Effect of Superovulation Treatment on Oocyte’s DNA Methylation
Abstract
1. Introduction
2. Results
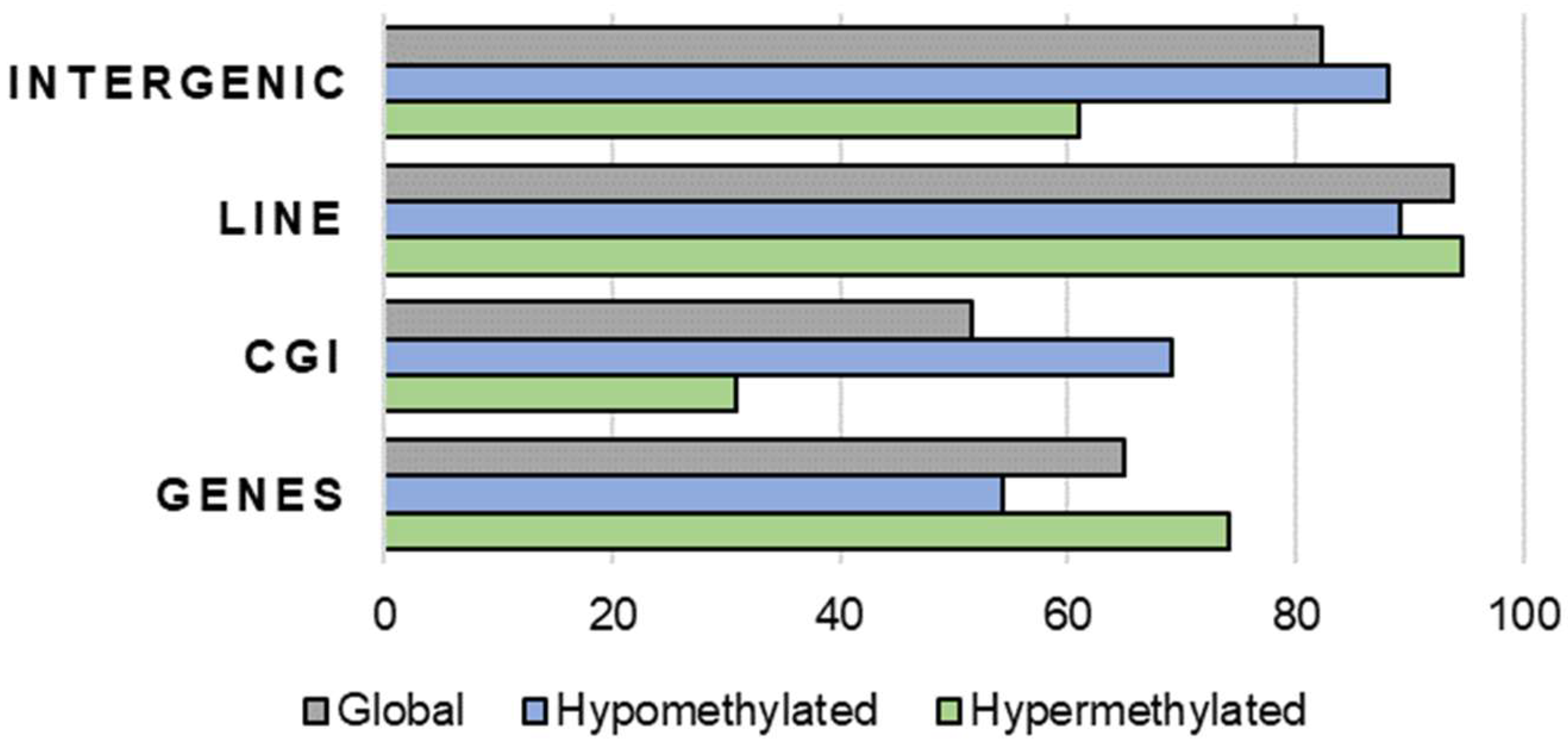
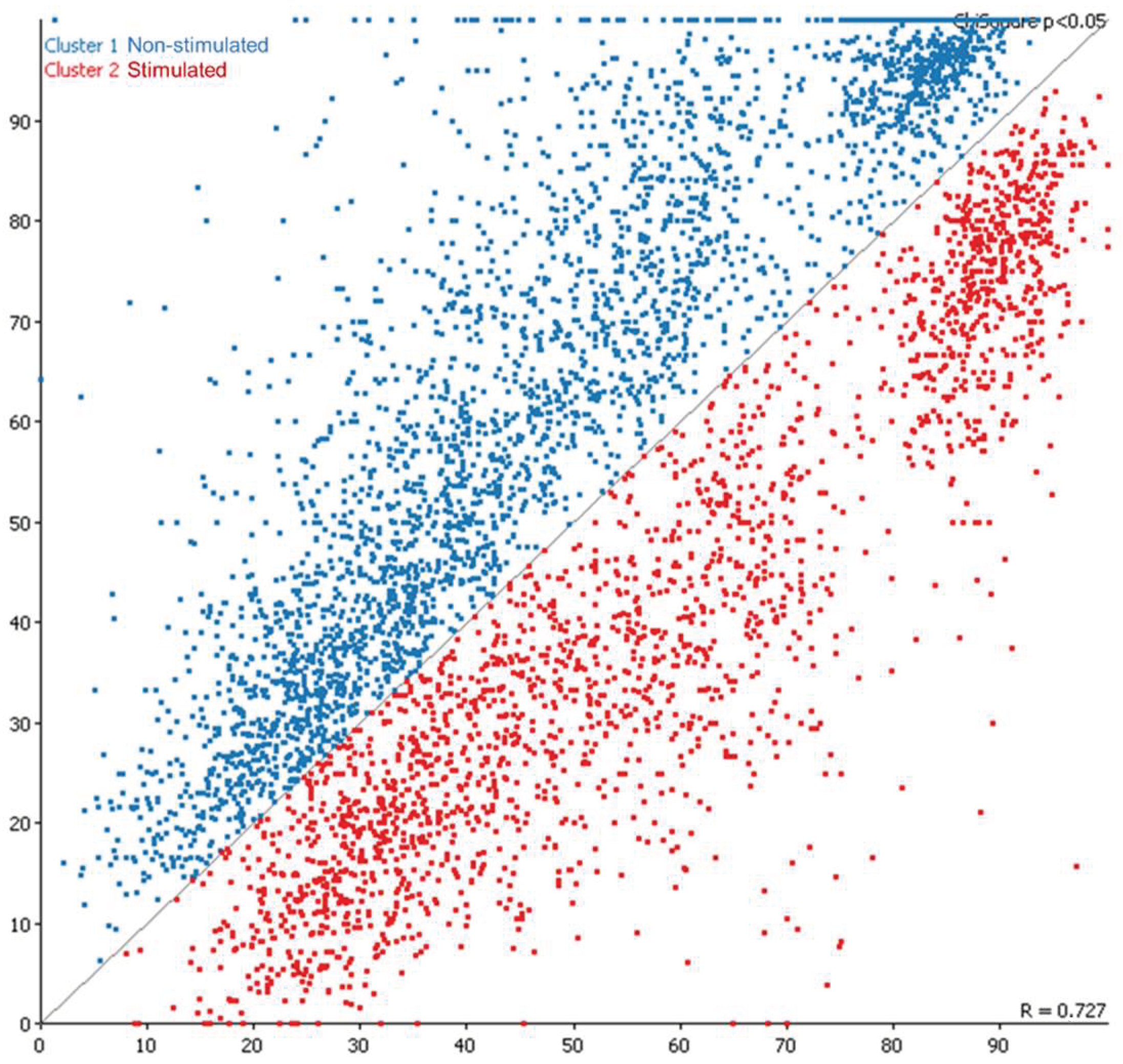
2.1. Global Methylation Profile of Oocytes from Ovarian-Stimulated versus Non-Stimulated Cows
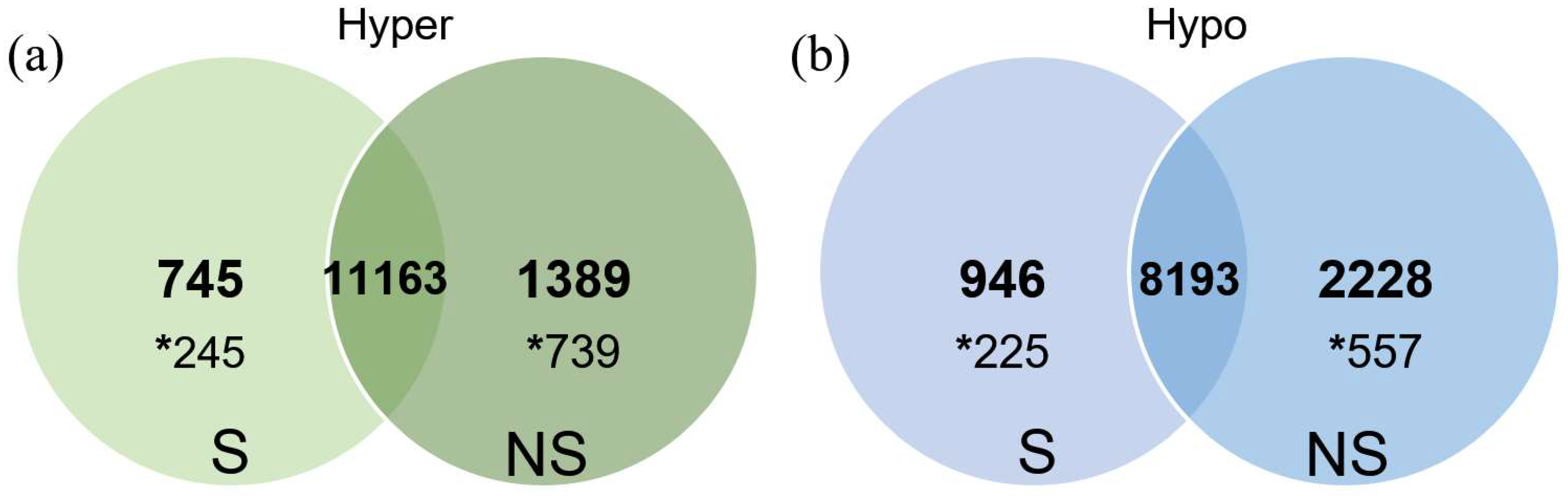
2.2. Identification of Differentially Methylated Regions (DMRs) in Oocytes from Ovarian Stimulated and Non-Stimulated Animals
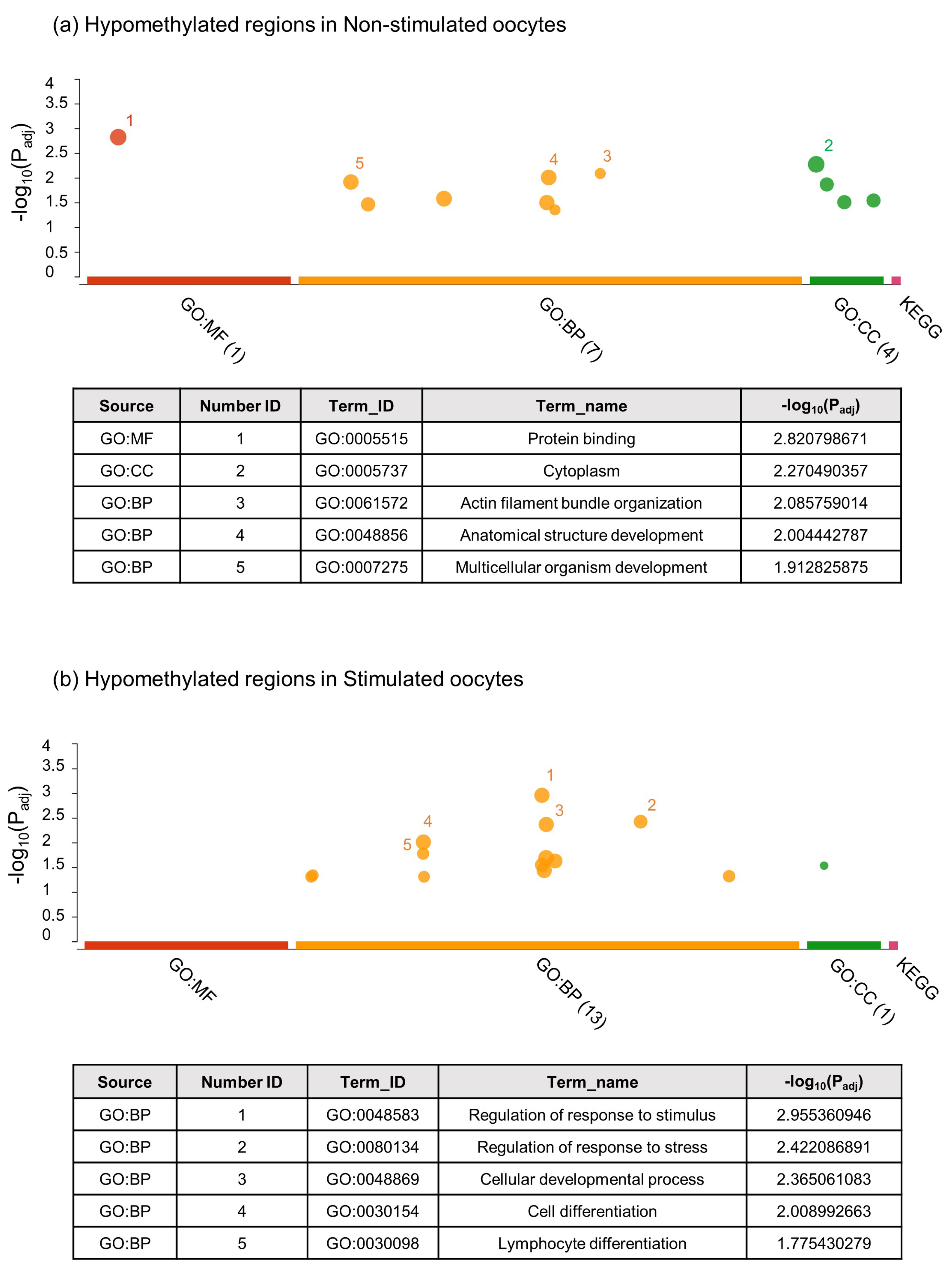
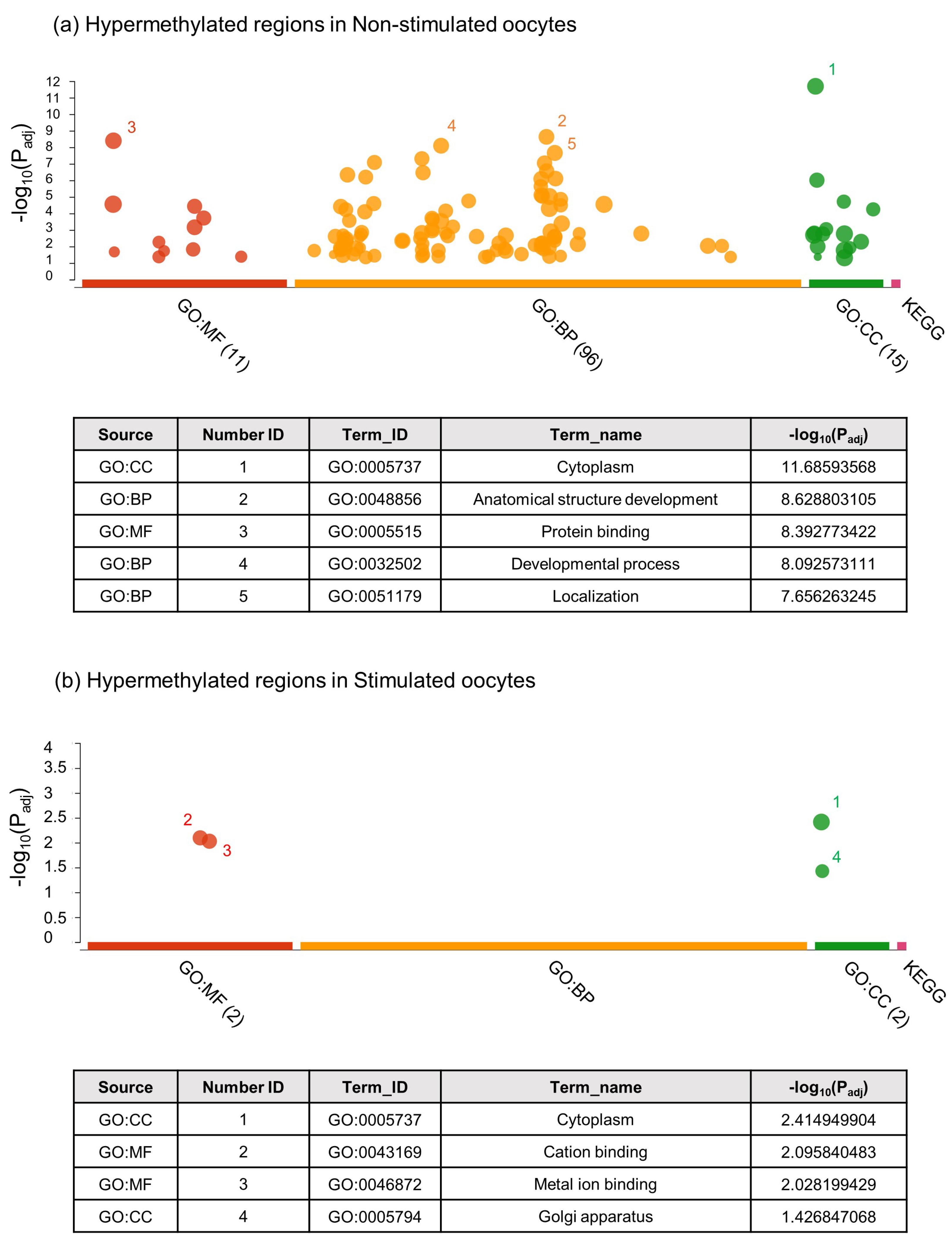
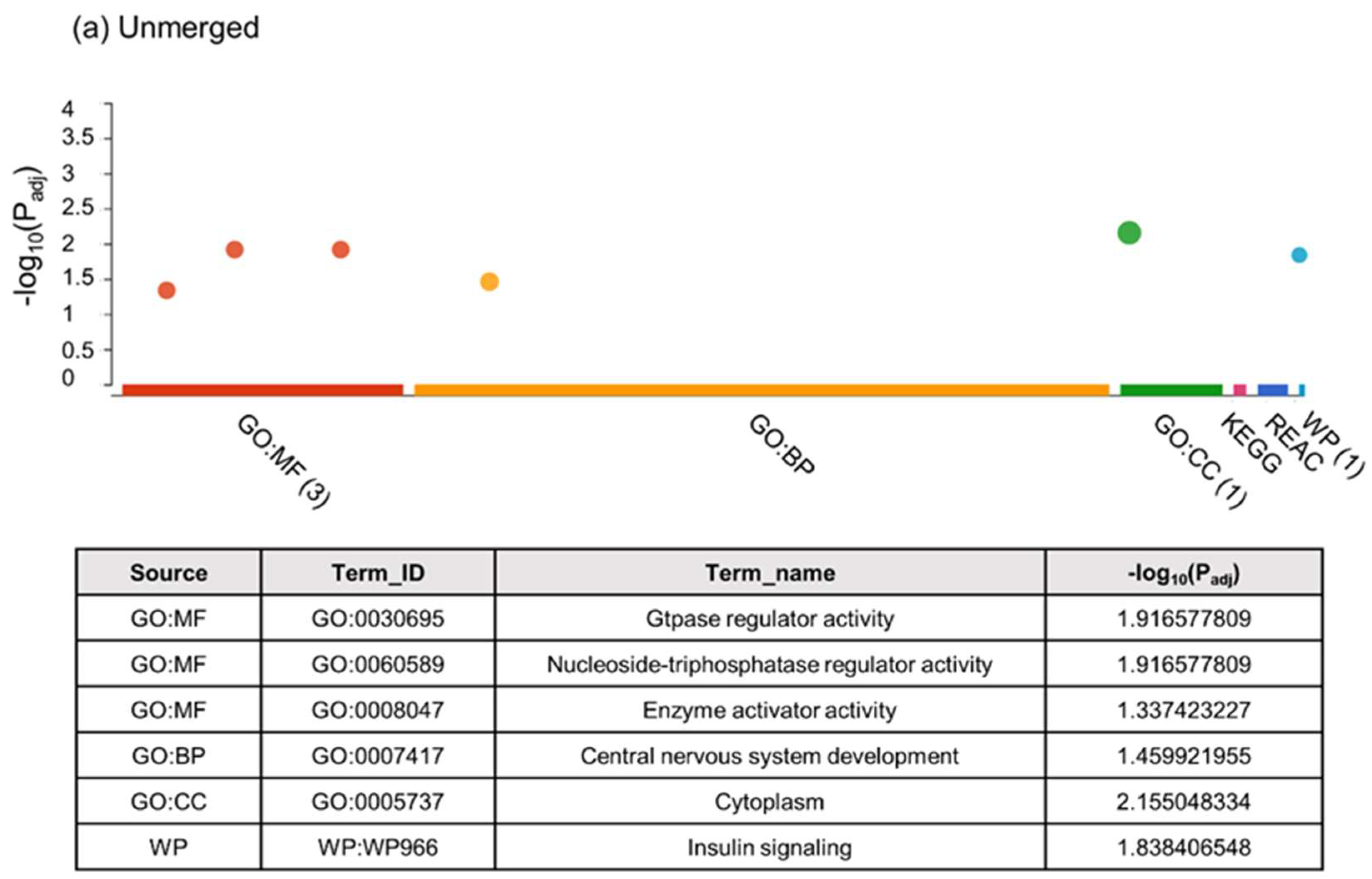
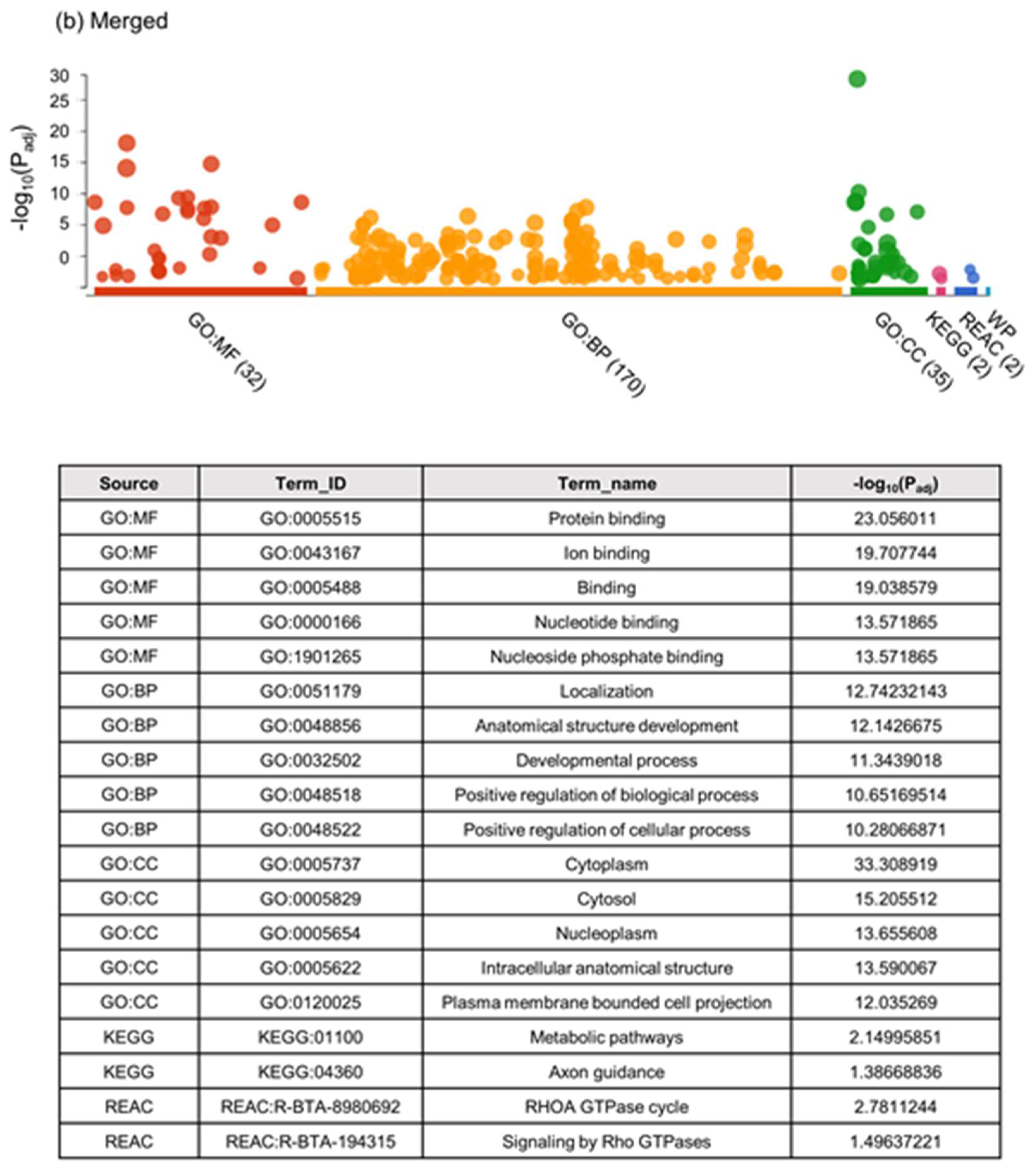
2.2.1. Unbiased Analysis of DNA Methylation in Oocytes for Ovarian Stimulated versus Non-Stimulated Animals
2.2.2. Targeted Analysis of Cow Oocyte DNA Methylation by Segmenting the Genome in Hypermethylated and Hypomethylated Domains for two Treatments (S versus NS animals)
2.2.3. Targeted Analysis of Cow Oocyte DNA Methylation against Imprinted Genes for Two Treatments (Ovarian S versus NS Animals)
3. Discussion
4. Materials and Methods
4.1. Ethics
4.2. Animals
4.3. Non-Stimulated (NS) Cows
4.4. Hormone-Stimulated (S) Cows
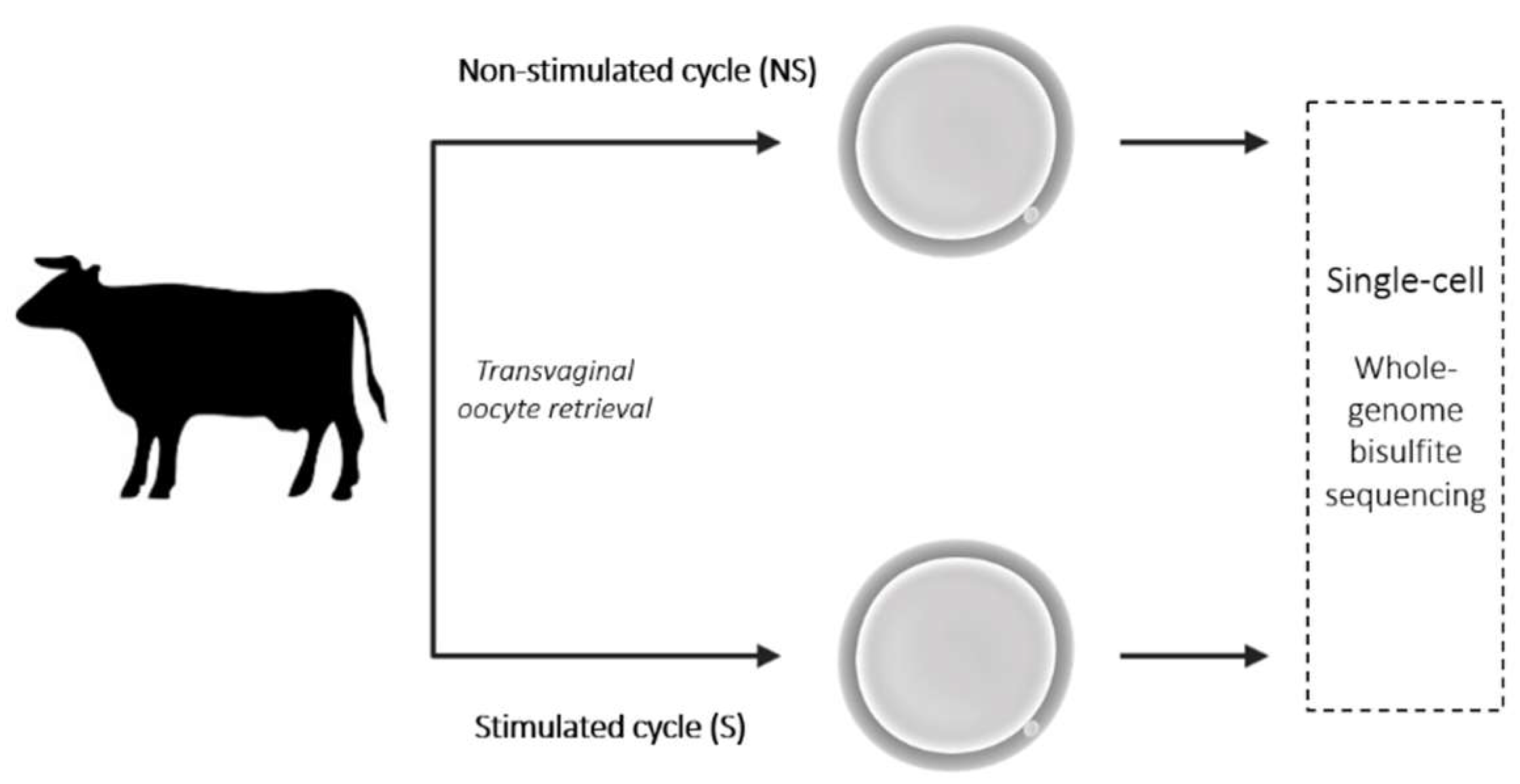
4.5. Transvaginal Oocyte Retrieval
4.6. Oocyte Retrieval and Storage
4.7. DNA Methylation: Single-Cell Methylome Analysis
4.8. Assessment of Data Quality
4.9. Statistics
4.10. Experimental Design
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Santos, M.A.; Kuijk, E.W.; Macklon, N.S. The impact of ovarian stimulation for IVF on the developing embryo. Reproduction 2010, 139, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Bloise, E.; Feuer, S.K.; Rinaudo, P.F. Comparative intrauterine development and placental function of ART concepti: Implications for human reproductive medicine and animal breeding. Hum. Reprod. Update 2014, 20, 822–839. [Google Scholar] [CrossRef] [PubMed]
- Canovas, S.; Ross, P.J.; Kelsey, G.; Coy, P. DNA Methylation in Embryo Development: Epigenetic Impact of ART (Assisted Reproductive Technologies). BioEssays 2017, 39, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Keck, C.; Bassett, R.; Ludwig, M. Factors influencing response to ovarian stimulation. Reprod. Biomed. Online 2005, 11, 562–569. [Google Scholar] [CrossRef]
- La Marca, A.; Sunkara, S.K. Individualization of controlled ovarian stimulation in IVF using ovarian reserve markers: From theory to practice. Hum. Reprod. Update 2014, 20, 124–140. [Google Scholar] [CrossRef]
- Senapati, S.; Wang, F.; Ord, T.; Coutifaris, C.; Feng, R.; Mainigi, M. Superovulation alters the expression of endometrial genes critical to tissue remodeling and placentation. J. Assist. Reprod. Genet. 2018, 35, 1799–1808. [Google Scholar] [CrossRef]
- Ertzeid, G.; Storeng, R. The impact of ovarian stimulation on implantation and fetal development in mice. Hum. Reprod. 2001, 16, 221–225. [Google Scholar] [CrossRef]
- Wang, X.; Wu, H.; He, X.; Jiang, H.; Wu, L.; Xu, Y.; Zhou, P.; Wei, Z.; Cao, Y. Retrospective Study to Compare Frozen-Thawed Embryo Transfer with Fresh Embryo Transfer on Pregnancy Outcome Following Intracytoplasmic Sperm Injection for Male Infertility. Med. Sci. Monit. 2018, 24, 2668–2674. [Google Scholar] [CrossRef]
- Kalra, S.K.; Ratcliffe, S.J.; Coutifaris, C.; Molinaro, T.; Barnhart, K.T. Ovarian Stimulation and Low Birth Weight in Newborns Conceived Through In Vitro Fertilization. Obstet. Gynecol. 2011, 118, 863–871. [Google Scholar] [CrossRef]
- Beall, S.A.; Decherney, A. History and challenges surrounding ovarian stimulation in the treatment of infertility. Fertil. Steril. 2012, 97, 795–801. [Google Scholar] [CrossRef]
- White, C.R.; Denomme, M.M.; Tekpetey, F.R.; Feyles, V.; Power, S.G.A.; Mann, M.R.W. High Frequency of Imprinted Methylation Errors in Human Preimplantation Embryos. Sci. Rep. 2015, 5, 17311. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Hagen, D.E.; Elsik, C.G.; Ji, T.; Morris, C.J.; Moon, L.E.; Rivera, R.M. Characterization of global loss of imprinting in fetal overgrowth syndrome induced by assisted reproduction. Proc. Natl. Acad. Sci. USA 2015, 112, 4618–4623. [Google Scholar] [CrossRef] [PubMed]
- el Hajj, N.; Haaf, T. Epigenetic disturbances in in vitro cultured gametes and embryos: Implications for human assisted reproduction. Fertil. Steril. 2013, 99, 632–641. [Google Scholar] [CrossRef] [PubMed]
- Turro, E.; Astle, W.J.; Megy, K.; Gräf, S.; Greene, D.; Shamardina, O.; Allen, H.L.; Sanchis-Juan, A.; Frontini, M.; Thys, C.; et al. Whole-genome sequencing of patients with rare diseases in a national health system. Nature 2020, 583, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Uysal, F.; Ozturk, S.; Akkoyunlu, G. Superovulation alters DNA methyltransferase protein expression in mouse oocytes and early embryos. J. Assist. Reprod. Genet. 2018, 35, 503–513. [Google Scholar] [CrossRef]
- Sato, A.; Otsu, E.; Negishi, H.; Utsunomiya, T.; Arima, T. Aberrant DNA methylation of imprinted loci in superovulated oocytes. Hum. Reprod. 2007, 22, 26–35. [Google Scholar] [CrossRef]
- Market-Velker, B.A.; Zhang, L.; Magri, L.S.; Bonvissuto, A.C.; Mann, M.R.W. Dual effects of superovulation: Loss of maternal and paternal imprinted methylation in a dose-dependent manner. Hum. Mol. Genet. 2010, 19, 36–51. [Google Scholar] [CrossRef]
- Saenz-de-Juano, M.D.; Ivanova, E.; Romero, S.; Lolicato, F.; Sánchez, F.; Van Ranst, H.; Krueger, F.; Segonds-Pichon, A.; De Vos, M.; Andrews, S.; et al. DNA methylation and mRNA expression of imprinted genes in blastocysts derived from an improved in vitro maturation method for oocytes from small antral follicles in polycystic ovary syndrome patients. Hum. Reprod. 2019, 34, 1640–1649. [Google Scholar] [CrossRef]
- Chu, T.; Dufort, I.; Sirard, M.A. Effect of ovarian stimulation on oocyte gene expression in cattle. Theriogenology 2012, 77, 1928–1938. [Google Scholar] [CrossRef] [PubMed]
- Van Soom, A.; Wydooghe, E.; Heras, S.; Vandaele, L. Alternative models for the study of embryo]maternal cross-talk and signaling molecules from fertilisation to implantation. Reprod. Fertil. Dev. 2011, 23, 10–12. [Google Scholar] [CrossRef]
- Bó, G.A.; Mapletoft, R.J. Historical perspectives and recent research on superovulation in cattle. Theriogenology 2014, 81, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Sirard, M.A. The Ovarian Follicle of Cows as a Model for Human. In Animal Models and Human Reproduction; Constantinescu, G., Schatten, H., Eds.; Wiley: Hoboken, NJ, USA, 2017; pp. 127–144. ISBN 9781118881286. [Google Scholar]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. G:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [PubMed]
- Galli, C.; Duchi, R.; Colleoni, S.; Lagutina, I.; Lazzari, G. Ovum pick up, intracytoplasmic sperm injection and somatic cell nuclear transfer in cattle, buffalo and horses: From the research laboratory to clinical practice. Theriogenology 2014, 81, 138–151. [Google Scholar] [CrossRef]
- Pavlok, A.; Lucas-Han, A.; Niemann, H. Fertilization and Developmental Competence of Bovine Oocytes Derived From Different Categories of Antral Follicles. Mol. Reprod. Dev. 1992, 31, 63–67. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Fernandez, J.; Herrera-Puerta, E.; Demond, H.; Clark, S.; Hanna, C.; Hemberger, M.; Kelsey, G. Increased transcriptome variation and localised DNA methylation changes in oocytes from aged mice revealed by parallel single-cell analysis. Aging Cell 2020, 19, e13278. [Google Scholar] [CrossRef]
- Kindsfather, A.J.; Czekalski, M.A.; Pressimone, C.A.; Erisman, M.P.; Mann, M.R.W. Perturbations in imprinted methylation from assisted reproductive technologies but not advanced maternal age in mouse preimplantation embryos. Clin. Epigenet. 2019, 11, 162. [Google Scholar] [CrossRef]
- Ertzeid, G.; Storeng, R. Adverse effects of gonadotrophin treatment on pre- and postimplantation development in mice. J. Reprod. Fertil. 1992, 96, 649–655. [Google Scholar] [CrossRef]
- Van der Auwera, I.; D’Hooghe, T. Superovulation of female mice delays embryonic and fetal development. Hum. Reprod. 2001, 16, 1237–1243. [Google Scholar] [CrossRef]
- Beaumont, H.M.; Smith, A.F. Embryonic mortality during the pre- and post-implantation periods of pregnancy in mature mice after superovulation. J. Reprod. Fertil. 1975, 45, 437–448. [Google Scholar] [CrossRef][Green Version]
- Olson, C.K.; Keppler-Noreuil, K.M.; Romitti, P.A.; Budelier, W.T.; Ryan, G.; Sparks, A.E.T.; Van Voorhis, B.J. In vitro fertilization is associated with an increase in major birth defects. Fertil. Steril. 2005, 84, 1308–1315. [Google Scholar] [CrossRef]
- Klemetti, R.; Sevón, T.; Gissler, M.; Hemminki, E. Health of children born after ovulation induction. Fertil. Steril. 2010, 93, 1157–1168. [Google Scholar] [CrossRef] [PubMed]
- Marshall, K.L.; Rivera, R.M. The effects of superovulation and reproductive aging on the epigenome of the oocyte and embryo. Mol. Reprod. Dev. 2017, 85, 90–105. [Google Scholar] [CrossRef]
- Huffman, S.R.; Pak, Y.; Rivera, R.M. Superovulation induces alterations in the epigenome of zygotes, and results in differences in gene expression at the blastocyst stage in mice. Mol. Reprod. Dev. 2015, 82, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Almamun, M.; Rivera, R.M. Size-Dependent Acquisition of Global DNA Methylation in Oocytes Is Altered by Hormonal Stimulation. Biol. Reprod. 2010, 83, 312. [Google Scholar] [CrossRef]
- Anderiesz, C.; Ferraretti, A.; Magli, C.; Fiorentino, A.; Fortini, D.; Gianaroli, L.; Jones, G.M.; Trounson, A.O. Effect of recombinant human gonadotrophins on human, bovine and murine oocyte meiosis, fertilization and embryonic development in vitro. Hum. Reprod. 2000, 15, 1140–1148. [Google Scholar] [CrossRef][Green Version]
- Liu, S.; Feng, H.L.; Marchesi, D.; Chen, Z.-J.; Hershlag, A. Effect of gonadotropins on dynamic events and global deoxyribonucleic acid methylation during in vitro maturation of oocytes: An animal model. Fertil. Steril. 2011, 95, 1503–1506. [Google Scholar] [CrossRef]
- Canovas, S.; Ivanova, E.; Romar, R.; García-Martínez, S.; Soriano-Úbeda, C.; García-Vázquez, F.A.; Saadeh, H.; Andrews, S.; Kelsey, G.; Coy, P. DNA methylation and gene expression changes derived from assisted reproductive technologies can be decreased by reproductive fluids. Elife 2017, 6, e23670. [Google Scholar] [CrossRef] [PubMed]
- De La Fuente, R.; Eppig, J.J. Transcriptional activity of the mouse oocyte genome: Companion granulosa cells modulate transcription and chromatin remodeling. Dev. Biol. 2001, 229, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Haaf, T. Aberrant methylation patterns at the two-cell stage as an indicator of early developmental failure. Mol. Reprod. Dev. 2002, 63, 329–334. [Google Scholar] [CrossRef]
- Liang, X.-W.; Cui, X.-S.; Sun, S.-C.; Jin, Y.-X.; Heo, Y.; Namgoong, S.; Kim, N.-H. Superovulation induces defective methylation in line-1 retrotransposon elements in blastocyst. Reprod. Biol. Endocrinol. 2013, 11, 69. [Google Scholar] [CrossRef]
- Denomme, M.M.; Zhang, L.; Mann, M.R.W. Embryonic imprinting perturbations do not originate from superovulation-induced defects in DNA methylation acquisition. Fertil. Steril. 2011, 96, 734–738. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Pausch, H.; Venhoranta, H.; Adamowicz, K.; Andersson, M.; Zwierzchowski, L.; Kind, A.; Schnieke, A.; Flisikowski, K. PEG3 domain gene expression in maternal and foetal placenta in intrauterine growth restricted bovine foetuses. Anim. Genet. 2016, 47, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Li, D.; Zhang, M.; Yang, W.; Wu, G.; Cui, Y.; Li, S. Biallelic expression of Tssc4, Nap1l4, Phlda2 and Osbpl5 in adult cattle. J. Genet. 2015, 94, 391–395. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.N.; Li, A.; Sun, S.M.; Liu, W.B.; Meng, T.G.; Guo, X.P.; Schatten, H.; Sun, Q.Y.; Ou, X.H. The methylation status in GNAS clusters May Be an epigenetic marker for oocyte quality. Biochem. Biophys. Res. Commun. 2020, 533, 586–591. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, E.; Canovas, S.; Garcia-Martínez, S.; Romar, R.; Lopes, J.S.; Rizos, D.; Sanchez-Calabuig, M.J.; Krueger, F.; Andrews, S.; Perez-Sanz, F.; et al. DNA methylation changes during preimplantation development reveal inter-species differences and reprogramming events at imprinted genes. Clin. Epigenet. 2020, 12, 64. [Google Scholar] [CrossRef]
- Clark, S.J.; Smallwood, S.A.; Lee, H.J.; Krueger, F.; Reik, W.; Kelsey, G. Genome-wide base-resolution mapping of DNA methylation in single cells using single-cell bisulfite sequencing (scBS-seq). Nat. Protoc. 2017, 12, 534–547. [Google Scholar] [CrossRef]
- Krueger, F.; Andrews, S.R. Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011, 27, 1571–1572. [Google Scholar] [CrossRef]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. Revigo summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef]
- Smallwood, S.A.; Lee, H.J.; Angermueller, C.; Krueger, F.; Saadeh, H.; Peat, J.; Andrews, S.R.; Stegle, O.; Reik, W.; Kelsey, G. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat. Methods 2014, 11, 817–820. [Google Scholar] [CrossRef]
- Tomizawa, S.; Kobayashi, H.; Watanabe, T.; Andrews, S.; Hata, K.; Kelsey, G.; Sasaki, H. Dynamic stage-specific changes in imprinted differentially methylated regions during early mammalian development and prevalence of non-CpG methylation in oocytes. Development 2011, 138, 811–820. [Google Scholar] [CrossRef]
- Shirane, K.; Toh, H.; Kobayashi, H.; Miura, F.; Chiba, H.; Ito, T.; Kono, T.; Sasaki, H. Mouse Oocyte Methylomes at Base Resolution Reveal Genome-Wide Accumulation of Non-CpG Methylation and Role of DNA Methyltransferases. PLoS Genet. 2013, 9, e1003439. [Google Scholar] [CrossRef] [PubMed]


| Global Mean Methylation (%) | ||
| Treatment | ||
| Non-stimulated | 52.91 | |
| Stimulated | 51.21 | |
| Animal | ||
| Cow 1 | 51.29 | |
| Cow 2 | 53.03 | |
| Cow 3 | 51.87 |
| Gene | p Value | FDR | Difference | NS | S | % Read CGs | |
|---|---|---|---|---|---|---|---|
| NS | S | ||||||
| APEG3 | 5.2618736 × 10−5 | 6.3142485 × 10−5 | 32.594048 | 29.62963 | 56.06027 | 40.37 | 69.14 |
| MEG3 | 1.0383111 × 10−8 | 1.5574667 × 10−8 | 17.314835 | 23.323172 | 32.191566 | 31.12 | 87.15 |
| MEG9 | 3.1143794 × 10−4 | 3.1143794 × 10−4 | 18.571428 | 46.951218 | 76.15186 | 39.81 | 93.20 |
| TSSC4 | 0.0 | 0.0 | 41.000954 | 81.801994 | 52.26771 | 55.56 | 90.85 |
| Probe | Gene | p-Value | FDR | Difference | NS | S | % Read CGs | |
|---|---|---|---|---|---|---|---|---|
| NS | S | |||||||
| oe = 0.66 | MEST (PEG1) | 0.010897397 | 0.012454168 | 17.783426 | 97.06 | 77.37 | 23.44 | 86.72 |
| oe = 0.85 | IGF2R | 5.962238 × 10−4 | 0.0011924476 | 11.239578 | 97.94 | 88.54 | 31.41 | 74.73 |
| oe = 1.07 | IGF2R | 9.842796 × 10−5 | 2.6247458× 10−4 | 32.3551 | 97.37 | 65.52 | 60.00 | 64.44 |
| oe = 0.75 | GNAS (SCG6) | 0.0020540599 | 0.0027387466 | 18.394482 | 97.30 | 81.26 | 15.43 | 70.86 |
| oe = 0.87 | KvDMR1 ICR UMD | 0.0 | 0.0 | 45.271866 | 100 | 58.57 | 28.99 | 98.07 |
| oe = 0.60 | IGF2 | 8.9613185× 10−4 | 0.0014338109 | 18.31723 | 2.63 | 10.92 | 36.19 | 100 |
| oe = 0.81 | IGF2 | 0.0 | 0.0 | 45.195435 | 44.91 | 5.43 | 35.00 | 100 |
| Cow 1 | Cow 2 | Cow 3 | ||||
|---|---|---|---|---|---|---|
| No. of oocytes | Collected | Sequenced | Collected | Sequenced | Collected | Sequenced |
| Non-stimulated (NS) | 6 | 6 | 6 | 4 | 8 | 5 |
| Stimulated (S) | 11 | 10 | 9 | 7 | 29 | 15 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lopes, J.S.; Ivanova, E.; Ruiz, S.; Andrews, S.; Kelsey, G.; Coy, P. Effect of Superovulation Treatment on Oocyte’s DNA Methylation. Int. J. Mol. Sci. 2022, 23, 16158. https://doi.org/10.3390/ijms232416158
Lopes JS, Ivanova E, Ruiz S, Andrews S, Kelsey G, Coy P. Effect of Superovulation Treatment on Oocyte’s DNA Methylation. International Journal of Molecular Sciences. 2022; 23(24):16158. https://doi.org/10.3390/ijms232416158
Chicago/Turabian StyleLopes, Jordana S., Elena Ivanova, Salvador Ruiz, Simon Andrews, Gavin Kelsey, and Pilar Coy. 2022. "Effect of Superovulation Treatment on Oocyte’s DNA Methylation" International Journal of Molecular Sciences 23, no. 24: 16158. https://doi.org/10.3390/ijms232416158
APA StyleLopes, J. S., Ivanova, E., Ruiz, S., Andrews, S., Kelsey, G., & Coy, P. (2022). Effect of Superovulation Treatment on Oocyte’s DNA Methylation. International Journal of Molecular Sciences, 23(24), 16158. https://doi.org/10.3390/ijms232416158






