Dysregulation in Multiple Transcriptomic Endometrial Pathways Is Associated with Recurrent Implantation Failure and Recurrent Early Pregnancy Loss
Abstract
1. Introduction
2. Results
2.1. Midluteal Phase Endometrial Tissue for RIF, REPL and Control Group Exhibited Distinct mRNA Expression Profiles
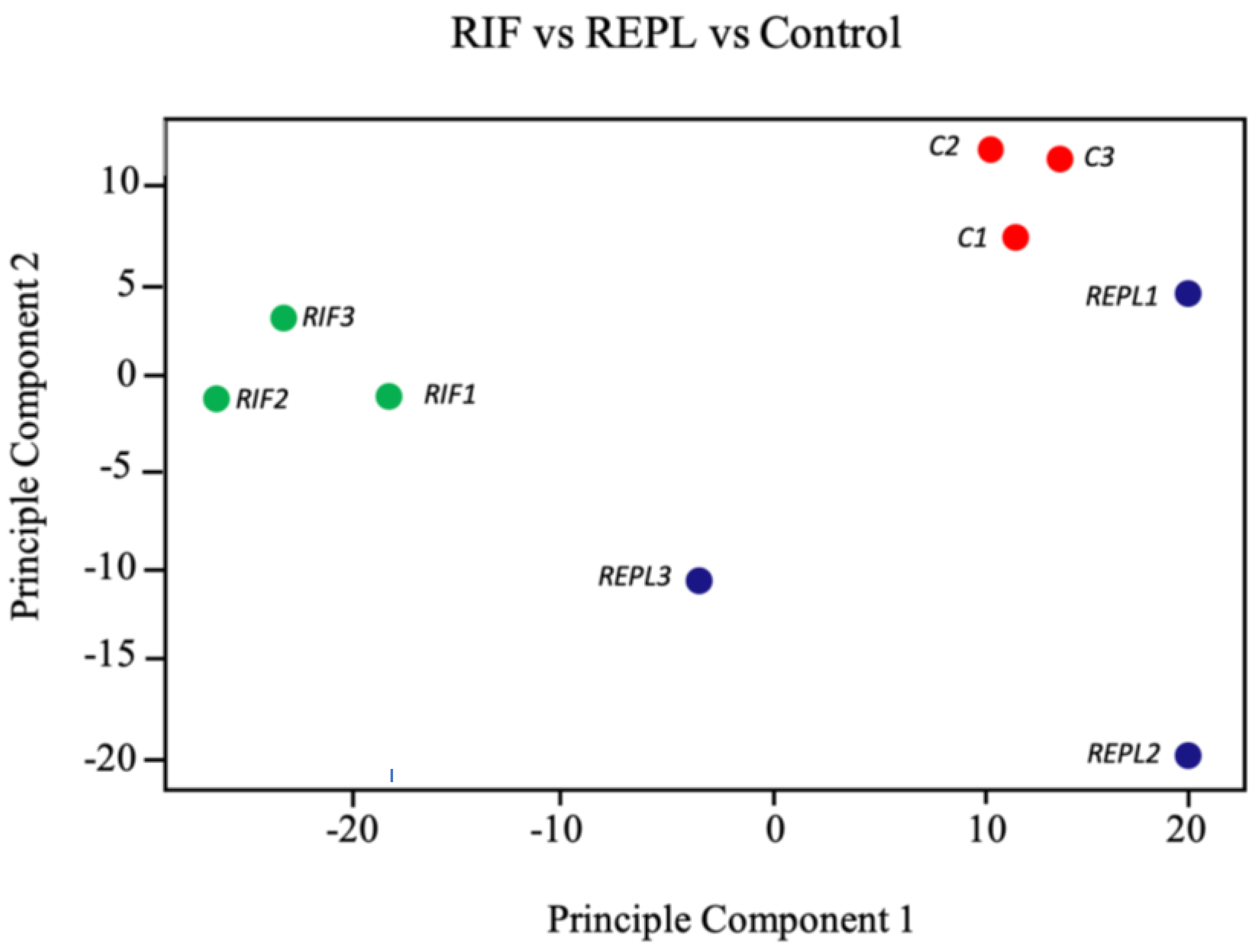
2.2. Unsupervised Clustering of Microarray Data Sets of the Endometrial Biopsies
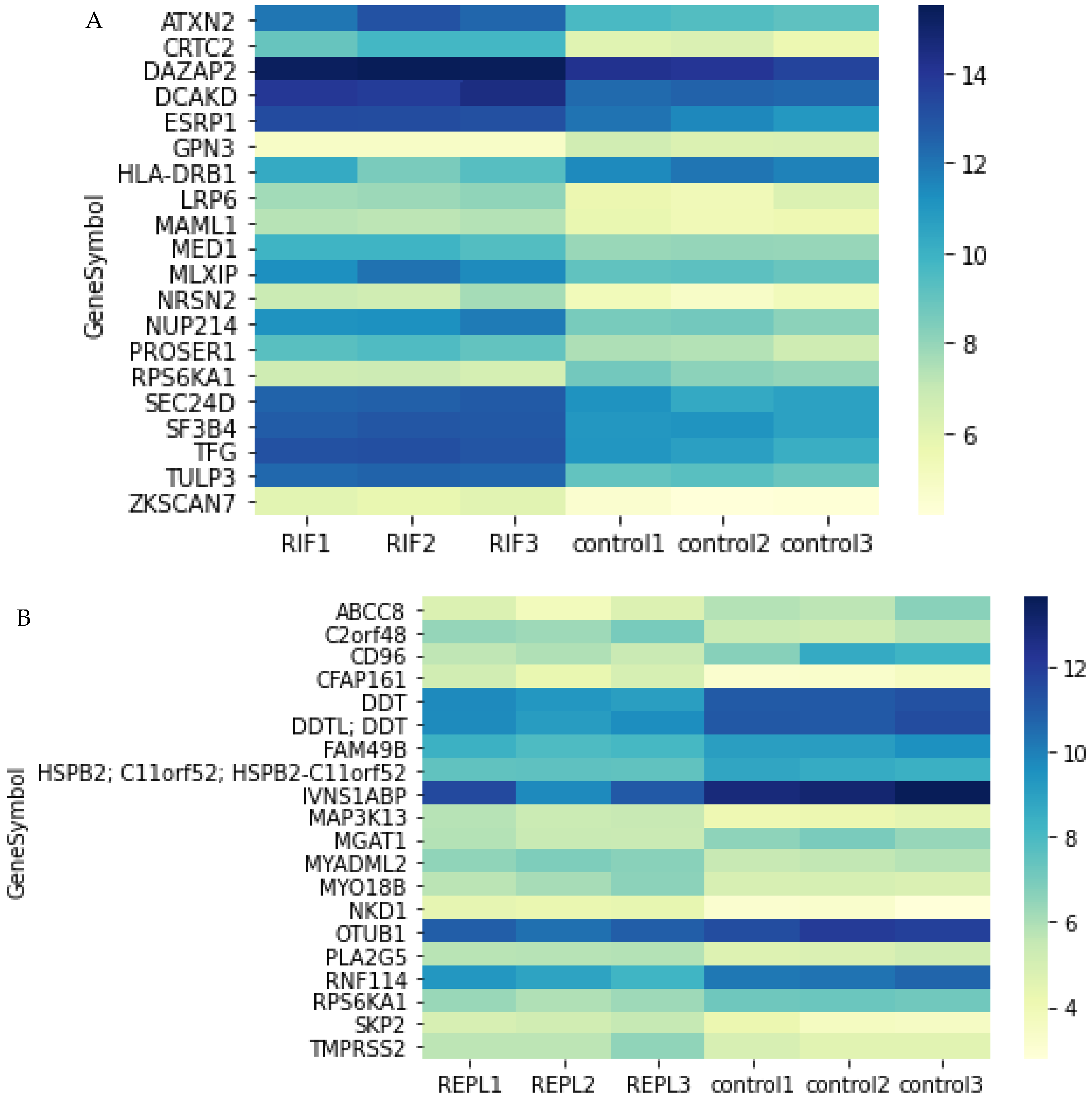
2.3. Top 20 DEGs in Endometrial Tissue of RIF and REPL Group of Patients
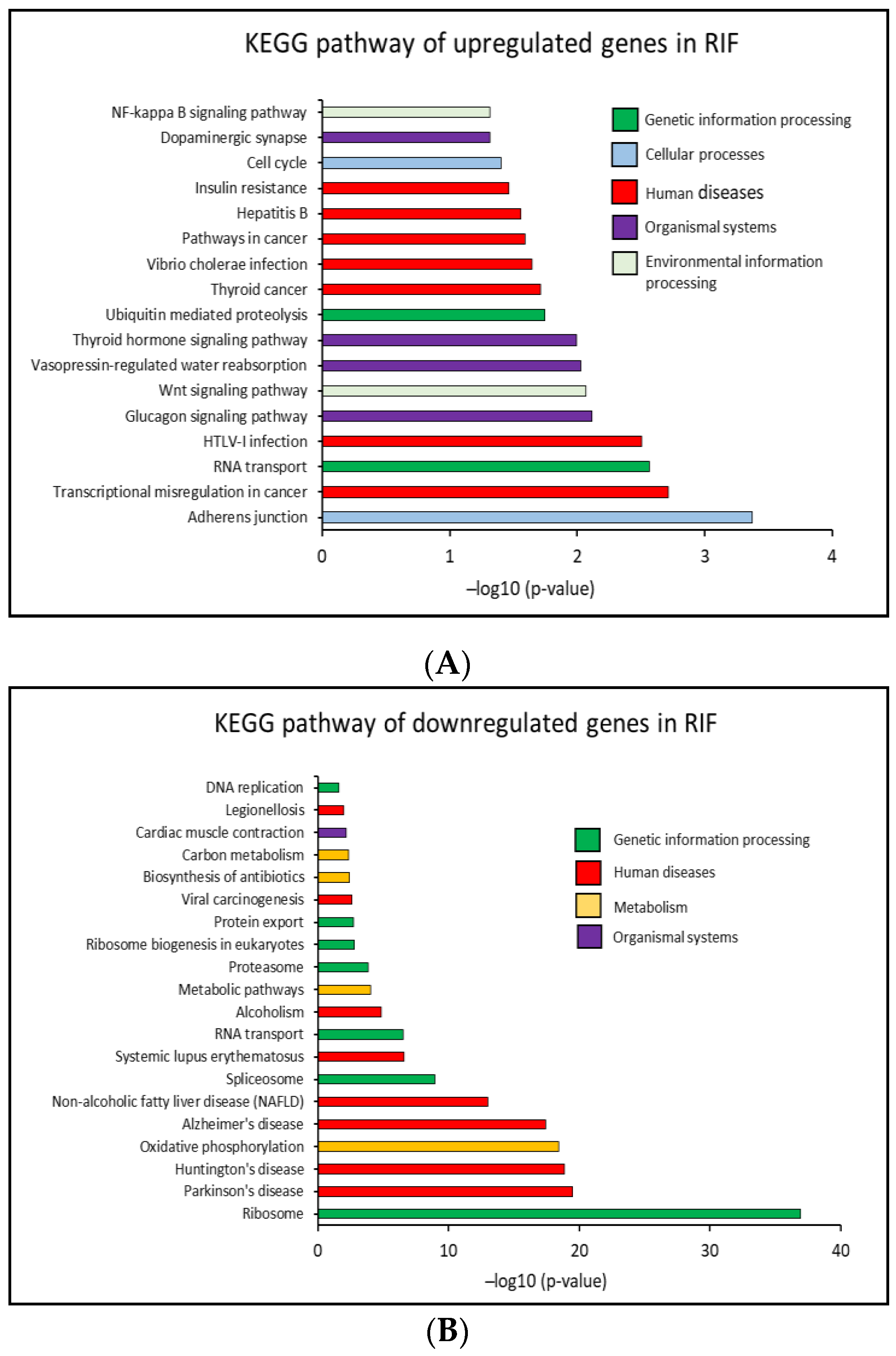
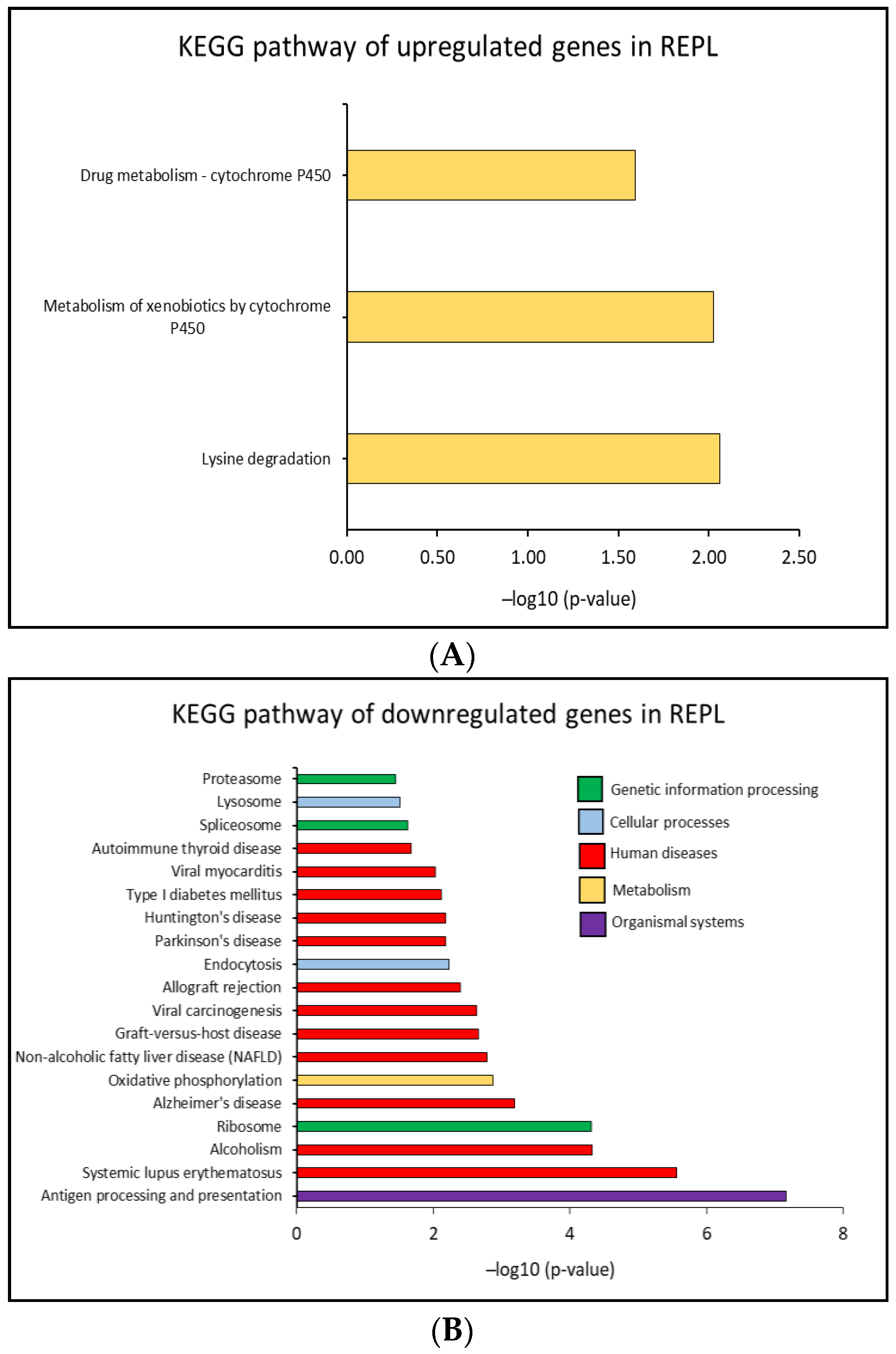
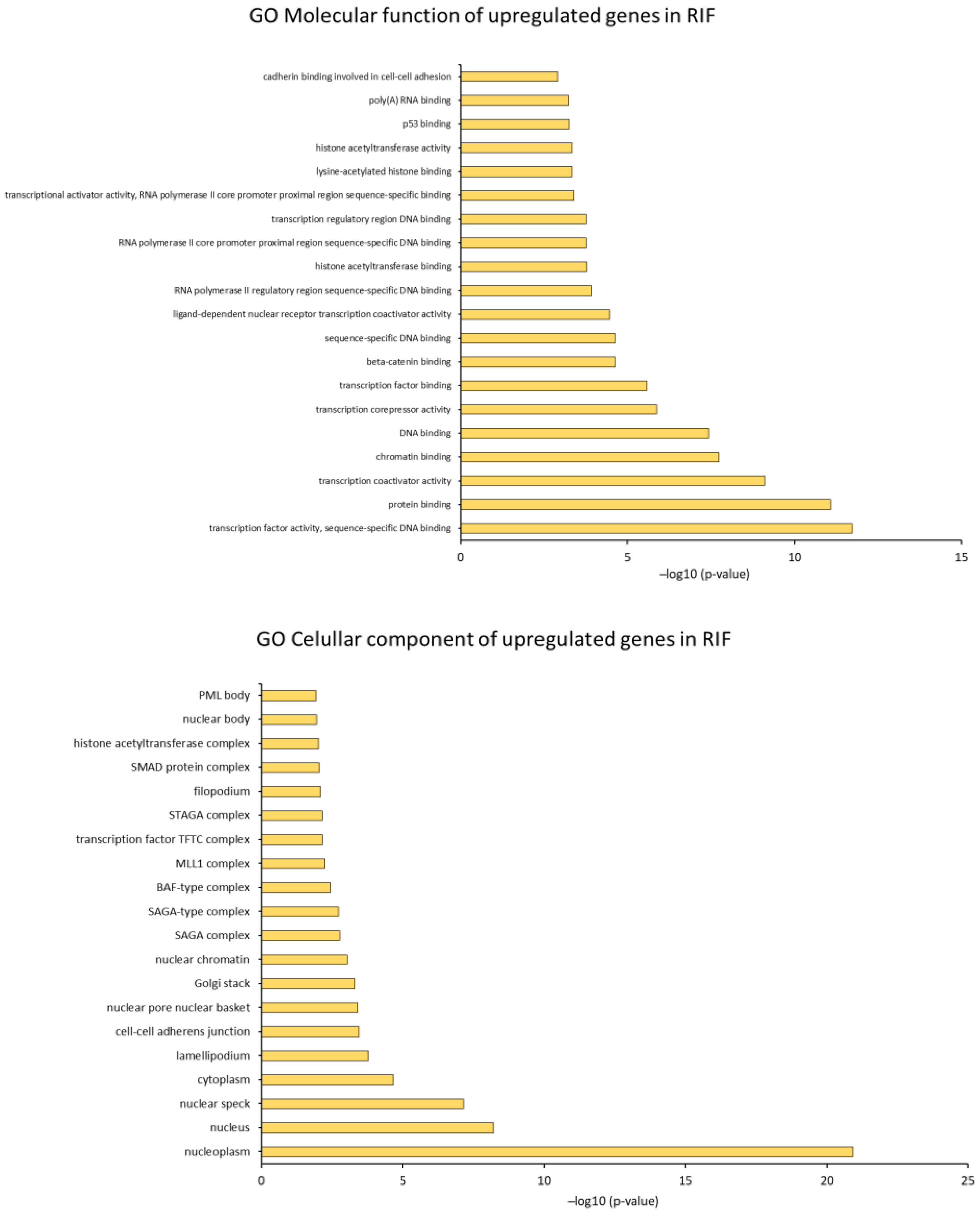
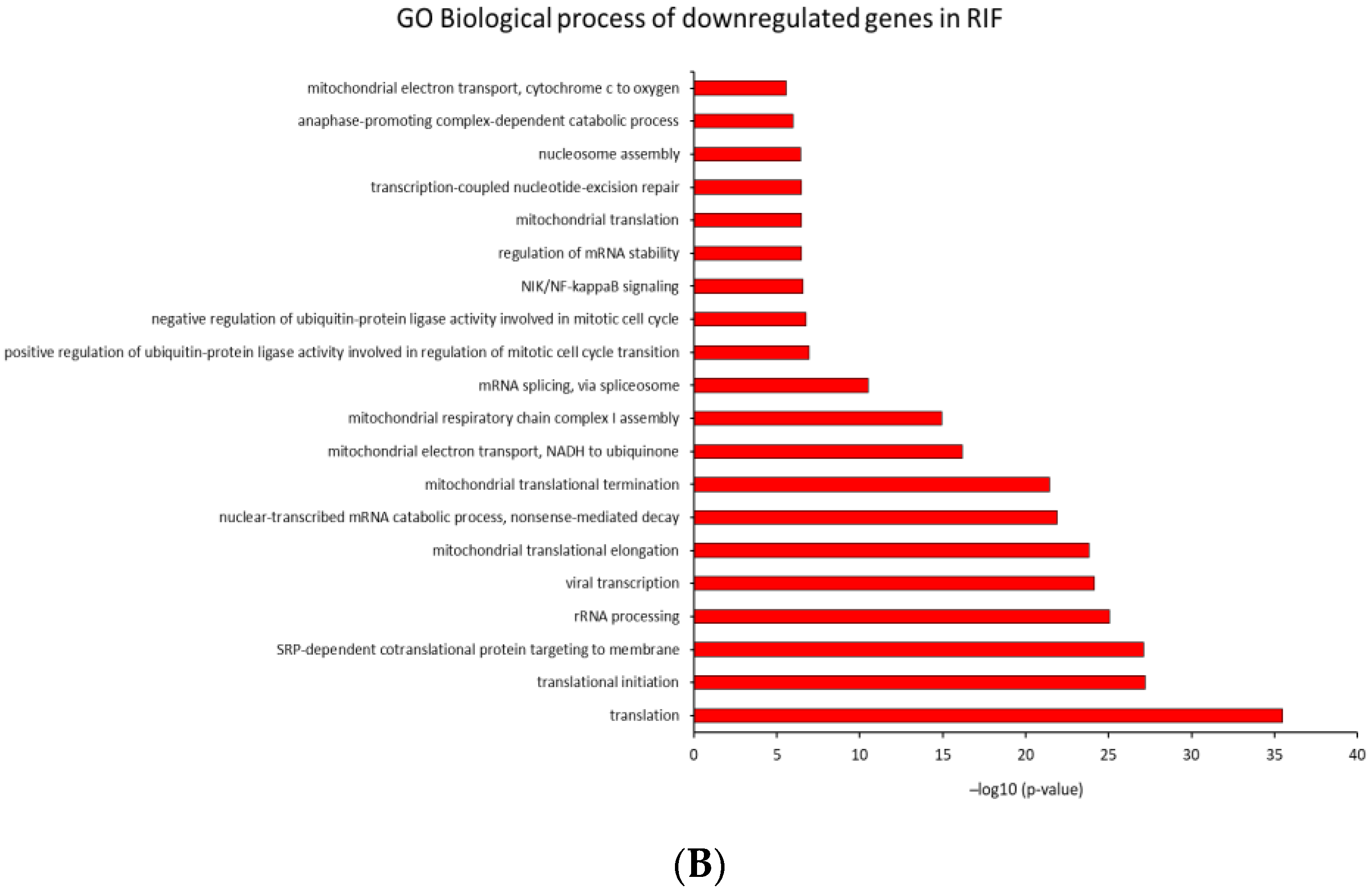
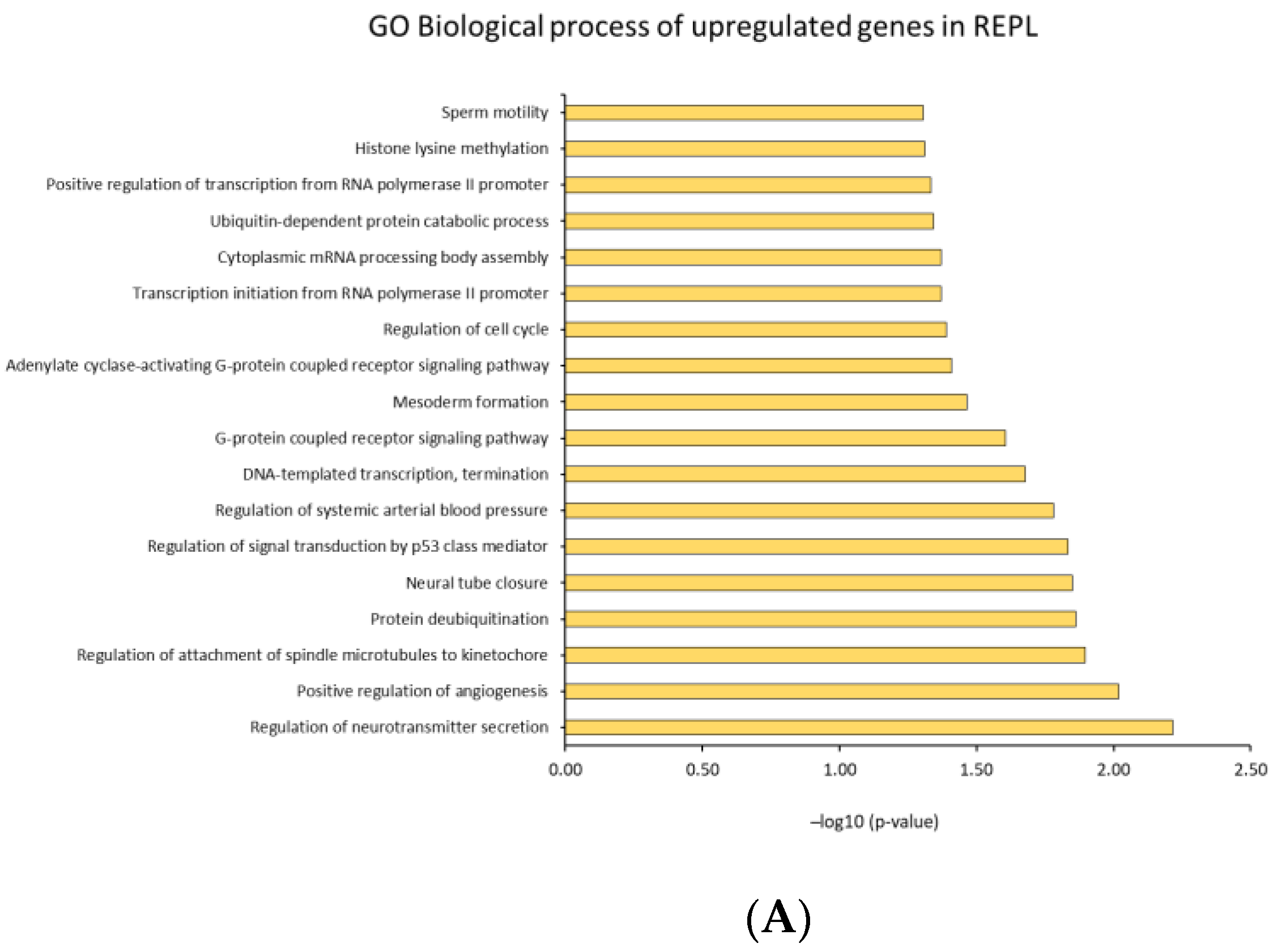
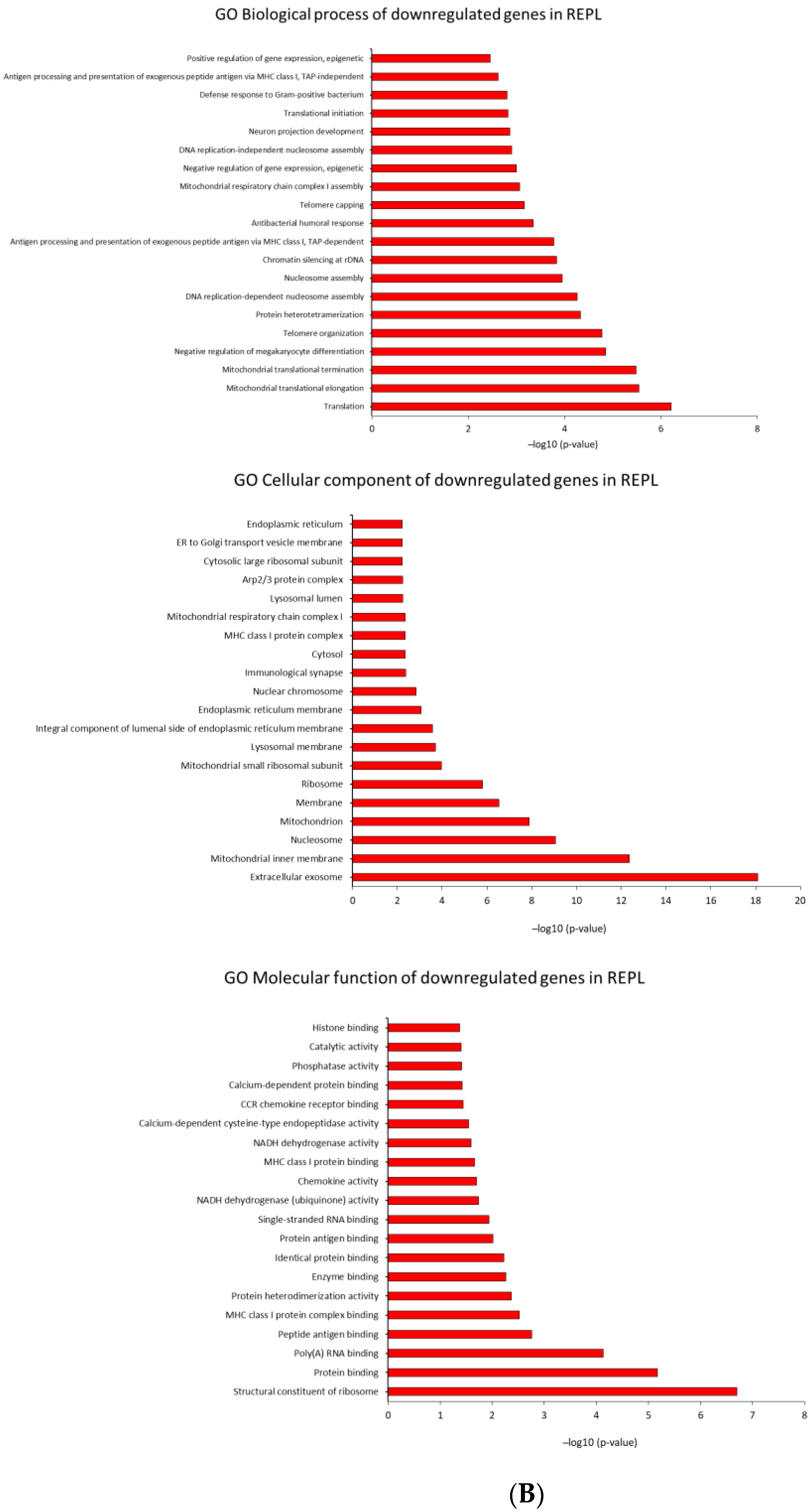
2.4. Identification of Biological Pathways
2.4.1. KEGG Pathway
2.4.2. GO Analysis
3. Discussion
4. Materials and Methods
4.1. Ethics Approval
4.2. Patient Selection
4.3. Patient Recruitment
4.4. Endometrial Tissue Collection/Biopsy
4.5. Total RNA Isolation
4.6. Microarray Analysis
4.7. Bioinformatic Analysis
4.8. Results Validation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yatsenko, S.A.; Rajkovic, A. Genetics of Human Female Infertility†. Biol. Reprod. 2019, 101, 549–566. [Google Scholar] [CrossRef]
- Rutstein, S.O.; Shah, I.H. Infecundity, Infertility, and Childlessness in Developing Countries; World Health Organization: Geneva, Switzerland, 2004. [Google Scholar]
- Mascarenhas, M.N.; Flaxman, S.R.; Boerma, T.; Vanderpoel, S.; Stevens, G.A. National, Regional, and Global Trends in Infertility Prevalence Since 1990: A Systematic Analysis of 277 Health Surveys. PLoS Med. 2012, 9, e1001356. [Google Scholar] [CrossRef] [PubMed]
- Afshani, S.A.; Mohammadi, S.M.R.G.; Khani, P.; Khosravi, A. Role of Resilience Training on Compromising of Infertile Couples’ Applicant for Divorce: A Cross-Sectional Study. Int. J. Reprod. Biomed. 2020, 18, 193–208. [Google Scholar] [CrossRef]
- Hasanpoor-Azghdy, S.B.; Simbar, M.; Vedadhir, A. The Emotional-Psychological Consequences of Infertility among Infertile Women Seeking Treatment: Results of a Qualitative Study. Iran. J. Reprod. Med. 2014, 12, 131–138. [Google Scholar]
- Lédée, N.; Petitbarat, M.; Chevrier, L.; Vitoux, D.; Vezmar, K.; Rahmati, M.; Dubanchet, S.; Gahéry, H.; Bensussan, A.; Chaouat, G. The Uterine Immune Profile May Help Women with Repeated Unexplained Embryo Implantation Failure after In Vitro Fertilization. Am. J. Reprod. Immunol. 2016, 75, 388–401. [Google Scholar] [CrossRef]
- Coughlan, C.; Ledger, W.; Wang, Q.; Liu, F.; Demirol, A.; Gurgan, T.; Cutting, R.; Ong, K.; Sallam, H.; Li, T.C. Recurrent Implantation Failure: Definition and Management. Reprod. Biomed. Online 2014, 28, 14–38. [Google Scholar] [CrossRef] [PubMed]
- Bashiri, A.; Halper, K.I.; Orvieto, R. Recurrent Implantation Failure-Update Overview on Etiology, Diagnosis, Treatment and Future Directions. Reprod. Biol. Endocrinol. 2018, 16, 121. [Google Scholar] [CrossRef] [PubMed]
- Practice Committee of American Society for Reproductive Medicine. Definitions of Infertility and Recurrent Pregnancy Loss. Fertil. Steril. 2008, 90 (Suppl. 5). [Google Scholar] [CrossRef]
- Toth, B.; Würfel, W.; Bohlmann, M.; Zschocke, J.; Rudnik-Schöneborn, S.; Nawroth, F.; Schleußner, E.; Rogenhofer, N.; Wischmann, T.; von Wolff, M.; et al. Recurrent Miscarriage: Diagnostic and Therapeutic Procedures. Guideline of the DGGG, OEGGG and SGGG (S2k-Level, AWMF Registry Number 015/050). Geburtshilfe Frauenheilkd 2018, 78, 364–381. [Google Scholar] [CrossRef]
- Baek, K.-H.; Lee, E.-J.; Kim, Y.-S. Recurrent Pregnancy Loss: The Key Potential Mechanisms. Trends. Mol. Med. 2007, 13, 310–317. [Google Scholar] [CrossRef]
- Ford, H.B.; Schust, D.J. Recurrent Pregnancy Loss: Etiology, Diagnosis, and Therapy. Rev. Obstet. Gynecol. 2009, 2, 76–83. [Google Scholar]
- Garrido-Gimenez, C.; Alijotas-Reig, J. Recurrent Miscarriage: Causes, Evaluation and Management. Postgrad. Med. J. 2015, 91, 151–162. [Google Scholar] [CrossRef]
- Huang, J.; Qin, H.; Yang, Y.; Chen, X.; Zhang, J.; Laird, S.; Wang, C.C.; Chan, T.F.; Li, T.C. A Comparison of Transcriptomic Profiles in Endometrium during Window of Implantation between Women with Unexplained Recurrent Implantation Failure and Recurrent Miscarriage. Reproduction 2017, 153, 749–758. [Google Scholar] [CrossRef]
- Wang, L.; Tang, H.; Xiong, Y.; Tang, L. Differential Expression Profile of Long Noncoding RNAs in Human Chorionic Villi of Early Recurrent Miscarriage. Clin. Chim. Acta 2017, 464, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Bastu, E.; Demiral, I.; Gunel, T.; Ulgen, E.; Gumusoglu, E.; Hosseini, M.K.; Sezerman, U.; Buyru, F.; Yeh, J. Potential Marker Pathways in the Endometrium That May Cause Recurrent Implantation Failure. Reprod. Sci. 2019, 26, 879–890. [Google Scholar] [CrossRef] [PubMed]
- Baek, K.-H. Aberrant Gene Expression Associated with Recurrent Pregnancy Loss. Mol. Hum. Reprod. 2004, 10, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Qiao, J.; Wang, L.; Li, R.; Zhang, X. Microarray Evaluation of Endometrial Receptivity in Chinese Women with Polycystic Ovary Syndrome. Reprod. Biomed. Online 2008, 17, 425–435. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Yao, G.; Wang, Y.; Xu, H.; Ji, X.; He, Y.; Zhu, Q.; Chen, Z.; Sun, Y. Transcriptomic Changes during the Pre-Receptive to Receptive Transition in Human Endometrium Detected by RNA-Seq. J. Clin. Endocrinol. Metab. 2014, 99, E2744–E2753. [Google Scholar] [CrossRef]
- Othman, R.; Omar, M.H.; Shan, L.P.; Shafiee, M.N.; Jamal, R.; Mokhtar, N.M. Microarray Profiling of Secretory-Phase Endometrium from Patients with Recurrent Miscarriage. Reprod. Biol. 2012, 12, 183–199. [Google Scholar] [CrossRef]
- Haouzi, D.; Assou, S.; Mahmoud, K.; Tondeur, S.; Rème, T.; Hedon, B.; De Vos, J.; Hamamah, S. Gene Expression Profile of Human Endometrial Receptivity: Comparison between Natural and Stimulated Cycles for the Same Patients. Hum. Reprod. 2009, 24, 1436–1445. [Google Scholar] [CrossRef]
- Lessey, B.A. Embryo Quality and Endometrial Receptivity: Lessons Learned from the ART Experience. J. Assist. Reprod. Genet. 1998, 15, 173–176. [Google Scholar] [CrossRef] [PubMed]
- Wilcox, A.J.; Baird, D.D.; Weinberg, C.R. Time of Implantation of the Conceptus and Loss of Pregnancy. N. Engl. J. Med. 1999, 340, 1796–1799. [Google Scholar] [CrossRef]
- Messaoudi, S.; El Kasmi, I.; Bourdiec, A.; Crespo, K.; Bissonnette, L.; Le Saint, C.; Bissonnette, F.; Kadoch, I.-J. 15 Years of Transcriptomic Analysis on Endometrial Receptivity: What Have We Learnt? Fertil. Res. Pract. 2019, 5, 9. [Google Scholar] [CrossRef] [PubMed]
- Coutifaris, C.; Myers, E.R.; Guzick, D.S.; Diamond, M.P.; Carson, S.A.; Legro, R.S.; McGovern, P.G.; Schlaff, W.D.; Carr, B.R.; Steinkampf, M.P.; et al. Histological Dating of Timed Endometrial Biopsy Tissue Is Not Related to Fertility Status. Fertil. Steril. 2004, 82, 1264–1272. [Google Scholar] [CrossRef] [PubMed]
- Giudice, L.C. Potential Biochemical Markers of Uterine Receptivity. Hum. Reprod. 1999, 14 (Suppl. 2), 3–16. [Google Scholar] [CrossRef]
- Simon, A.; Laufer, N. Assessment and Treatment of Repeated Implantation Failure (RIF). J. Assist. Reprod. Genet. 2012, 29, 1227–1239. [Google Scholar] [CrossRef]
- Horcajadas, J.; Díaz-Gimeno, P.; Pellicer, A.; Simón, C. Uterine Receptivity and the Ramifications of Ovarian Stimulation on Endometrial Function. Semin. Reprod. Med. 2007, 25, 454–460. [Google Scholar] [CrossRef]
- Choi, Y.; Kim, H.-R.; Lim, E.J.; Park, M.; Yoon, J.A.; Kim, Y.S.; Kim, E.-K.; Shin, J.-E.; Kim, J.H.; Kwon, H.; et al. Integrative Analyses of Uterine Transcriptome and MicroRNAome Reveal Compromised LIF-STAT3 Signaling and Progesterone Response in the Endometrium of Patients with Recurrent/Repeated Implantation Failure (RIF). PLoS ONE 2016, 11, e0157696. [Google Scholar] [CrossRef]
- Koot, Y.E.M.; van Hooff, S.R.; Boomsma, C.M.; van Leenen, D.; Groot Koerkamp, M.J.A.; Goddijn, M.; Eijkemans, M.J.C.; Fauser, B.C.J.M.; Holstege, F.C.P.; Macklon, N.S. An Endometrial Gene Expression Signature Accurately Predicts Recurrent Implantation Failure after IVF. Sci. Rep. 2016, 6, 19411. [Google Scholar] [CrossRef]
- He, A.; Zou, Y.; Wan, C.; Zhao, J.; Zhang, Q.; Yao, Z.; Tian, F.; Wu, H.; Huang, X.; Fu, J.; et al. The Role of Transcriptomic Biomarkers of Endometrial Receptivity in Personalized Embryo Transfer for Patients with Repeated Implantation Failure. J. Transl. Med. 2021, 19, 176. [Google Scholar] [CrossRef]
- Ramakrishnan, V. The Ribosome Emerges from a Black Box. Cell 2014, 159, 979–984. [Google Scholar] [CrossRef]
- Calamita, P.; Gatti, G.; Miluzio, A.; Scagliola, A.; Biffo, S. Translating the Game: Ribosomes as Active Players. Front. Genet. 2018, 9, 533. [Google Scholar] [CrossRef]
- Huang, G.; Li, H.; Zhang, H. Abnormal Expression of Mitochondrial Ribosomal Proteins and Their Encoding Genes with Cell Apoptosis and Diseases. Int. J. Mol. Sci. 2020, 21, 8879. [Google Scholar] [CrossRef]
- Sõber, S.; Rull, K.; Reiman, M.; Ilisson, P.; Mattila, P.; Laan, M. RNA Sequencing of Chorionic Villi from Recurrent Pregnancy Loss Patients Reveals Impaired Function of Basic Nuclear and Cellular Machinery. Sci. Rep. 2016, 6, 38439. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Feng, Y.; Zhou, W.-J.; Cheng, Z.-J.; Jiang, M.-Y.; Zhou, Y.; Fei, X.-Y. Screening and Identification of Endometrial Proteins as Novel Potential Biomarkers for Repeated Implantation Failure. PeerJ 2021, 9, e11009. [Google Scholar] [CrossRef] [PubMed]
- Xin, L.; Xu, B.; Ma, L.; Hou, Q.; Ye, M.; Meng, S.; Ding, X.; Ge, W. Proteomics Study Reveals That the Dysregulation of Focal Adhesion and Ribosome Contribute to Early Pregnancy Loss. Prot. Clin. Appl. 2016, 10, 554–563. [Google Scholar] [CrossRef] [PubMed]
- Lyu, S.W.; Song, H.; Yoon, J.A.; Chin, M.-U.; Sung, S.R.; Kim, Y.S.; Lee, W.S.; Yoon, T.K.; Cha, D.H.; Shim, S.H. Transcriptional Profiling with a Pathway-Oriented Analysis in the Placental Villi of Unexplained Miscarriage. Placenta 2013, 34, 133–140. [Google Scholar] [CrossRef]
- Wu, M.; Gu, J.; Zong, S.; Guo, R.; Liu, T.; Yang, M. Research Journey of Respirasome. Protein Cell 2020, 11, 318–338. [Google Scholar] [CrossRef]
- Hroudová, J.; Fišar, Z. Control Mechanisms in Mitochondrial Oxidative Phosphorylation. Neural Regen. Res. 2013, 8, 363–375. [Google Scholar] [CrossRef]
- Pieczenik, S.R.; Neustadt, J. Mitochondrial Dysfunction and Molecular Pathways of Disease. Exp. Mol. Pathol. 2007, 83, 84–92. [Google Scholar] [CrossRef]
- Yin, X.-J.; Hong, W.; Tian, F.-J.; Li, X.-C. Proteomic Analysis of Decidua in Patients with Recurrent Pregnancy Loss (RPL) Reveals Mitochondrial Oxidative Stress Dysfunction. Clin. Proteom. 2021, 18, 9. [Google Scholar] [CrossRef] [PubMed]
- Bi, R.; Zhang, W.; Zhang, D.-F.; Xu, M.; Fan, Y.; Hu, Q.-X.; Jiang, H.-Y.; Tan, L.; Li, T.; Fang, Y.; et al. Genetic Association of the Cytochrome c Oxidase-Related Genes with Alzheimer’s Disease in Han Chinese. Neuropsychopharmacology 2018, 43, 2264–2276. [Google Scholar] [CrossRef] [PubMed]
- Antsiferova, Y.S. Apoptosis and Endometrial Receptivity: Relationship with In Vitro Fertilization Treatment Outcome. WJOG 2016, 5, 87. [Google Scholar] [CrossRef]
- Zaninovic, N.; Berrios, R.; Clarke, R.N.; Bodine, R.; Ye, Z.; Veeck, L.L. Blastocyst expansion, inner cell mass (ICM) formation, and trophectoderm (TM) quality: Is one more important for implantation? Fertil. Steril. 2001, 76, S8. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]


| Patient Selection | Control Group (n = 3) | RIF (n = 3) | REPL (n = 3) |
|---|---|---|---|
| Inclusion criteria | Age under 40 years | Age 40 years or less | Age 40 years or less |
| Undergoing an IVF cycle that result in an ongoing clinical pregnancy as defined above | Fulfilling the definition of RIF | Fulfilling the definition of REPL | |
| Consent to participate in this study | Normal results obtained on routine RIF investigations (thrombophilia screening, Karyotyping, hormonal profile, and detailed pelvic scan) | Normal results obtained on routine REPL investigations (thrombophilia screening, Karyotyping, hormonal profile, and detailed pelvic scan) | |
| Consent to participate in this study | Consent to participation in this study | ||
| Exclusion criteria | Women aged 40 years or older | Women aged 40 years or older | Women aged 40 years or older |
| Known uterine or endometrial pathology. | Known uterine or endometrial pathology | Known uterine or endometrial pathology | |
| History of RIF | Known cause of RIF | Known cause of REPL | |
| Low ovarian reserve defined by a baseline serum FSH level above 10 IU/ML | |||
| Known uncorrected endocrinological pathologies |
| Gene Symbol | Gene Name | Fold Change | |
|---|---|---|---|
| MLXIP | MLX interacting protein | 1.53 | Upregulated genes |
| LRP6 | LDL receptor related protein 6 | 1.63 | |
| ZKSCAN7 | zinc finger with KRAB and SCAN domains 7 | 2.13 | |
| DAZAP2 | DAZ Associated Protein 2 | 2.73 | |
| NRSN2 | neurensin 2 | 2.77 | |
| DCAKD | dephospho-CoA kinase domain containing | 2.96 | |
| ESRP1 | epithelial splicing regulatory protein 1 | 3.43 | |
| MAML1 | mastermind-like transcriptional coactivator 1 | 3.51 | |
| PROSER1 | proline and serine rich 1 | 3.57 | |
| SF3B4 | splicing factor 3b, subunit 4, 49 kDa | 3.6 | |
| SEC24D | SEC24 Homolog D, COPII Coat Complex Component | 3.7 | |
| MED1 | mediator complex subunit 1 | 3.87 | |
| TFG | TRK-fused gene | 5.36 | |
| NUP214 | nucleoporin 214 kDa | 6.15 | |
| ATXN2 | ataxin 2 | 8.22 | |
| TULP3 | tubby like protein 3 | 10.26 | |
| CRTC2 | CREB regulated transcription coactivator 2 | 13.24 | |
| HLA-DRB1 | major histocompatibility complex, class II, DR beta 1 | −5.04 | Downregulated genes |
| RPS6KA1 | Ribosomal Protein S6 Kinase A1 | −2.9 | |
| GPN3 | GPN-Loop GTPase 3 | −2.75 |
| Gene Symbol | Gene Name | Fold Change | |
|---|---|---|---|
| SKP2 | Transcript Identified by AceView, Entrez Gene ID(s) 6502 | 1.9 | Upregulated genes |
| PLA2G5 | phospholipase A2, group V | 1.93 | |
| MYADML2 | myeloid-associated differentiation marker-like 2 | 2.08 | |
| C2orf48 | chromosome 2 open reading frame 48 | 2.09 | |
| TMPRSS2 | transmembrane protease, serine 2 | 2.15 | |
| MYO18B | myosin XVIIIB | 2.3 | |
| NKD1 | NKD Inhibitor of WNT Signaling Pathway 1 | 2.3 | |
| MAP3K13 | mitogen-activated protein kinase kinase kinase 13 | 2.38 | |
| CFAP161 | cilia and flagella associated protein 161 | 3.33 | |
| CD96 | CD96 molecule | −6.41 | Downregulated genes |
| IVNS1ABP | influenza virus NS1A binding protein | −4.29 | |
| DDT | D-dopachrome tautomerase | −3.2 | |
| RNF114 | ring finger protein 114 | −2.91 | |
| DDTL; DDT | D-dopachrome tautomerase-like; D-dopachrome tautomerase | −2.65 | |
| MGAT1 | mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase | −2.3 | |
| HSPB2; C11orf52; HSPB2-C11orf52 | heat shock 27kDa protein 2; chromosome 11 open reading frame 52; HSPB2-C11orf52 readthrough (NMD candidate) | −2.16 | |
| FAM49B | family with sequence similarity 49, member B | −1.98 | |
| ABCC8 | ATP binding cassette subfamily C member 8 | −1.98 | |
| OTUB1 | OTU deubiquitinase, ubiquitin aldehyde binding 1 | −1.93 | |
| RPS6KA1 | ribosomal protein S6 kinase, 90 kDa, polypeptide 1 | −1.93 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liaqat Ali Khan, N.; Nafee, T.; Shao, T.; Hart, A.R.; Elliott, S.; Ola, B.; Heath, P.R.; Fazeli, A. Dysregulation in Multiple Transcriptomic Endometrial Pathways Is Associated with Recurrent Implantation Failure and Recurrent Early Pregnancy Loss. Int. J. Mol. Sci. 2022, 23, 16051. https://doi.org/10.3390/ijms232416051
Liaqat Ali Khan N, Nafee T, Shao T, Hart AR, Elliott S, Ola B, Heath PR, Fazeli A. Dysregulation in Multiple Transcriptomic Endometrial Pathways Is Associated with Recurrent Implantation Failure and Recurrent Early Pregnancy Loss. International Journal of Molecular Sciences. 2022; 23(24):16051. https://doi.org/10.3390/ijms232416051
Chicago/Turabian StyleLiaqat Ali Khan, Norhayati, Tamer Nafee, Tingting Shao, Amber Rose Hart, Sarah Elliott, Bolarinde Ola, Paul Roy Heath, and Alireza Fazeli. 2022. "Dysregulation in Multiple Transcriptomic Endometrial Pathways Is Associated with Recurrent Implantation Failure and Recurrent Early Pregnancy Loss" International Journal of Molecular Sciences 23, no. 24: 16051. https://doi.org/10.3390/ijms232416051
APA StyleLiaqat Ali Khan, N., Nafee, T., Shao, T., Hart, A. R., Elliott, S., Ola, B., Heath, P. R., & Fazeli, A. (2022). Dysregulation in Multiple Transcriptomic Endometrial Pathways Is Associated with Recurrent Implantation Failure and Recurrent Early Pregnancy Loss. International Journal of Molecular Sciences, 23(24), 16051. https://doi.org/10.3390/ijms232416051






