Anp32a Promotes Neuronal Regeneration after Spinal Cord Injury of Zebrafish Embryos
Abstract
1. Introduction
2. Results
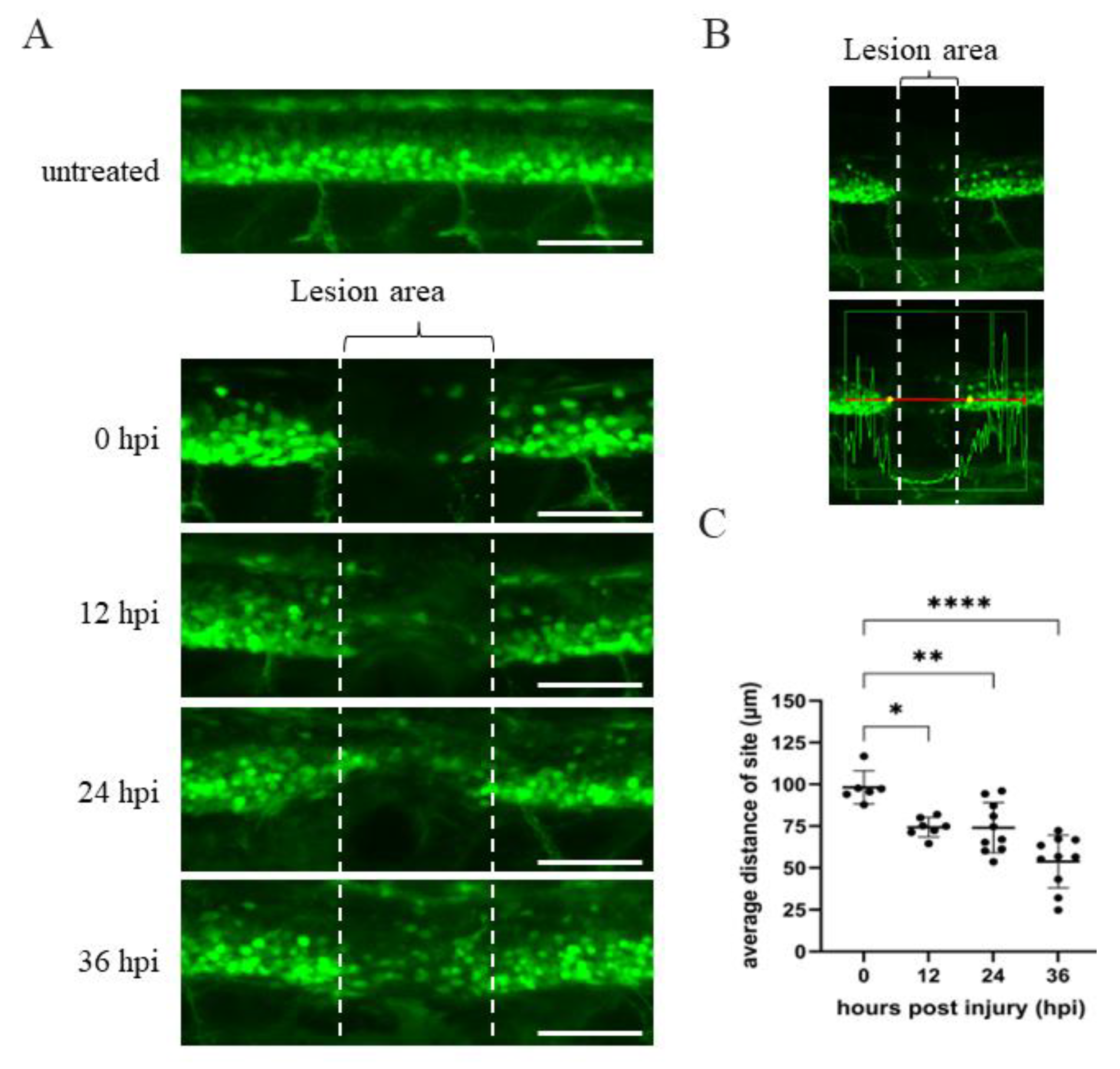
2.1. Optimal Recovery Time after SCI as a Baseline to Study Spinal Cord Regeneration of SCI Embryos
2.2. Effect of Knockdown or Overexpression of ANP32a mRNA on the Expression of ANP32a Protein in Zebrafish Embryos
2.3. Using Gain- and Loss-of-Function Strategies to Analyze the Effects of ANP32a on Axonal Regeneration of SCI-Zebrafish Embryos
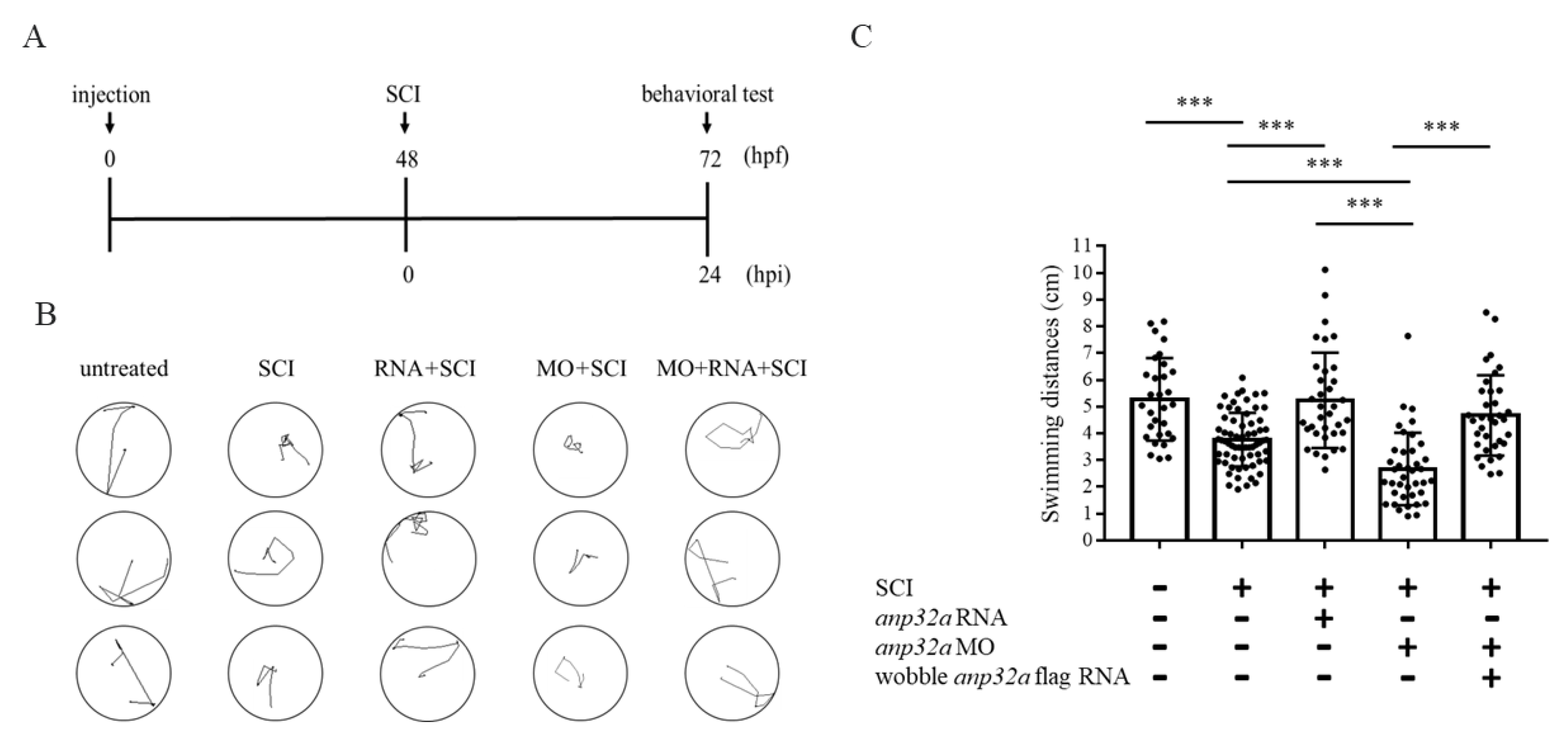
2.4. Using Gain- and Loss-of-Function Strategies to Analyze the Effects of ANP32a on Swimming Capability of SCI Zebrafish Embryos
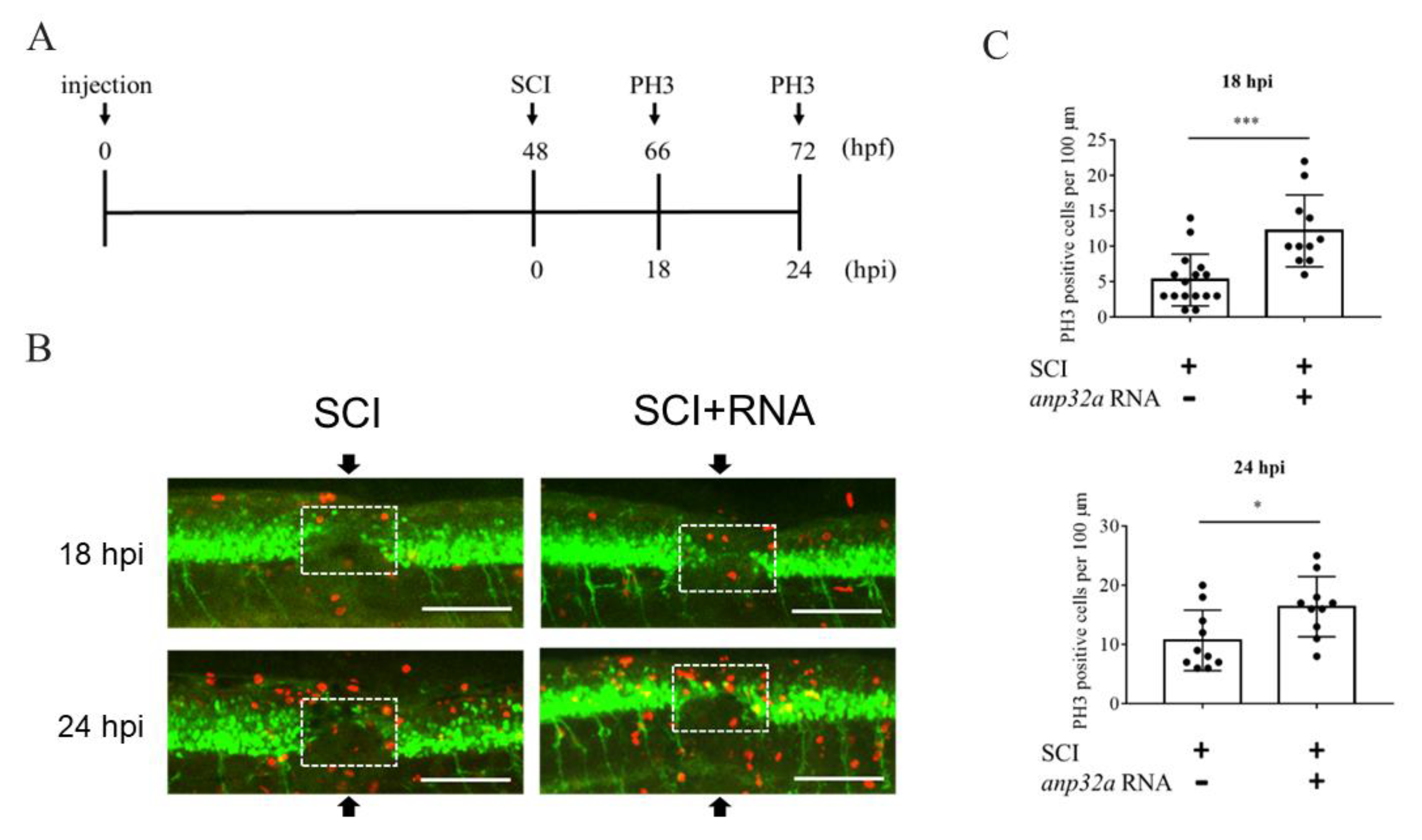
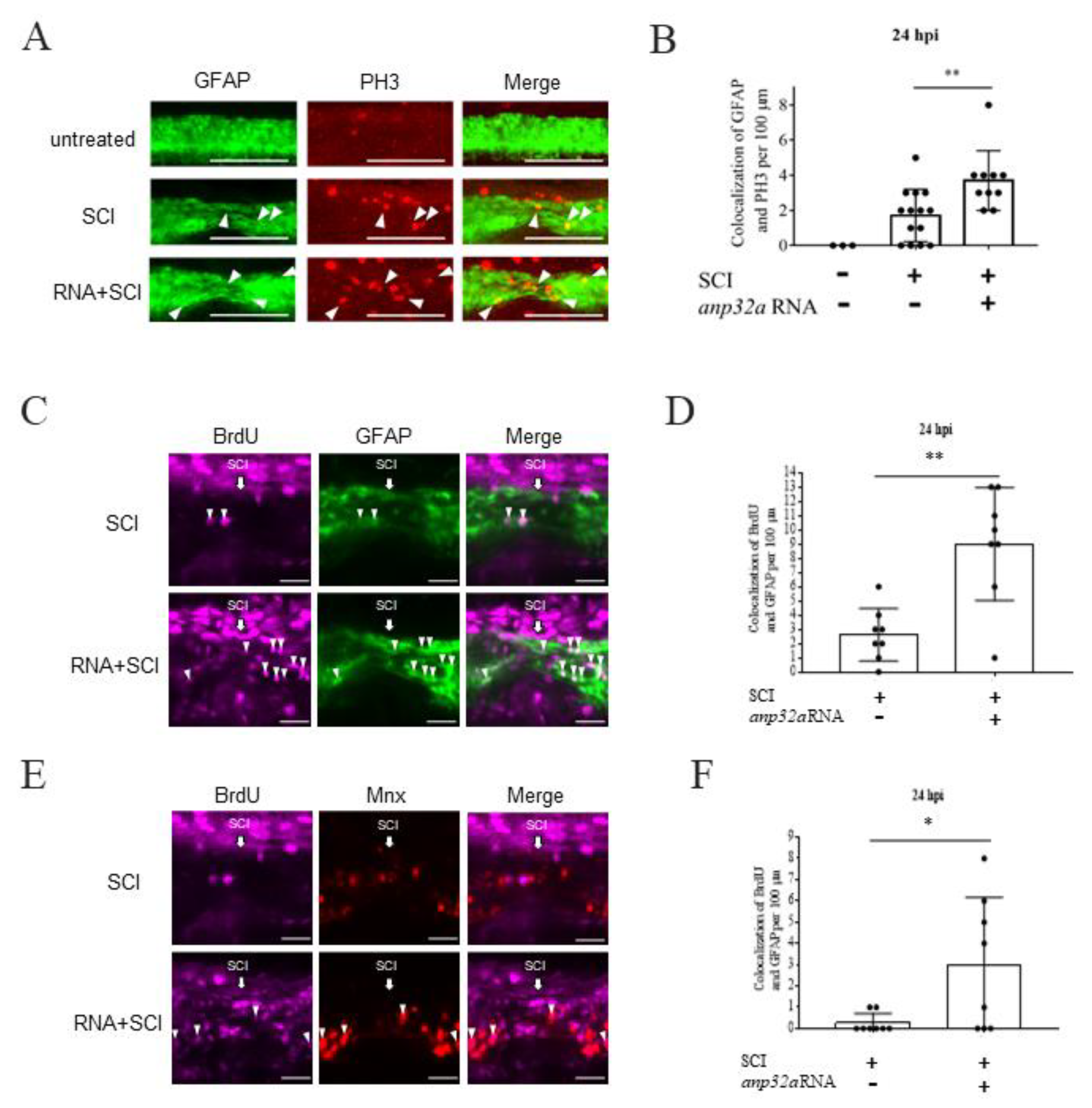
2.5. ANP32a Played a Positive Role in the Proliferation of Neural Cells at the SCI Site during Regeneration of SCI Embryos
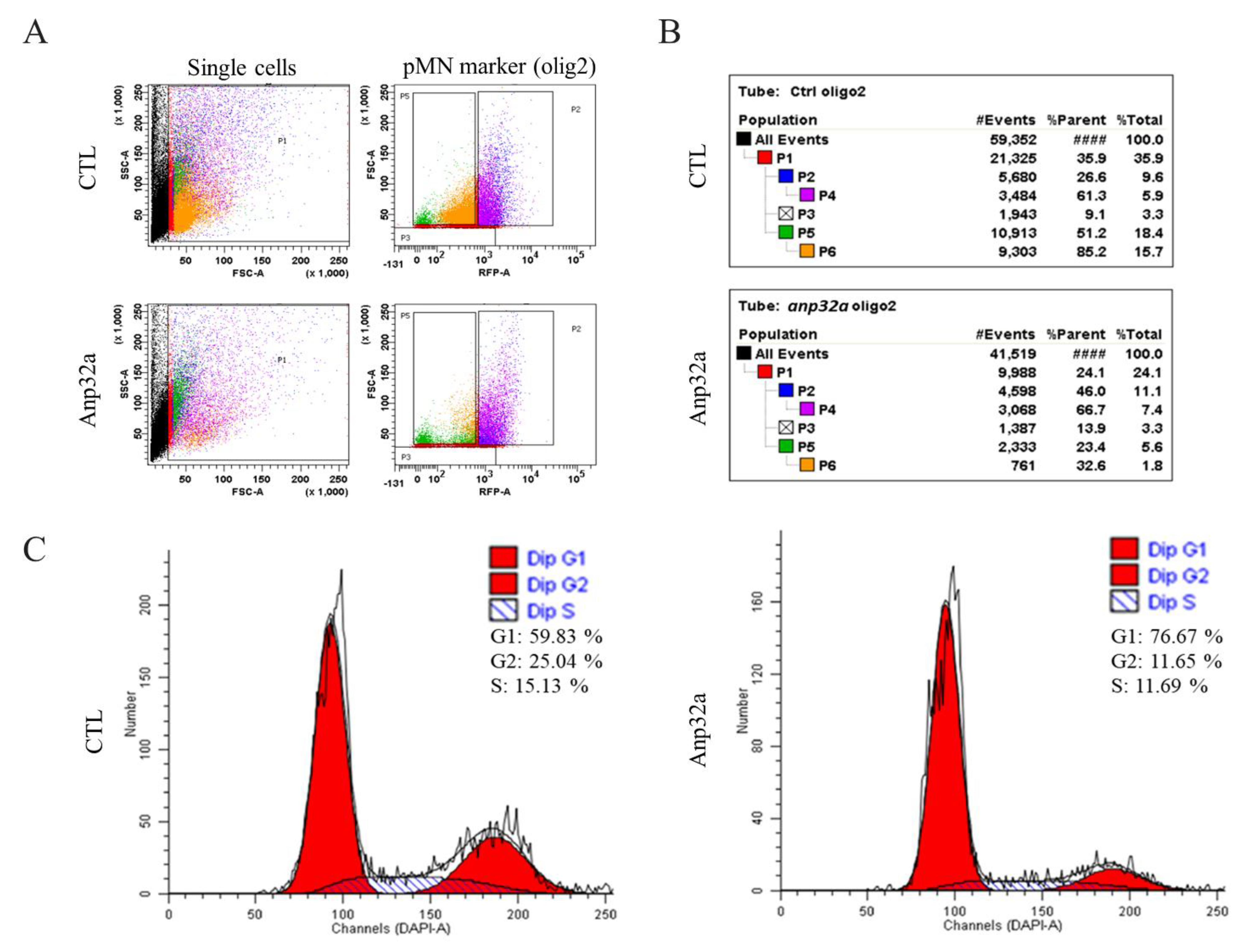
2.6. ANP32a Improved the Proliferation of Motor Neuron Progenitor Cells and Radial Glial Cells at the SCI Site of SCI Embryos
3. Discussion
3.1. ANP32a Promotes Cell Proliferation in Some Cellular Processes, including Regeneration of SCI Zebrafish Embryos
3.2. The Multifunctional Protein ANP32a Is Also Involved in Neurogenesis, including Neuronal Development and Neural Regeneration of Spinal Cord after SCI
3.3. Cell Signaling Pathways with Possible Involvement in Promoting Cell Proliferation Mediated by ANP32a
3.4. Specific Phenotypes Induced by MO Knockdown in Zebrafish Embryos
3.5. Conclusions
4. Materials and Methods
4.1. Zebrafish
4.2. Mechanical Crush Injury to Induce Spinal Cord Injury of Zebrafish Larvae
4.3. Confocal Microscopy and Image Processing
4.4. Microinjection and mRNA Synthesis
4.5. FACS
4.6. Immunohistochemistry
4.7. Western Blot Analysis
4.8. Swimming Capability Assay
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Azevedo, G.R.; Santos, V.L. (Handicapped) caregiver: The social representations of family members about the caregiving process. Rev. Lat. Am. Enfermagem. 2006, 14, 770–780. [Google Scholar] [CrossRef]
- Rahimi-Movaghar, V.; Sayyah, M.K.; Akbari, H.; Khorramirouz, R.; Rasouli, M.R.; Moradi-Lakeh, M.; Shokraneh, F.; Vaccaro, A.R. Epidemiology of Traumatic Spinal Cord Injury in Developing Countries: A Systematic Review. Neuroepidemiology 2013, 41, 65–85. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, C.S.; Nori, S.; Tetreault, L.; Wilson, J.; Kwon, B.; Harrop, J.; Choi, D.; Fehlings, M.G. Traumatic Spinal Cord Injury—Repair and Regeneration. Neurosurgery 2017, 80, S9–S22. [Google Scholar] [CrossRef] [PubMed]
- New, P.W.; Marshall, R. International Spinal Cord Injury Data Sets for non-traumatic spinal cord injury. Spinal Cord 2014, 52, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Atrice, B.S.; Morrison, S.A.; McDowell, S.L.; Ackerman, P.M.; Foy, T.A. Traumatic spinal cord injury. In Neurological Rehabilitation, 5th ed.; Umphred, D.A., Ed.; Mosby Elsevier: Maryland Heights, MO, USA, 2007; pp. 605–657. [Google Scholar]
- Stahel, P.F.; VanderHeiden, T.; Finn, M.A. Management strategies for acute spinal cord injury: Current options and future perspectives. Curr. Opin. Crit. Care 2012, 18, 651–660. [Google Scholar] [CrossRef]
- Wilson, J.R.; Forgione, N.; Fehlings, M.G. Emerging therapies for acute traumatic spinal cord injury. CMAJ 2013, 185, 485–492, Correction in CMAJ 2014, 4, 186–294. [Google Scholar] [CrossRef]
- Grossman, S.; Rosenberg, L.; Wrathall, J. Temporal–Spatial Pattern of Acute Neuronal and Glial Loss after Spinal Cord Contusion. Exp. Neurol. 2001, 168, 273–282. [Google Scholar] [CrossRef]
- Bradbury, E.J.; Burnside, E.R. Moving beyond the glial scar for spinal cord repair. Nat. Commun. 2019, 10, 3879. [Google Scholar] [CrossRef]
- Courtine, G.; Sofroniew, M.V. Spinal cord repair: Advances in biology and technology. Nat. Med. 2019, 25, 898–908. [Google Scholar] [CrossRef]
- Thuret, S.; Moon, L.D.; Gage, F.H. Therapeutic interventions after spinal cord injury. Nat. Rev. Neurosci. 2006, 7, 628–643, Correction in Nat. Rev. Neurosci. 2006, 7, 902. [Google Scholar] [CrossRef]
- Cajal, S.R. Degeneration and Regeneration of the Nervous System; Oxford University Press: New York, NY, USA, 1928. [Google Scholar]
- Cigliola, V.; Becker, C.J.; Poss, K.D. Building bridges, not walls: Spinal cord regeneration in zebrafish. Dis. Model. Mech. 2020, 13, dmm044131. [Google Scholar] [CrossRef]
- Becker, C.G.; Becker, T. Model Organisms in Spinal Cord Regeneration; Wiley: Weinheim, Germany, 2007. [Google Scholar]
- Vajn, K.; Suler, D.; Plunkett, J.A.; Oudega, M. Temporal Profile of Endogenous Anatomical Repair and Functional Recovery following Spinal Cord Injury in Adult Zebrafish. PLoS ONE 2014, 9, e105857. [Google Scholar] [CrossRef]
- Ghosh, S.; Hui, S.P. Axonal regeneration in zebrafish spinal cord. Regeneration 2018, 5, 43–60. [Google Scholar] [CrossRef]
- Ma, L.; Shen, Y.-Q.; Khatri, H.P.; Schachner, M. The Asparaginyl Endopeptidase Legumain Is Essential for Functional Recovery after Spinal Cord Injury in Adult Zebrafish. PLoS ONE 2014, 9, e95098. [Google Scholar] [CrossRef]
- Reimer, M.M.; Sörensen, I.; Kuscha, V.; Frank, R.E.; Liu, C.; Becker, C.G.; Becker, T. Motor Neuron Regeneration in Adult Zebrafish. J. Neurosci. 2008, 28, 8510–8516. [Google Scholar] [CrossRef]
- Mokalled, M.H.; Patra, C.; Dickson, A.L.; Endo, T.; Stainier, D.Y.R.; Poss, K.D. Injury-induced ctgfa directs glial bridging and spinal cord regeneration in zebrafish. Science 2016, 354, 630–634. [Google Scholar] [CrossRef]
- Wehner, D.; Tsarouchas, T.M.; Michael, A.; Haase, C.; Weidinger, G.; Reimer, M.M.; Becker, T.; Becker, C.G. Wnt signaling controls pro-regenerative Collagen XII in functional spinal cord regeneration in zebrafish. Nat. Commun. 2017, 8, 126. [Google Scholar] [CrossRef]
- Zeng, C.-W.; Kamei, Y.; Shigenobu, S.; Sheu, J.-C.; Tsai, H.-J. Injury-induced Cavl-expressing cells at lesion rostral side play major roles in spinal cord regeneration. Open Biol. 2021, 11, 200304. [Google Scholar] [CrossRef]
- Kadkol, S.S.; El Naga, G.A.; Brody, J.R.; Bai, J.; Gusev, Y.; Dooley, W.C.; Pasternack, G.R. Expression of pp32 gene family members in breast cancer. Breast Cancer Res. Treat. 2001, 68, 65–73. [Google Scholar] [CrossRef]
- Santa-Coloma, T.A. Anp32e (Cpd1) and related protein phosphatase 2 inhibitors. Cerebellum 2003, 2, 310–320. [Google Scholar] [CrossRef]
- Shen, L.-F.; Cheng, H.; Tsai, M.-C.; Kuo, H.-S.; Chak, K.F. PAL31 may play an important role as inflammatory modulator in the repair process of the spinal cord injury rat. J. Neurochem. 2009, 108, 1187–1197. [Google Scholar] [CrossRef] [PubMed]
- Imamachi, K.; Higashino, F.; Kitamura, T.; Kakuguchi, W.; Yanagawa-Matsuda, A.; Ishikawa, M.; Kitagawa, Y.; Totsuka, Y.; Shindoh, M. pp32r1 controls the decay of the RNA-binding protein HuR. Oncol. Rep. 2014, 31, 1103–1108. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huyton, T.; Wolberger, C. The crystal structure of the tumor suppressor protein pp32 (Anp32a): Structural insights into Anp32 family of proteins. Protein Sci. 2007, 16, 1308–1315. [Google Scholar] [CrossRef] [PubMed]
- Theodosiou, A.; Ashworth, A. MAP kinase phosphatases. Genome Biol. 2002, 3, reviews3009. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kadota, S.; Nagata, K. pp32, an INHAT component, is a transcription machinery recruiter for maximal induction of IFN-stimulated genes. J. Cell Sci. 2011, 124, 988. [Google Scholar] [CrossRef][Green Version]
- Opal, P.; Garcia, J.J.; Propst, F.; Matilla-Dueñas, A.; Orr, H.; Zoghbi, H. Mapmodulin/Leucine-rich Acidic Nuclear Protein Binds the Light Chain of Microtubule-associated Protein 1B and Modulates Neuritogenesis. J. Biol. Chem. 2003, 278, 34691–34699. [Google Scholar] [CrossRef]
- Beresford, P.J.; Zhang, D.; Oh, D.Y.; Fan, Z.; Greer, E.L.; Russo, M.L.; Jaju, M.; Lieberman, J. Granzyme A Activates an Endoplasmic Reticulum-associated Caspase-independent Nuclease to Induce Single-stranded DNA Nicks. J. Biol. Chem. 2001, 276, 43285–43293. [Google Scholar] [CrossRef]
- Fan, Z.; Beresford, P.J.; Oh, D.Y.; Zhang, D.; Lieberman, J. Tumor Suppressor NM23-H1 Is a Granzyme A-Activated DNase during CTL-Mediated Apoptosis, and the Nucleosome Assembly Protein SET Is Its Inhibitor. Cell 2003, 112, 659–672. [Google Scholar] [CrossRef]
- Chen, T.-H.; Brody, J.R.; Romantsev, F.E.; Yu, J.G.; Kayler, A.E.; Voneiff, E.; Kuhajda, F.P.; Pasternack, G.R. Structure of pp32, an acidic nuclear protein which inhibits oncogene-induced formation of transformed foci. Mol. Biol. Cell. 1996, 7, 2045–2056. [Google Scholar] [CrossRef][Green Version]
- Brody, J.R.; Kadkol, S.S.; Mahmoud, M.A.; Rebel, J.M.; Pasternack, G.R. Identification of sequences required for inhibition of oncogene-mediated transformation by pp32. J. Biol. Chem. 1999, 274, 20053–20055. [Google Scholar] [CrossRef]
- Bai, J.; Brody, J.R.; Kadkol, S.S.; Pasternack, G.R. Tumor suppression and potentiation by manipulation of pp32 expression. Oncogene 2001, 20, 2153–2160. [Google Scholar] [CrossRef]
- Yan, W.; Bai, Z.; Wang, J.; Li, X.; Chi, B.; Chen, X. ANP32A modulates cell growth by regulating p38 and Akt activity in colorectal cancer. Oncol. Rep. 2017, 38, 1605–1612. [Google Scholar] [CrossRef]
- Tian, Z.; Liu, Z.; Fang, X.; Cao, K.; Zhang, B.; Wu, R.; Wen, X.; Wen, Q.; Shi, H.; Wang, R. ANP32A promotes the proliferation, migration and invasion of hepatocellular carcinoma by modulating the HMGA1/STAT3 pathway. Carcinogenesis 2021, 42, 493–506. [Google Scholar] [CrossRef]
- Khan, M.Z.; Vaidya, A.; Meucci, O. CXCL12-Mediated Regulation of ANP32A/Lanp, A Component of the Inhibitor of Histone Acetyl Transferase (INHAT) Complex, in Cortical Neurons. J. Neuroimmune Pharmacol. 2011, 6, 163–170. [Google Scholar] [CrossRef]
- Sánchez, I.; Piñol, P.; Corral-Juan, M.; Pandolfo, M.; Matilla-Dueñas, A. A novel function of Ataxin-1 in the modulation of PP2A activity is dysregulated in the spinocerebellar ataxia type 1. Hum. Mol. Genet. 2013, 22, 3425–3437. [Google Scholar] [CrossRef]
- Chen, S.; Li, B.; Grundke-Iqbal, I.; Iqbal, K. I PP2A 1 Affects Tau Phosphorylation via Association with the Catalytic Subunit of Protein Phosphatase 2A. J. Biol. Chem. 2008, 283, 10513–10521. [Google Scholar] [CrossRef]
- Chai, G.-S.; Feng, Q.; Wang, Z.-H.; Hu, Y.; Sun, D.-S.; Li, X.-G.; Ke, D.; Li, H.-L.; Liu, G.-P.; Wang, J.-Z. Downregulating ANP32A rescues synapse and memory loss via chromatin remodeling in Alzheimer model. Mol. Neurodegener. 2017, 12, 34. [Google Scholar] [CrossRef]
- Bhatt, D.H.; Otto, S.J.; Depoister, B.; Fetcho, J.R. Cyclic AMP-Induced Repair of Zebrafish Spinal Circuits. Science 2004, 305, 254–258. [Google Scholar] [CrossRef]
- Briona, L.K.; Dorsky, R.I. Radial glial progenitors repair the zebrafish spinal cord following transection. Exp. Neurol. 2014, 256, 81–92. [Google Scholar] [CrossRef]
- Ohnmacht, J.; Yang, Y.-J.; Maurer, G.W.; Barreiro-Iglesias, A.; Tsarouchas, T.M.; Wehner, D.; Sieger, D.; Becker, C.G.; Becker, T. Spinal motor neurons are regenerated after mechanical lesion and genetic ablation in larval zebrafish. Development 2016, 143, 1464–1474. [Google Scholar] [CrossRef]
- Saint-Amant, L.; Drapeau, P. Time course of the development of motor behaviors in the zebrafish embryo. J. Neurobiol. 1998, 37, 622–632. [Google Scholar] [CrossRef]
- O’Shea, T.M.; Burda, J.E.; Sofroniew, M.V. Cell biology of spinal cord injury and repair. J. Clin. Investig. 2017, 127, 3259–3270. [Google Scholar] [CrossRef] [PubMed]
- Reimer, M.M.; Norris, A.; Ohnmacht, J.; Patani, R.; Zhong, Z.; Dias, T.B.; Kuscha, V.; Scott, A.L.; Chen, Y.-C.; Rozov, S.; et al. Dopamine from the Brain Promotes Spinal Motor Neuron Generation during Development and Adult Regeneration. Dev. Cell 2013, 25, 478–491. [Google Scholar] [CrossRef]
- Brody, J.R.; Kadkol, S.S.; Hauer, M.C.; Rajaii, F.; Lee, J.; Pasternack, G.R. pp32 reduction induces differentiation of TSU-Pr1 cells. Am. J. Pathol. 2004, 164, 273–283. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Williams, T.K.; Costantino, C.L.; Bildzukewicz, N.A.; Richards, N.G.; Rittenhouse, D.W.; Einstein, L.; Cozzitorto, J.A.; Keen, J.C.; Dasgupta, A.; Gorospe, M.; et al. pp32 (ANP32A) Expression Inhibits Pancreatic Cancer Cell Growth and Induces Gemcitabine Resistance by Disrupting HuR Binding to mRNAs. PLoS ONE 2010, 5, e15455. [Google Scholar] [CrossRef]
- Brody, J.R.; Witkiewicz, A.; Williams, T.K.; Kadkol, S.S.; Cozzitorto, J.; Durkan, B.; Pasternack, G.R.; Yeo, C.J. Reduction of pp32 expression in poorly differentiated pancreatic ductal adenocarcinomas and intraductal papillary mucinous neoplasms with moderate dysplasia. Mod. Pathol. 2007, 20, 1238–1244. [Google Scholar] [CrossRef][Green Version]
- Hoffarth, S.; Zitzer, A.; Wiewrodt, R.; Hähnel, P.S.; Beyer, V.; Kreft, A.; Biesterfeld, S.; Schuler, M. pp32/PHAPI determines the apoptosis response of non-small-cell lung cancer. Cell Death Differ. 2008, 15, 161–170. [Google Scholar] [CrossRef]
- Sun, X.; Lu, B.; Han, C.; Qiu, W.; Jin, Q.; Li, D.; Li, Q.; Yang, Q.; Wen, Q.; Opal, P.; et al. ANP32A dysregulation contributes to abnormal megakaryopoiesis in acute megakaryoblastic leukemia. Blood Cancer J. 2017, 7, 661. [Google Scholar] [CrossRef]
- Xie, M.; Ji, Z.; Bao, Y.; Zhu, Y.; Xu, Y.; Wang, L.; Gao, S.; Liu, Z.; Tian, Z.; Meng, Q.; et al. PHAP1 promotes glioma cell proliferation by regulating the Akt/p27/stathmin pathway. J. Cell Mol. Med. 2018, 22, 3595–3604. [Google Scholar] [CrossRef]
- Yang, X.; Lu, B.; Sun, X.; Han, C.; Fu, C.; Xu, K.; Wang, M.; Li, D.; Chen, Z.; Opal, P.; et al. ANP32A regulates histone H3 acetylation and promotes leukemogenesis. Leukemia 2018, 32, 1587–1597. [Google Scholar] [CrossRef]
- Huang, S.; Huang, Z.; Ma, C.; Luo, L.; Li, Y.-F.; Wu, Y.-L.; Ren, Y.; Feng, C. Acidic leucine-rich nuclear phosphoprotein-32A expression contributes to adverse outcome in acute myeloid leukemia. Ann. Transl. Med. 2020, 8, 345. [Google Scholar] [CrossRef]
- Zeng, C.-W.; Sheu, J.-C.; Tsai, H.-J. The Neuronal Regeneration of Adult Zebrafish After Spinal Cord Injury Is Enhanced by Transplanting Optimized Number of Neural Progenitor Cells. Cell Transplant. 2020, 29, 963689720903679. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, Y.; Lu, Q.; Liu, X.; Wang, F.; Ma, X.; Cui, C.; Shi, C.; Li, J.; Zhang, D. The Expression and Distributions of ANP32A in the Developing Brain. BioMed Res. Int. 2015, 2015, 207347. [Google Scholar] [CrossRef]
- Seo, S.-B.; McNamara, P.; Heo, S.; Turner, A.; Lane, W.S.; Chakravarti, D. Regulation of Histone Acetylation and Transcription by INHAT, a Human Cellular Complex Containing the Set Oncoprotein. Cell 2001, 104, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Kular, R.K.; Cvetanovic, M.; Siferd, S.; Kini, A.R.; Opal, P. Neuronal Differentiation Is Regulated by Leucine-rich Acidic Nuclear Protein (LANP), a Member of the Inhibitor of Histone Acetyltransferase Complex. J. Biol. Chem. 2009, 284, 7783–7792. [Google Scholar] [CrossRef]
- Kisseleva, T.; Bhattacharya, S.; Braunstein, J.; Schindler, C. Signaling through the JAK/STAT pathway, recent advances and future challenges. Gene 2002, 285, 1–24. [Google Scholar] [CrossRef]
- Pineau, I.; Lacroix, S. Proinflammatory cytokine synthesis in the injured mouse spinal cord: Multiphasic expression pattern and identification of the cell types involved. J. Comp. Neurol. 2007, 500, 267–285. [Google Scholar] [CrossRef]
- Slaets, H.; Nelissen, S.; Janssens, K.; Vidal, P.M.; Lemmens, E.; Stinissen, P.; Hendrix, S.; Hellings, N. Oncostatin M Reduces Lesion Size and Promotes Functional Recovery and Neurite Outgrowth After Spinal Cord Injury. Mol. Neurobiol. 2014, 50, 1142–1151. [Google Scholar] [CrossRef]
- Tripathi, R.B.; McTigue, D.M. Chronically increased ciliary neurotrophic factor and fibroblast growth factor-2 expression after spinal contusion in rats. J. Comp. Neurol. 2008, 510, 129–144. [Google Scholar] [CrossRef]
- Schwaiger, F.W.; Schmitt, G.H.A.B.; Horvat, A.; Hager, G.; Streif, R.; Spitzer, C.; Gamal, S.; Breuer, S.; Brook, G.A.; Nacimiento, W.; et al. Peripheral but not central axotomy induces changes in Janus kinases (JAK) and signal transducers and activators of transcription (STAT). Eur. J. Neurosci. 2000, 12, 1165–1176. [Google Scholar] [CrossRef]
- Dominguez, E.; Rivat, C.; Pommier, B.; Mauborgne, A.; Pohl, M. JAK/STAT3 pathway is activated in spinal cord microglia after peripheral nerve injury and contributes to neuropathic pain development in rat. J. Neurochem. 2008, 107, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Okada, S.; Nakamura, M.; Katoh, H.; Miyao, T.; Shimazaki, T.; Ishii, K.; Yamane, J.; Yoshimura, A.; Iwamoto, Y.; Toyama, Y.; et al. Conditional ablation of Stat3 or Socs3 discloses a dual role for reactive astrocytes after spinal cord injury. Nat. Med. 2006, 12, 829–834. [Google Scholar] [CrossRef]
- Hall, E.D.; Yonkers, A.P.; Andrus, P.K.; Cox, J.W.; Anderson, D.K. Biochemistry and pharmacology of lipid antioxidants in acute brain and spinal cord injury. J. Neurotrauma 1992, 9, S425–S442. [Google Scholar]
- Hamann, K.; Durkes, A.; Ouyang, H.; Uchida, K.; Pond, A.; Shi, R. Critical role of acrolein in secondary injury following ex vivo spinal cord trauma. J. Neurochem. 2008, 107, 712–721. [Google Scholar] [CrossRef]
- Cornelis, F.M.F.; Monteagudo, S.; Guns, L.-A.K.A.; Hollander, W.D.; Nelissen, R.G.H.H.; Storms, L.; Peeters, T.; Jonkers, I.; Meulenbelt, I.; Lories, R.J. ANP32A regulates ATM expression and prevents oxidative stress in cartilage, brain, and bone. Sci. Transl. Med. 2018, 10, eaar8426. [Google Scholar] [CrossRef]
- Lin, C.-Y.; Lee, H.-C.; Fu, C.-Y.; Ding, Y.-Y.; Chen, J.-S.; Lee, M.-H.; Huang, W.-J.; Tsai, H.-J. miR-1 and miR-206 target different genes to have opposing roles during angiogenesis in zebrafish embryos. Nat. Commun. 2013, 4, 2829. [Google Scholar] [CrossRef]
- Lee, H.-C.; Lo, H.-C.; Lo, D.-M.; Su, M.-Y.; Hu, J.-R.; Wu, C.-C.; Chang, S.-N.; Dai, M.-S.; Tsai, C.-T.; Tsai, H.-J. Amiodarone Induces Overexpression of Similar to Versican b to Repress the EGFR/Gsk3b/Snail Signaling Axis during Cardiac Valve Formation of Zebrafish Embryos. PLoS ONE 2015, 10, e0144751. [Google Scholar] [CrossRef]
- Fu, C.-Y.; Wang, P.-C.; Tsai, H.-J. Competitive binding between Seryl-tRNA synthetase/YY1 complex and NFKB1 at the distal segment results in differential regulation of human vegfa promoter activity during angiogenesis. Nucleic Acids Res. 2017, 45, 2423–2437. [Google Scholar] [CrossRef]
- Lin, C.-Y.; Lin, H.-Y.; Chuang, C.-K.; Zhang, P.-H.; Tu, Y.-R.; Lin, S.-P.; Tsai, H.-J. Quantification of Idua Enzymatic Activity Combined with Observation of Phenotypic Change in Zebrafish Embryos Provide a Preliminary Assessment of Mutated idua Correlated with Mucopolysaccharidosis Type I. J. Pers. Med. 2022, 12, 1199. [Google Scholar] [CrossRef]
- Bernardos, R.L.; Raymond, P.A. GFAP transgenic zebrafish. Gene Expr. Patterns 2006, 6, 1007–1013. [Google Scholar] [CrossRef]
- Flanagan-Steet, H.; Fox, M.; Meyer, D.; Sanes, J.R. Neuromuscular synapses can form in vivo by incorporation of initially aneural postsynaptic specializations. Development 2005, 132, 4471–4481. [Google Scholar] [CrossRef] [PubMed]
- Jao, L.-E.; Appel, B.; Wente, S.R. A zebrafish model of lethal congenital contracture syndrome 1 reveals Gle1 function in spinal neural precursor survival and motor axon arborization. Development 2012, 139, 1316–1326. [Google Scholar] [CrossRef] [PubMed]
- Westerfield, M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio); University of Oregon Press: Eugene, OR, USA, 2000. [Google Scholar]
- Lee, H.-C.; Fu, C.-Y.; Zeng, C.-W.; Tsai, H.-J. Embryonic expression patterns of Eukaryotic EndoU ribonuclease family gene endouC in zebrafish. Gene Expr. Patterns 2017, 25–26, 66–70. [Google Scholar] [CrossRef]
- Lee, H.-C.; Fu, C.-Y.; Lin, C.-Y.; Hu, J.-R.; Huang, T.-Y.; Lo, K.-Y.; Tsai, H.-Y.; Sheu, J.-C.; Tsai, H.-J. Poly(U)-specific endoribonuclease ENDOU promotes translation of human CHOP mRNA by releasing uORF element-mediated inhibition. EMBO J. 2021, 40, e104123. [Google Scholar] [CrossRef]
- Dobson, J.T.; Da’as, S.; McBride, E.R.; Berman, J.N. Fluorescence-activated cell sorting (FACS) of whole mount in situ hybridization (WISH) labelled haematopoietic cell populations in the zebrafish. Br. J. Haematol. 2009, 144, 732–735. [Google Scholar] [CrossRef]
- Zeng, C.-W.; Kamei, Y.; Wang, C.-T.; Tsai, H.-J. Subtypes of hypoxia-responsive cells differentiate into neurons in spinal cord of zebrafish embryos after hypoxic stress. Biol. Cell 2016, 108, 357–377. [Google Scholar] [CrossRef]
- Graeden, E.; Sive, H. Live Imaging of the Zebrafish Embryonic Brain by Confocal Microscopy. J. Vis. Exp. 2009, 26, e1217. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, H.-C.; Lai, W.-L.; Lin, C.-Y.; Zeng, C.-W.; Sheu, J.-C.; Chou, T.-B.; Tsai, H.-J. Anp32a Promotes Neuronal Regeneration after Spinal Cord Injury of Zebrafish Embryos. Int. J. Mol. Sci. 2022, 23, 15921. https://doi.org/10.3390/ijms232415921
Lee H-C, Lai W-L, Lin C-Y, Zeng C-W, Sheu J-C, Chou T-B, Tsai H-J. Anp32a Promotes Neuronal Regeneration after Spinal Cord Injury of Zebrafish Embryos. International Journal of Molecular Sciences. 2022; 23(24):15921. https://doi.org/10.3390/ijms232415921
Chicago/Turabian StyleLee, Hung-Chieh, Wei-Lin Lai, Cheng-Yung Lin, Chih-Wei Zeng, Jin-Chuan Sheu, Tze-Bin Chou, and Huai-Jen Tsai. 2022. "Anp32a Promotes Neuronal Regeneration after Spinal Cord Injury of Zebrafish Embryos" International Journal of Molecular Sciences 23, no. 24: 15921. https://doi.org/10.3390/ijms232415921
APA StyleLee, H.-C., Lai, W.-L., Lin, C.-Y., Zeng, C.-W., Sheu, J.-C., Chou, T.-B., & Tsai, H.-J. (2022). Anp32a Promotes Neuronal Regeneration after Spinal Cord Injury of Zebrafish Embryos. International Journal of Molecular Sciences, 23(24), 15921. https://doi.org/10.3390/ijms232415921




