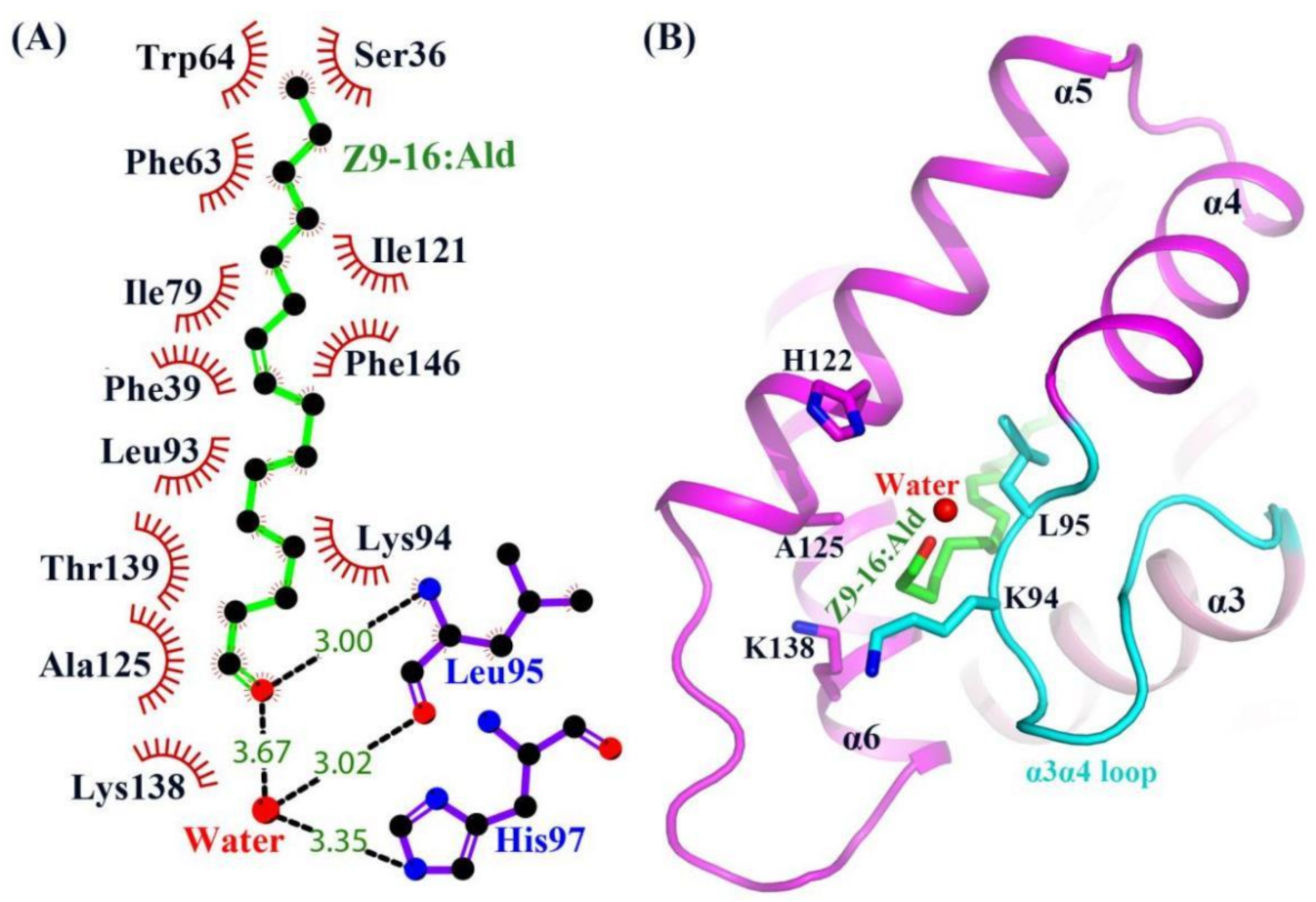
2.3. The Binding of Ligand in the HarmPBP1 Binding Cavity
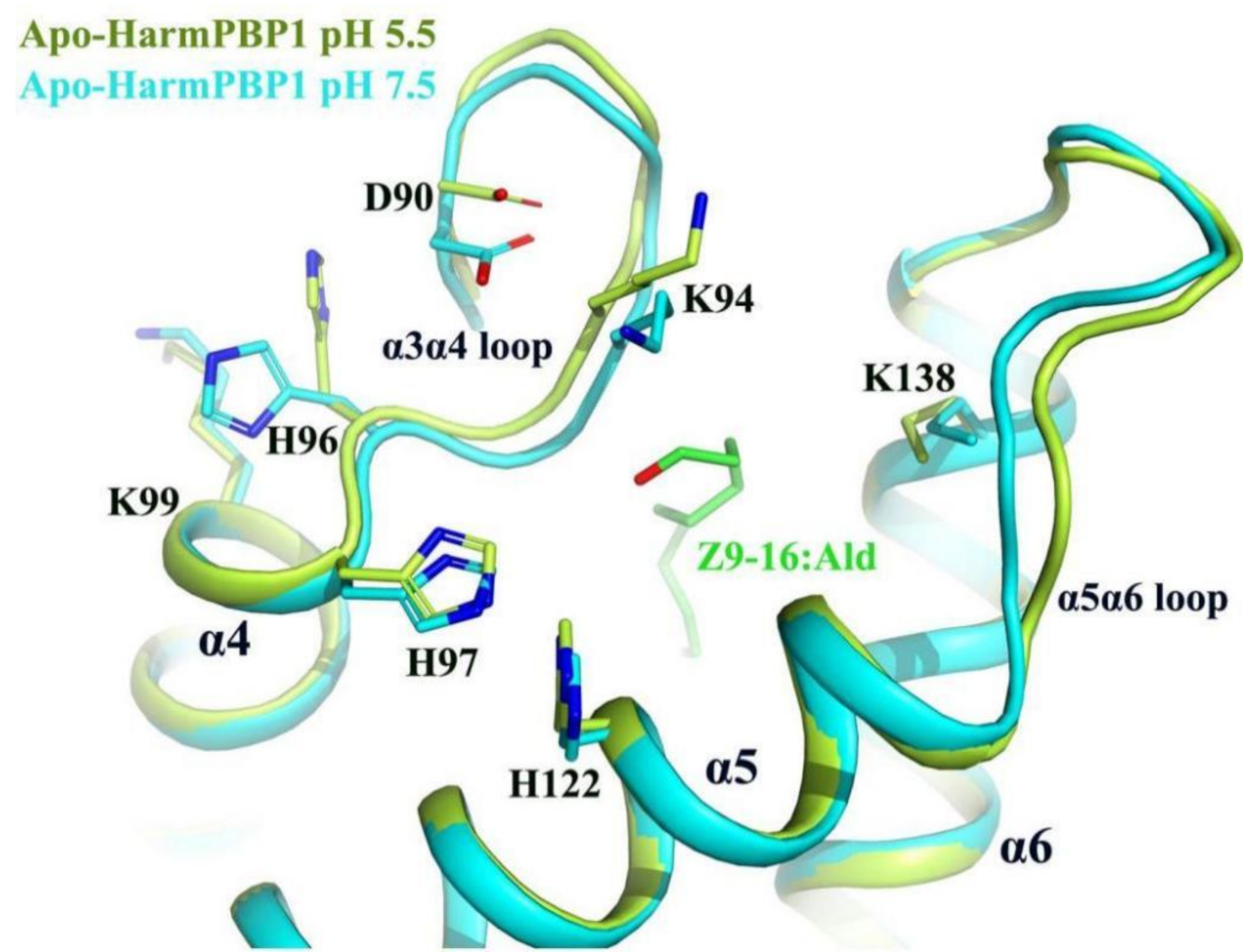
2.4. A New Releasing Mechanism at Low pH Conditions
3. Discussion
4. Materials and Methods
4.1. Expression and Purification of Recombinant HarmPBP1
4.2. Protein Crystallization and Data Collection
4.3. Structure Determination and Refinement
4.4. Fluorescence Binding Assays
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Leal, W.S.; Ishida, Y.; Pelletier, J.; Xu, W.; Rayo, J.; Xu, X.; Ames, J.B. Olfactory proteins mediating chemical communication in the navel orangeworm moth, Amyelois transitella. PLoS ONE 2009, 4, e7235. [Google Scholar] [CrossRef] [PubMed]
- Leal, W.S.; Barbosa, R.M.; Xu, W.; Ishida, Y.; Syed, Z.; Latte, N.; Chen, A.M.; Morgan, T.I.; Cornel, A.J.; Furtado, A. Reverse and conventional chemical ecology approaches for the development of oviposition attractants for Culex mosquitoes. PLoS ONE 2008, 3, e3045. [Google Scholar] [CrossRef] [PubMed]
- Vogt, R.G.; Prestwich, G.D.; Lerner, M.R. Odorant-binding-protein subfamilies associate with distinct classes of olfactory receptor neurons in insects. J. Neurobiol. 1991, 22, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Tegoni, M.; Campanacci, V.; Cambillau, C. Structural aspects of sexual attraction and chemical communication in insects. Trends Biochem. Sci. 2004, 29, 257–264. [Google Scholar] [CrossRef]
- Sandler, B.H.; Nikonova, L.; Leal, W.S.; Clardy, J. Sexual attraction in the silkworm moth: Structure of the pheromone-binding-protein-bombykol complex. Chem. Biol. 2000, 7, 143–151. [Google Scholar] [CrossRef]
- Xu, X.; Xu, W.; Rayo, J.; Ishida, Y.; Leal, W.S.; Ames, J.B. NMR structure of navel orangeworm moth pheromone-binding protein (AtraPBP1): Implications for pH-sensitive pheromone detection. Biochemistry 2010, 49, 1469–1476. [Google Scholar] [CrossRef]
- Zhou, J.J.; Robertson, G.; He, X.; Dufour, S.; Hooper, A.M.; Pickett, J.A.; Keep, N.H.; Field, L.M. Characterisation of Bombyx mori odorant-binding proteins reveals that a general odorant-binding protein discriminates between sex pheromone components. J. Mol. Biol. 2009, 389, 529–545. [Google Scholar] [CrossRef]
- Zubkov, S.; Gronenborn, A.M.; Byeon, I.J.; Mohanty, S. Structural consequences of the pH-induced conformational switch in A.polyphemus pheromone-binding protein: Mechanisms of ligand release. J. Mol. Biol. 2005, 354, 1081–1090. [Google Scholar] [CrossRef]
- Lautenschlager, C.; Leal, W.S.; Clardy, J. Coil-to-helix transition and ligand release of Bombyx mori pheromone-binding protein. Biochem. Biophys. Res. Commun. 2005, 335, 1044–1050. [Google Scholar] [CrossRef]
- Damberger, F.F.; Ishida, Y.; Leal, W.S.; Wuthrich, K. Structural basis of ligand binding and release in insect pheromone-binding proteins: NMR structure of Antheraea polyphemus PBP1 at pH 4.5. J. Mol. Biol. 2007, 373, 811–819. [Google Scholar] [CrossRef]
- Mohanty, S.; Zubkov, S.; Gronenborn, A.M. The solution NMR structure of Antheraea polyphemus PBP provides new insight into pheromone recognition by pheromone-binding proteins. J. Mol. Biol. 2004, 337, 443–451. [Google Scholar] [CrossRef]
- Zhang, T.T.; Mei, X.D.; Feng, J.N.; Berg, B.G.; Zhang, Y.J.; Guo, Y.Y. Characterization of three pheromone-binding proteins (PBPs) of Helicoverpa armigera (Hubner) and their binding properties. J. Insect Physiol. 2012, 58, 941–948. [Google Scholar] [CrossRef]
- Di Luccio, E.; Ishida, Y.; Leal, W.S.; Wilson, D.K. Crystallographic observation of pH-induced conformational changes in the Amyelois transitella pheromone-binding protein AtraPBP1. PLoS ONE 2013, 8, e53840. [Google Scholar] [CrossRef]
- Lautenschlager, C.; Leal, W.S.; Clardy, J. Bombyx mori pheromone-binding protein binding nonpheromone ligands: Implications for pheromone recognition. Structure 2007, 15, 1148–1154. [Google Scholar] [CrossRef]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995, 8, 127–134. [Google Scholar] [CrossRef]
- Dong, K.; Duan, H.X.; Liu, J.T.; Sun, L.; Gu, S.H.; Yang, R.N.; Dhiloo, K.H.; Gao, X.W.; Zhang, Y.J.; Guo, Y.Y. Key site residues of pheromone-binding protein 1 involved in interacting with sex pheromone components of Helicoverpa armigera. Sci. Rep. 2017, 7, 16859. [Google Scholar] [CrossRef]
- Dong, K.; Sun, L.; Liu, J.-T.; Gu, S.-H.; Zhou, J.-J.; Yang, R.-N.; Dhiloo, K.H.; Gao, X.-W.; Guo, Y.-Y.; Zhang, Y.-J. RNAi-induced electrophysiological and behavioral changes reveal two pheromone binding proteins of Helicoverpa armigera involved in the perception of the main sex pheromone component Z11–16:Ald. J. Chem. Ecol. 2017, 43, 207–214. [Google Scholar] [CrossRef]
- Ma, J.; Chen, Y.; Wu, W.; Chen, Z. Structure and function of N-terminal zinc finger domain of SARS-CoV-2 NSP2. Virol. Sin. 2021, 36, 1104–1112. [Google Scholar] [CrossRef]
- Pesenti, M.E.; Spinelli, S.; Bezirard, V.; Briand, L.; Pernollet, J.C.; Campanacci, V.; Tegoni, M.; Cambillau, C. Queen bee pheromone binding protein ph-induced domain swapping favors pheromone release. J. Mol. Biol. 2009, 390, 981–990. [Google Scholar] [CrossRef]
- Pesenti, M.E.; Spinelli, S.; Bezirard, V.; Briand, L.; Pernollet, J.C.; Tegoni, M.; Cambillau, C. Structural basis of the honey bee pbp pheromone and ph-induced conformational change. J. Mol. Biol. 2008, 380, 158–169. [Google Scholar] [CrossRef]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. In Method Enzymol; Carter, C.W., Sweet, R.M., Eds.; Macromolecular Crystallography, part A; Academic Press: New York, NY, USA, 1997; Volume 276, pp. 307–326. [Google Scholar]
- Long, F.; Vagin, A.A.; Young, P.; Murshudov, G.N. BALBES: A molecular-replacement pipeline. Acta Crystallogr. Sect. D Biol. Crystallogr. 2008, 64, 125–132. [Google Scholar] [CrossRef]
- Murshudov, G.N.; Skubak, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 355–367. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Kleywegt, G.J.; Jones, T.A. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994, 50, 178–185. [Google Scholar] [CrossRef] [PubMed]

| Apo-HarmPBP1 at pH 7.5 (PDB ID:7VW8) | Apo-HarmPBP1 at pH 5.5 (PDB ID:7VW9) | HarmPBP1/Z9-16:Ald Complex at pH 8.5 (PDB ID: 7VWA) | |
|---|---|---|---|
| Wavelength, Å | 0.9792 | 0.9792 | 1.0000 |
| Space group | P21 | P21 | P21 |
| Cell dimensions | |||
| a, b, c, Å | 32.03, 32.60, 54.79 | 32.43, 33.38, 55.38 | 32.91, 33.36, 56.04 |
| α, β, γ, ° | 90, 97.88, 90 | 90, 99.66, 90 | 90, 98.79, 90 |
| Resolution, Å | 50-1.30 (1.32–1.30) a | 50-2.05 (2.09–2.05) a | 50-2.09 (2.13–2.09) a |
| Rmerge, % | 6.6 (59.9) | 5.0 (21.4) | 5.7 (30.2) |
| I/σI | 17.7 (1.8) | 41.2 (7.6) | 31.1 (3.0) |
| Completeness, % | 93.1 (97.5) | 95.6 (93.5) | 98.2 (81.0) |
| Redundancy | 4.3 (3.9) | 3.5 (3.1) | 4.9 (3.7) |
| Refinement | |||
| Resolution, Å | 50-1.3 (1.33–1.30) | 50-2.05 (2.10–2.05) | 50-2.10 (2.15–2.10) |
| No. of unique reflections | 21224 (736) | 6834 (465) | 6682 (432) |
| Rwork/Rfree, % | 17.9/19.8 (22.3/24.9) | 22.2/26.5 (28.4/36.7) | 19.9/23.6 (24.2/37.1) |
| No. of atoms (protein/ligand/water) | 1048/0/62 | 1015/0/56 | 1059/17/40 |
| Average B factor (Å2) (protein/ligand/water) | 21.44/0/30.79 | 43.69/0/47.31 | 31.07/37.11/36.18 |
| rms deviations | |||
| Bond lengths, Å | 0.008 | 0.007 | 0.009 |
| Bond angles, ° | 1.378 | 0.994 | 1.515 |
| Ramachandran Plot, % b | 95.9/4.1/0/0 | 91.1/8.9/0/0 | 92.9/7.1/0/0 |
| Ligand | pH 7.5 | pH 5.5 |
|---|---|---|
| (Z)-11-Hexadecenal | 0.67 ± 0.05 | 13.09 ± 1.78 |
| (Z)-9-Hexadecenal | 0.56 ± 0.05 | 13.61 ± 1.33 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, J.; Yang, M.; Dong, K.; Zhang, J.; Wang, H.; Xie, M.; Wu, W.; Zhang, Y.-J.; Chen, Z. Structural Insights into the Ligand-Binding and -Releasing Mechanism of Helicoverpa armigera Pheromone-Binding Protein PBP1. Int. J. Mol. Sci. 2022, 23, 1190. https://doi.org/10.3390/ijms23031190
Zheng J, Yang M, Dong K, Zhang J, Wang H, Xie M, Wu W, Zhang Y-J, Chen Z. Structural Insights into the Ligand-Binding and -Releasing Mechanism of Helicoverpa armigera Pheromone-Binding Protein PBP1. International Journal of Molecular Sciences. 2022; 23(3):1190. https://doi.org/10.3390/ijms23031190
Chicago/Turabian StyleZheng, Jiangge, Meiting Yang, Kun Dong, Jianbo Zhang, Huali Wang, Mengjia Xie, Wei Wu, Yong-Jun Zhang, and Zhongzhou Chen. 2022. "Structural Insights into the Ligand-Binding and -Releasing Mechanism of Helicoverpa armigera Pheromone-Binding Protein PBP1" International Journal of Molecular Sciences 23, no. 3: 1190. https://doi.org/10.3390/ijms23031190
APA StyleZheng, J., Yang, M., Dong, K., Zhang, J., Wang, H., Xie, M., Wu, W., Zhang, Y.-J., & Chen, Z. (2022). Structural Insights into the Ligand-Binding and -Releasing Mechanism of Helicoverpa armigera Pheromone-Binding Protein PBP1. International Journal of Molecular Sciences, 23(3), 1190. https://doi.org/10.3390/ijms23031190







