Conserved Double Translation Initiation Site for Δ160p53 Protein Hints at Isoform’s Key Role in Mammalian Physiology
Abstract
1. Introduction
2. Results
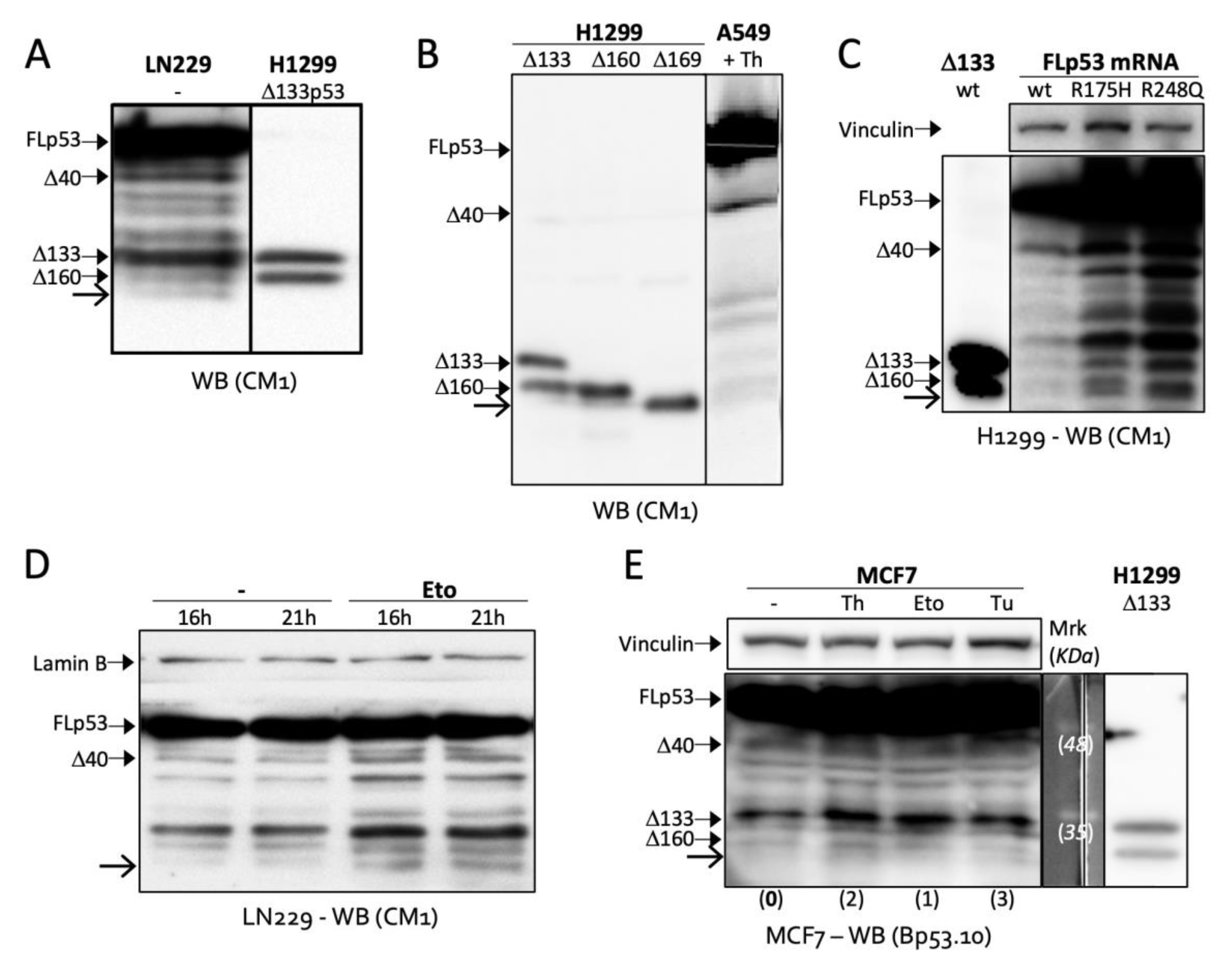
2.1. Toeprinting Identifies Translation Initiation Site Downstream of Codon 160
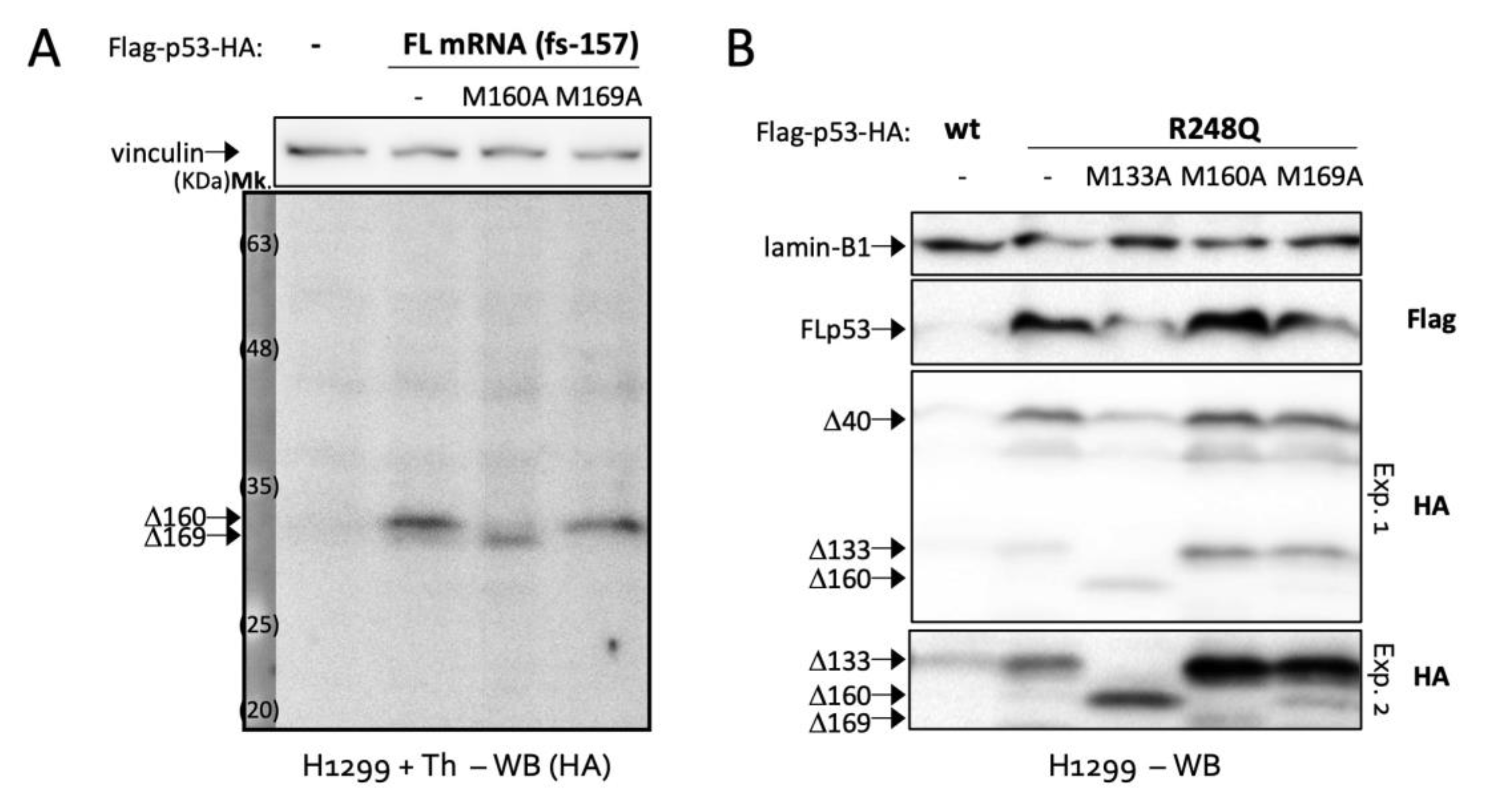
2.2. Δ169. p53 Is a Newly Identified Translation Product of Full-Length p53 mRNA
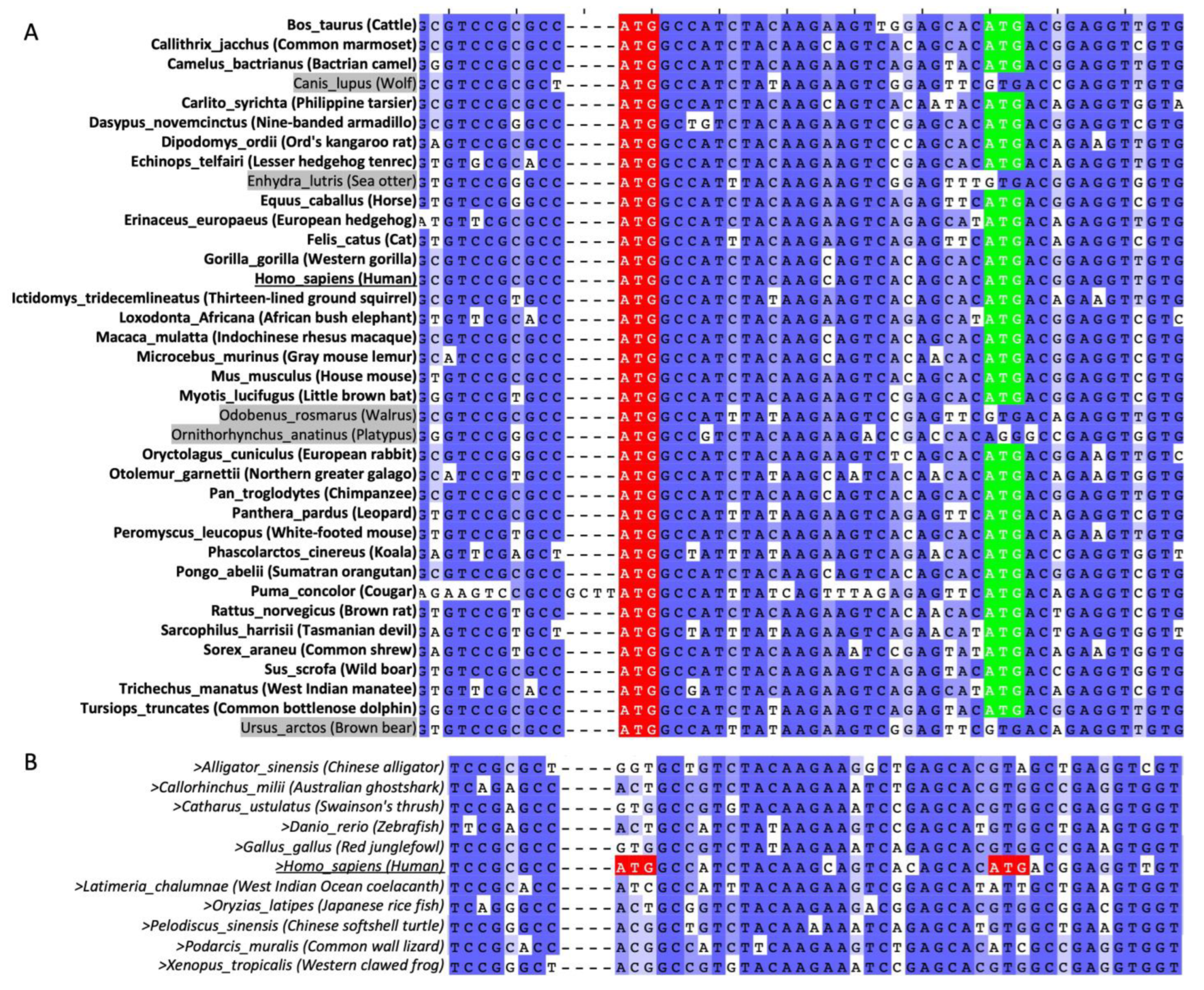
2.3. AUG160 and AUG169 May Have Co-Evolved in Mammals
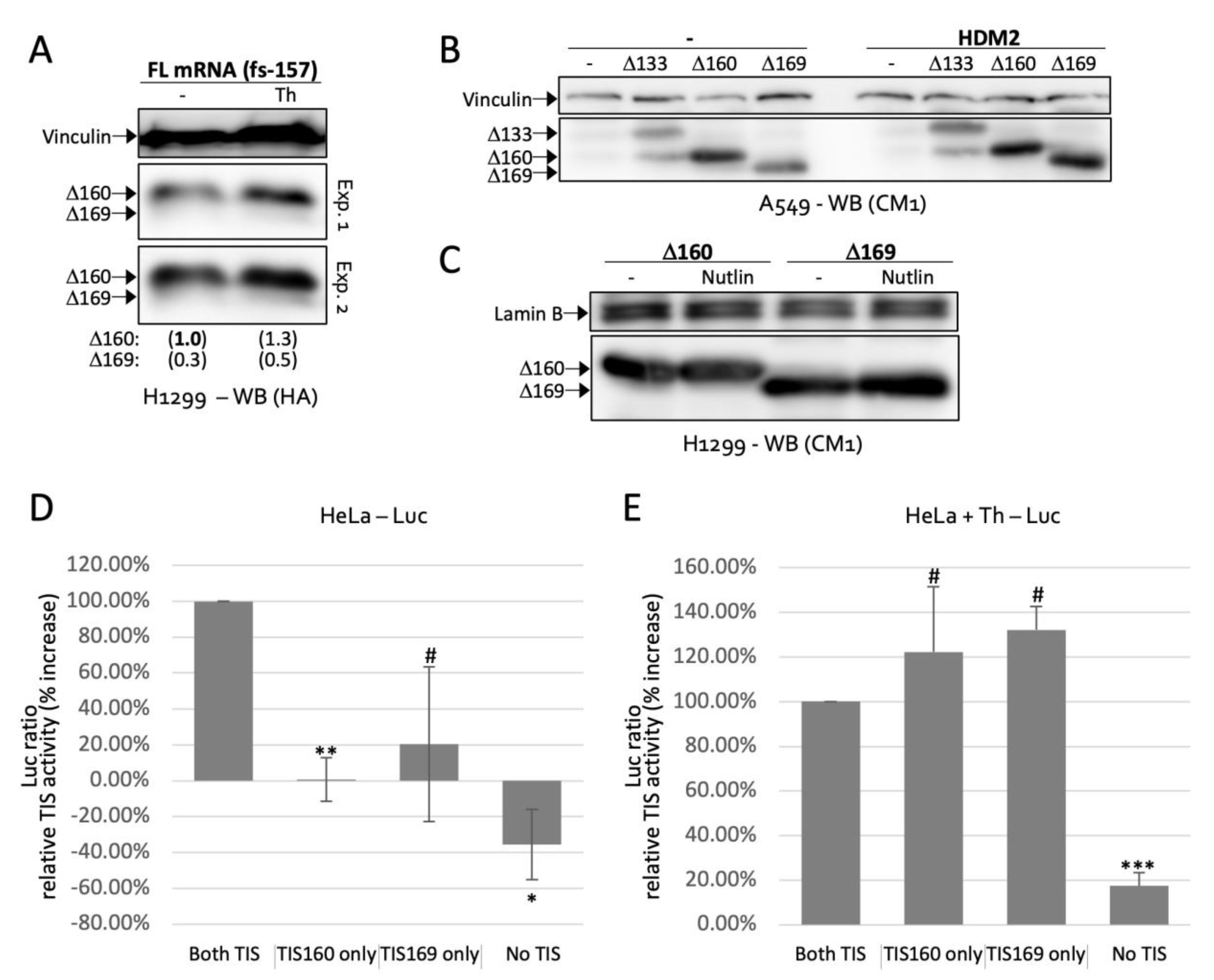
2.4. TIS160 Is Redundant during Stress Response
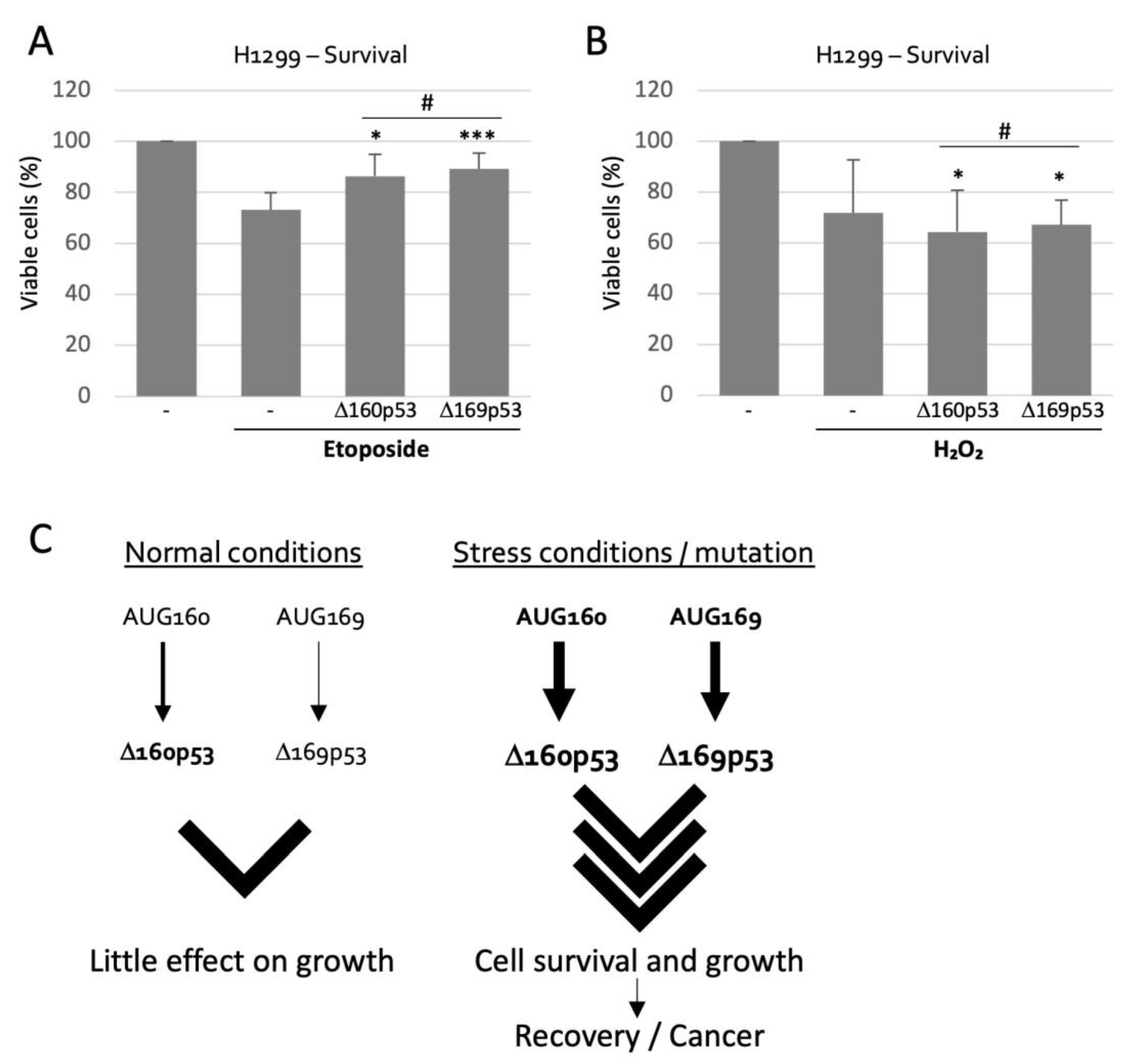
2.5. Δ160p53 and Δ169p53 Have Similar Survival Capacities
3. Discussion
4. Materials and Methods
4.1. Cellular Assays and Reagents
4.2. Sequence Analyses
- Alligator sinensis (Chinese alligator) XM_006038654.3
- Bos taurus (cattle) NM_174201.2
- Callithrix jacchus (marmoset) XM_002747948.4
- Callorhinchus milii (elephant shark) JN794073.1
- Camelus bactrianus (camel) XM_010965924.1
- Canis lupus familiaris (dog) NM_001003210.1
- Carlito syrichta (tarsier) XM_008062341.2
- Catharus ustulatus (thrush) XM_033084255.1
- Danio rerio (zebrafish) NM_001271820.1
- Dasypus novemcinctus (armadillo) XM_012529094.2
- Dipodomys ordii (kangaroo rat) XM_013013490.1
- Echinops telfairi (lesser tenrec) XM_013007322.2
- Enhydra lutris kenyoni (sea otter) XM_022524640.1
- Equus caballus (horse) XM_023651624.1
- Erinaceus europaeus (hedgehog) XM_007523372.2
- Felis catus (cat) NM_001009294.1
- Gallus gallus (chicken) NM_205264.1
- Gorilla gorilla (gorilla) XM_004058511.3
- Homo sapiens (human) NM_001126112.3
- lctidomys tridecemlineatus (squirrel) XM_005332819.3
- Latimeria chalumnae (coelacanth) XM_005999799.2
- Loxodonta africana (elephant) XM_010596586.2
- Macaca mulatta (Rhesus monkey) NM_001047151.2
- Microcebus murinus (mouse lemur) XM_012776058.2
- Mus musculus (mouse) NM_011640.3
- Myotis lucifugus (bat) XM_006102578.3
- Odobenus rosmarus (walrus) XM_004398491.1
- Ornithorhynchus anatinus (platypus) ENSOANT00000053152.1
- Oryctolagus cuniculus (rabbit) NM_001082404.1
- Oryzias latipes (medaka) NM_001104742.1
- Otolemur garnettii (galago) XM_012806041.2
- Pan troglodytes (chimpanzee) XM_001172077.5
- Panthera pardus (leopard) XM_019413568.1
- Pelodiscus sinensis (turtle) XM_006112136.3
- Peromyscus leucopus (white-footed mouse) XM_028869449.1
- Phascolarctos cinereus (koala) XM_020966468.1
- Podarcis muralis (lizard) XM_028752002.1
- Pongo abelii (orangutan) XM_002826974.4
- Puma concolor (puma) XM_025920288.1
- Rattus norvegicus (rat) NM_030989.3
- Sarcophilus harrisii (Tasmanian devil) XM_031965445.1
- Sorex araneu (shrew) XM_004604858.1
- Sus scrota (pig) NM_213824.3
- Trichechus manatus (manatee) XM_004376021.2
- Tursiops truncatus (dolphin) XM_019944223.2
- Ursus arctos horribilis (grizzly) XM_026520889.1
- Xenopus tropicalis (frog) NM_001001903.1
4.3. Toeprinting
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Lane, D.P.; Cheok, C.F.; Brown, C.; Madhumalar, A.; Ghadessy, F.J.; Verma, C. Mdm2 and p53 are highly conserved from placozoans to man. Cell Cycle 2010, 9, 540–547. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Donehower, L.A.; Harvey, M.; Slagle, B.L.; McArthur, M.J.; Montgomery, C.A.; Butel, J.S.; Bradley, A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 1992, 356, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Maier, B.; Gluba, W.; Bernier, B.; Turner, T.; Mohammad, K.; Guise, T.; Sutherland, A.; Thorner, M.; Scrable, H. Modulation of mammalian life span by the short isoform of p53. Genes Dev. 2004, 18, 306–319. [Google Scholar] [CrossRef]
- Anbarasan, T.; Bourdon, J.-C. The Emerging Landscape of p53 Isoforms in Physiology, Cancer and Degenerative Diseases. Int. J. Mol. Sci. 2019, 20, 6257. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.L.; Chen, Y.M.; Bookstein, R.; Lee, W.H. Genetic mechanisms of tumor suppression by the human p53 gene. Science 1990, 250, 1576–1580. [Google Scholar] [CrossRef] [PubMed]
- Bourougaa, K.; Naski, N.; Boularan, C.; Mlynarczyk, C.; Candeias, M.M.; Marullo, S.; Fahraeus, R. Endoplasmic reticulum stress induces G2 cell-cycle arrest via mRNA translation of the p53 isoform p53/47. Mol. Cell 2010, 38, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Candeias, M.M.; Malbert-Colas, L.; Powell, D.J.; Daskalogianni, C.; Maslon, M.M.; Naski, N.; Bourougaa, K.; Calvo, F.; Fahraeus, R. P53 mRNA controls p53 activity by managing Mdm2 functions. Nat. Cell Biol. 2008, 10, 1098–1105. [Google Scholar] [CrossRef] [PubMed]
- Candeias, M.M.; Hagiwara, M.; Matsuda, M. Cancer-specific mutations in p53 induce the translation of Δ160p53 promoting tumorigenesis. EMBO Rep. 2016, 17, 1542–1551. [Google Scholar] [CrossRef] [PubMed]
- Niu, G.; Hellmuth, I.; Flisikowska, T.; Pausch, H.; Rieblinger, B.; Carrapeiro, A.; Schade, B.; Böhm, B.; Kappe, E.; Fischer, K.; et al. Porcine model elucidates function of p53 isoform in carcinogenesis and reveals novel circTP53 RNA. Oncogene 2021, 40, 1896–1908. [Google Scholar] [CrossRef] [PubMed]
- Candeias, M.M. Mutant p53 loses “gain-of-functions” after losing isoform expression. In Proceedings of the 6th International Mutant p53 Workshop; Cell Death and Differentiation Conferences, Toronto, ON, USA, 12–14 September 2013; p. 1. [Google Scholar]
- Lacerda, R.; Menezes, J.; Candeias, M.M. Alternative Mechanisms of mRNA Translation Initiation in Cellular Stress Response and Cancer. In The mRNA Metabolism in Human Disease; Springer: Berlin/Heidelberg, Germany, 2019; pp. 117–132. [Google Scholar]
- Sachs, M.S.; Wang, Z.; Gaba, A.; Fang, P.; Belk, J.; Ganesan, R.; Amrani, N.; Jacobson, A. Toeprint analysis of the positioning of translation apparatus components at initiation and termination codons of fungal mRNAs. Methods 2002, 26, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Levav-Cohen, Y.; Goldberg, Z.; Tan, K.H.; Alsheich-Bartok, O.; Zuckerman, V.; Haupt, S.; Haupt, Y. The p53-Mdm2 loop: A critical juncture of stress response. Subcell. Biochem. 2014, 85, 161–186. [Google Scholar] [PubMed]
- Marques-Ramos, A.; Candeias, M.M.; Menezes, J.; Lacerda, R.; Willcocks, M.; Teixeira, A.; Locker, N.; Romão, L. Cap-independent translation ensures mTOR expression and function upon protein synthesis inhibition. RNA 2017, 23, 1712–1728. [Google Scholar] [CrossRef] [PubMed]
- Bazykin, G.A.; Kochetov, A.V. Alternative translation start sites are conserved in eukaryotic genomes. Nucleic Acids Res. 2011, 39, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Candeias, M.M.; Powell, D.J.; Roubalova, E.; Apcher, S.; Bourougaa, K.; Vojtesek, B.; Bruzzoni-Giovanelli, H.; Fahraeus, R. Expression of p53 and p53/47 are controlled by alternative mechanisms of messenger RNA translation initiation. Oncogene 2006, 25, 6936–6947. [Google Scholar] [CrossRef] [PubMed]
- Midgley, C.A.; Fisher, C.J.; Bartek, J.; Vojtesek, B.; Lane, D.; Barnes, D.M. Analysis of p53 expression in human tumours: An antibody raised against human p53 expressed in Escherichia coli. J. Cell Sci. 1992, 101, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Gould, P.S.; Bird, H.; Easton, A.J. Translation toeprinting assays using fluorescently labeled primers and capillary electrophoresis. Biotechniques 2005, 38, 397–400. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Iniesta, M.J.; Parkar, S.N.; Ramalho, A.C.; Lacerda, R.; Costa, I.F.; Zhao, J.; Romão, L.; Candeias, M.M. Conserved Double Translation Initiation Site for Δ160p53 Protein Hints at Isoform’s Key Role in Mammalian Physiology. Int. J. Mol. Sci. 2022, 23, 15844. https://doi.org/10.3390/ijms232415844
López-Iniesta MJ, Parkar SN, Ramalho AC, Lacerda R, Costa IF, Zhao J, Romão L, Candeias MM. Conserved Double Translation Initiation Site for Δ160p53 Protein Hints at Isoform’s Key Role in Mammalian Physiology. International Journal of Molecular Sciences. 2022; 23(24):15844. https://doi.org/10.3390/ijms232415844
Chicago/Turabian StyleLópez-Iniesta, Maria José, Shrutee N. Parkar, Ana Catarina Ramalho, Rafaela Lacerda, Inês F. Costa, Jingyuan Zhao, Luísa Romão, and Marco M. Candeias. 2022. "Conserved Double Translation Initiation Site for Δ160p53 Protein Hints at Isoform’s Key Role in Mammalian Physiology" International Journal of Molecular Sciences 23, no. 24: 15844. https://doi.org/10.3390/ijms232415844
APA StyleLópez-Iniesta, M. J., Parkar, S. N., Ramalho, A. C., Lacerda, R., Costa, I. F., Zhao, J., Romão, L., & Candeias, M. M. (2022). Conserved Double Translation Initiation Site for Δ160p53 Protein Hints at Isoform’s Key Role in Mammalian Physiology. International Journal of Molecular Sciences, 23(24), 15844. https://doi.org/10.3390/ijms232415844






