The Mutual Relationship between Glycosylation and Non-Coding RNAs in Cancer and Other Physio-Pathological Conditions
Abstract
1. Introduction
2. The Essentials of Non-Coding RNAs
2.1. miRNA
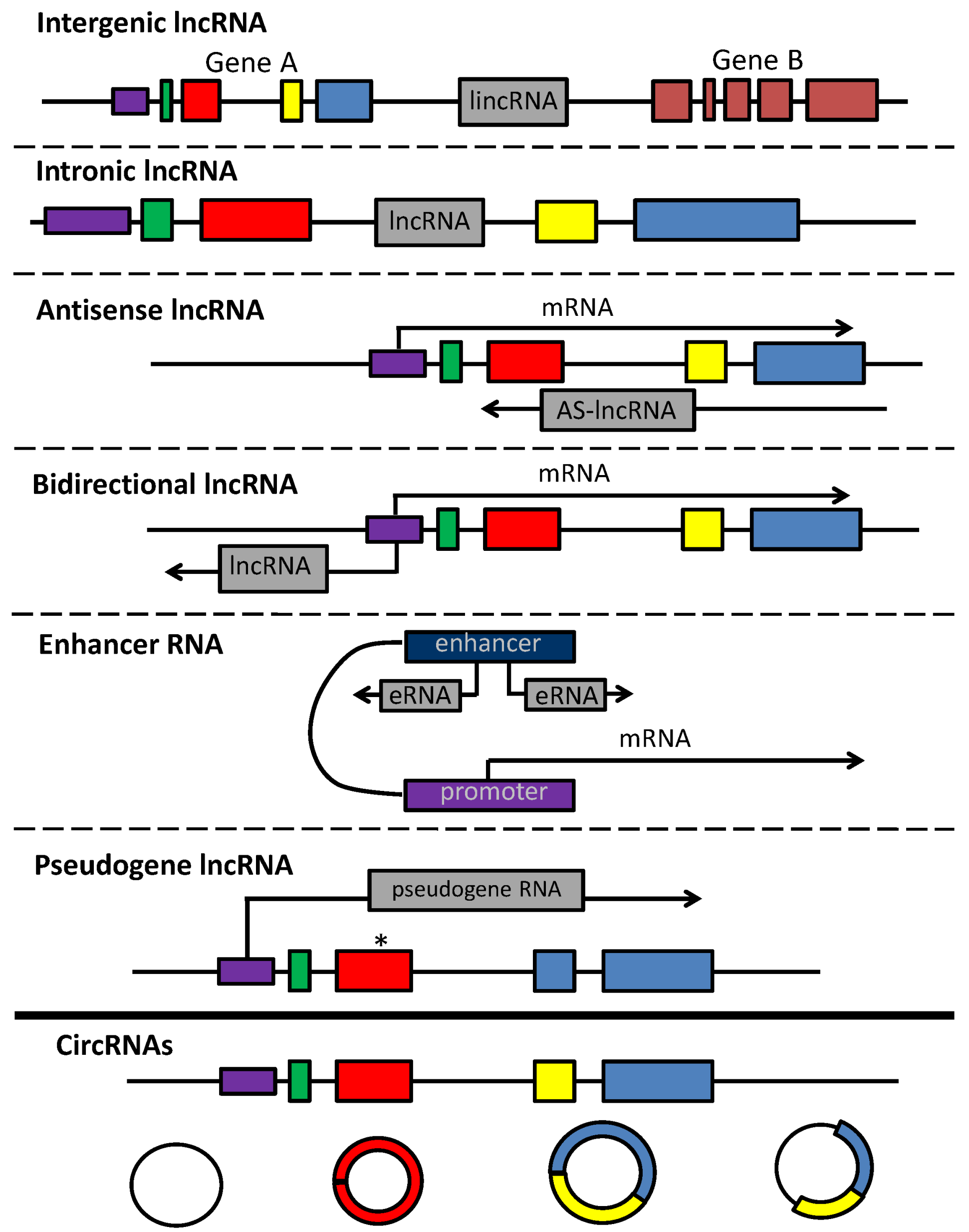
2.2. lncRNAs
3. The Essentials of Glycosylation
3.1. N-Glycosylation
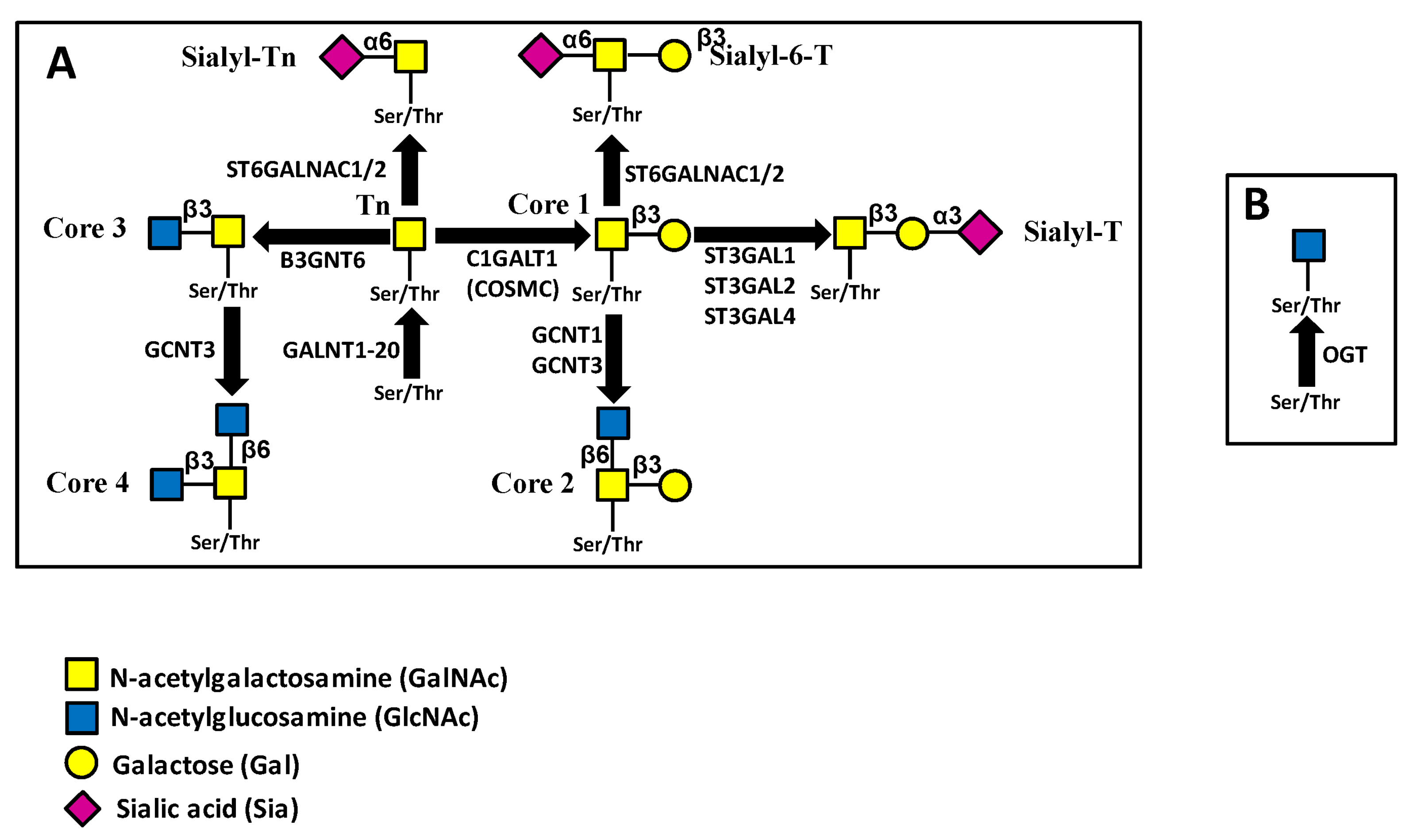
3.2. O-Glycosylation
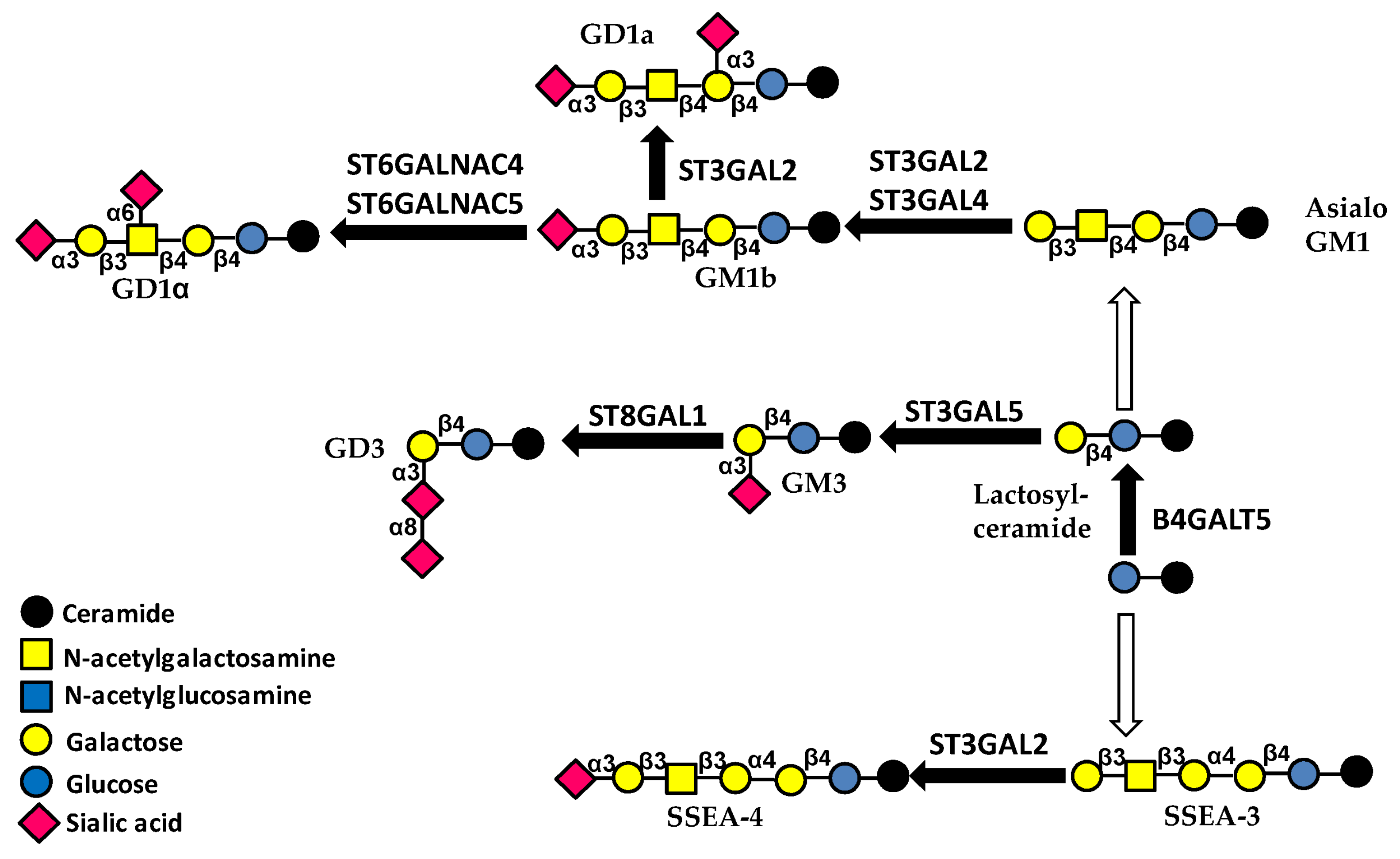
3.3. Glycolipids
4. Regulation of Glycosylation by ncRNAs
4.1. Initiating Glycosyltransferases
4.1.1. N-Linked Chains
4.1.2. Mucin-Type O-Glycosylation
4.1.3. O-Linked GlcNAc
4.1.4. Glycolipids
4.2. Core-Extending Glycosyltransferases
4.2.1. N-Linked Chains
4.2.2. O-Linked Chains
4.2.3. Glycolipids
4.3. Elongating Glycosyltransferases
4.3.1. GlcNAc Transferases
4.3.2. Gal Transferases
4.4. Capping Glycosyltransferases
4.4.1. Fucosyltransferases
4.4.2. Sialyltransferases
4.4.3. AB0 Glycosyltransferases
5. Regulation of Sugar-Binding Molecules by ncRNAs
5.1. Galectins
5.2. Siglecs
6. Non-Coding RNAs Derived from Glycosyltransferase Genes but Not Involved in Glycogene Regulation
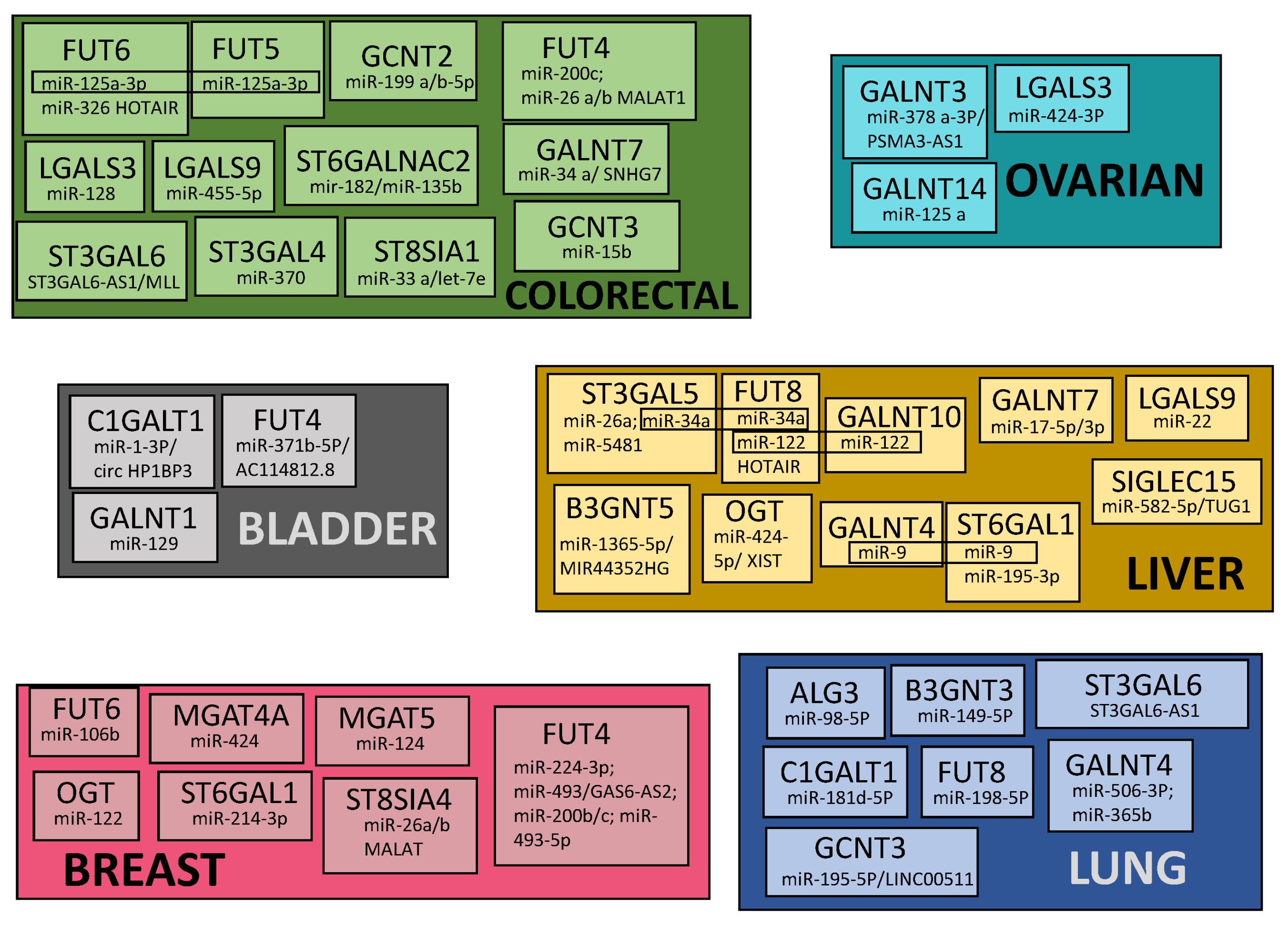
7. Common Patterns of Glycogene Modulation by ncRNA in Cancers
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| APAL | aurora A/polo kinase 1 |
| AP-2γ | activator protein 2γ |
| circRNA | circular RNA |
| DANCR | differentiation antagonizing non-protein-coding RNA |
| DLGAP1 | discs large-homolog-associated protein 1 |
| DPAGT1 | dolichyl-phosphate N-acetylglucosaminephosphotransferase 1 |
| EMT | epithelial to mesenchymal transition |
| EBV | Epstein–Barr virus |
| HNRPA2B1 | Heterogeneous Nuclear Ribonucleoprotein A2/B1 |
| HOTAIR | HOX transcript antisense RNA |
| LEF1 | Lymphoid enhancer binding factor 1 |
| Lex | Lewisx |
| lncRNA | long non-coding RNA |
| MALAT1 | metastasis-associated lung adenocarcinoma 1 |
| miRNA | micro RNA |
| MUC1 | mucin 1 |
| NCAM | neural cell adhesion molecule |
| ncRNA | non-coding RNA |
| NSCLC | non-small-cell lung cancer |
| OGT | O-GlcNActransferase |
| PSMA3 | proteasome 20S subunit alpha 3 |
| RER | rough endoplasmic reticulum |
| RISC | RNA-induced silencing complex |
| RYR1 | ryanodine receptor 1 |
| sLex | sialyl Lewisx |
| SNHG1 | small nuclear RNA host gene 1 |
| TINCR | TINCR ubiquitin domain containing |
| TUG1 | Taurine up-regulated 1 |
| XIST | X-inactivated specific transcript |
References
- Dall’Olio, F.; Malagolini, N.; Trinchera, M.; Chiricolo, M. Mechanisms of cancer-associated glycosylation changes. Front. Biosci. 2012, 17, 670–699. [Google Scholar] [CrossRef] [PubMed]
- Dall’Olio, F.; Malagolini, N. Immunoglobulin G Glycosylation Changes in Aging and Other Inflammatory Conditions. Exp. Suppl. 2021, 112, 303–340. [Google Scholar] [CrossRef] [PubMed]
- Dall’Olio, F.; Vanhooren, V.; Chen, C.C.; Slagboom, P.E.; Wuhrer, M.; Franceschi, C. N-glycomic biomarkers of biological aging and longevity: A link with inflammaging. Ageing Res. Rev. 2013, 12, 685–698. [Google Scholar] [CrossRef] [PubMed]
- Dall’Olio, F. Glycobiology of Aging. Subcell. Biochem. 2018, 90, 505–526. [Google Scholar] [CrossRef]
- Flynn, R.A.; Pedram, K.; Malaker, S.A.; Batista, P.J.; Smith, B.A.H.; Johnson, A.G.; George, B.M.; Majzoub, K.; Villalta, P.W.; Carette, J.E.; et al. Small RNAs are modified with N-glycans and displayed on the surface of living cells. Cell 2021, 184, 3109–3124. [Google Scholar] [CrossRef]
- Fernandes, J.C.R.; Acuna, S.M.; Aoki, J.I.; Floeter-Winter, L.M.; Muxel, S.M. Long Non-Coding RNAs in the Regulation of Gene Expression: Physiology and Disease. Non-Coding RNA 2019, 5, 17. [Google Scholar] [CrossRef]
- Yan, H.; Bu, P. Non-coding RNA in cancer. Essays Biochem. 2021, 65, 625–639. [Google Scholar] [CrossRef]
- Bridges, M.C.; Daulagala, A.C.; Kourtidis, A. LNCcation: lncRNA localization and function. J. Cell Biol. 2021, 220, e202009045. [Google Scholar] [CrossRef]
- Larkin, A.; Imperiali, B. The expanding horizons of asparagine-linked glycosylation. Biochemistry 2011, 50, 4411–4426. [Google Scholar] [CrossRef]
- Schwarz, F.; Aebi, M. Mechanisms and principles of N-linked protein glycosylation. Curr. Opin. Struct. Biol. 2011, 21, 576–582. [Google Scholar] [CrossRef]
- Gupta, R.; Leon, F.; Rauth, S.; Batra, S.K.; Ponnusamy, M.P. A Systematic Review on the Implications of O-linked Glycan Branching and Truncating Enzymes on Cancer Progression and Metastasis. Cells 2020, 9, 446. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Hansen, L.; Clausen, H. Polypeptide N-acetylgalactosaminyltransferase-Associated Phenotypes in Mammals. Molecules 2021, 26, 5504. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.B.; Pyo, K.H.; Kim, H.R. Role and Function of O-GlcNAcylation in Cancer. Cancers 2021, 13, 5365. [Google Scholar] [CrossRef] [PubMed]
- Gambetta, M.C.; Oktaba, K.; Muller, J. Essential role of the glycosyltransferase sxc/Ogt in polycomb repression. Science 2009, 325, 93–96. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, D.A.; Syrzycka, M.; Macauley, M.S.; Rastgardani, T.; Komljenovic, I.; Vocadlo, D.J.; Brock, H.W.; Honda, B.M. Drosophila O-GlcNAc transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc). Proc. Nat. Acad. Sci. USA 2009, 106, 13427–13432. [Google Scholar] [CrossRef]
- Kurcon, T.; Liu, Z.; Paradkar, A.V.; Vaiana, C.A.; Koppolu, S.; Agrawal, P.; Mahal, L.K. miRNA proxy approach reveals hidden functions of glycosylation. Proc. Nat. Acad. Sci. USA 2015, 112, 7327–7332. [Google Scholar] [CrossRef]
- Thu, C.T.; Mahal, L.K. Sweet Control: MicroRNA Regulation of the Glycome. Biochemistry 2020, 59, 3098–3110. [Google Scholar] [CrossRef]
- Kronstein-Wiedemann, R.; Nowakowska, P.; Milanov, P.; Gubbe, K.; Seifried, E.; Bugert, P.; Chavakis, T.; Tonn, T. Regulation of ABO blood group antigen expression by miR-331-3p and miR-1908-5p during hematopoietic stem cell differentiation. Stem Cells 2020, 38, 1348–1362. [Google Scholar] [CrossRef]
- Ke, S.B.; Qiu, H.; Chen, J.M.; Shi, W.; Han, C.; Gong, Y.; Chen, Y.S. ALG3 contributes to the malignancy of non-small cell lung cancer and is negatively regulated by MiR-98-5p. Pathol. Res. Pract. 2020, 216, 152761. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, T.; Xian, L.; Liu, W.; Liu, J.; Zhou, H. B3GNT3, a Direct Target of miR-149-5p, Promotes Lung Cancer Development and Indicates Poor Prognosis of Lung Cancer. Cancer Manag. Res. 2020, 12, 2381–2391. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, B.; Xu, G.; Han, C.; Xing, G. lncRNA MIR44352HG promotes the progression of liver cancer by upregulating B3GNT5 expression. Mol. Med. Rep. 2022, 25, 38. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Yang, X.; Liu, M.; Tang, H. B4GALT3 up-regulation by miR-27a contributes to the oncogenic activity in human cervical cancer cells. Cancer Lett. 2016, 375, 284–292. [Google Scholar] [CrossRef] [PubMed]
- Bi, C.; Shan, J.; Li, M.; Zhang, Q.; Li, C.; Tong, J.; Huang, Q. Long noncoding RNA differentiation antagonizing nonprotein coding RNA promotes the proliferation, invasion and migration of neuroblastoma cells via targeting β-1, 4-galactosyltransferase III by sponging miR-338-3p. Neuroreport 2021, 32, 965–974. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhao, B.; Chen, X.; Geng, X.; Zhang, Z. Circ_0009910 sponges miR-491-5p to promote acute myeloid leukemia progression through modulating B4GALT5 expression and PI3K/AKT signaling pathway. Int. J. Lab. Hematol. 2022, 44, 320–332. [Google Scholar] [CrossRef]
- Dong, X.; Liu, Y.; Deng, X.; Shao, J.; Tian, S.; Chen, S.; Huang, R.; Lin, Z.; Chen, C.; Shen, L. C1GALT1, Negatively Regulated by miR-181d-5p, Promotes Tumor Progression via Upregulating RAC1 in Lung Adenocarcinoma. Front. Cell Dev. Biol. 2021, 9, 707970. [Google Scholar] [CrossRef]
- Tan, Z.; Jiang, Y.; Liang, L.; Wu, J.; Cao, L.; Zhou, X.; Song, Z.; Ye, Z.; Zhao, Z.; Feng, H.; et al. Dysregulation and prometastatic function of glycosyltransferase C1GALT1 modulated by cHP1BP3/ miR-1-3p axis in bladder cancer. J. Exp. Clin. Cancer Res. 2022, 41, 228. [Google Scholar] [CrossRef]
- Huang, L.; Sun, T.Y.; Hu, L.J.; Hu, S.L.; Sun, H.M.; Zhao, F.Q.; Wu, B.; Yang, S.; Ji, F.Q.; Zhou, D.S. Elevated miR-124-3p in the aging colon disrupts mucus barrier and increases susceptibility to colitis by targeting T-synthase. Aging Cell 2020, 19, e13252. [Google Scholar] [CrossRef]
- Hu, S.; Bao, H.; Xu, X.; Zhou, X.; Qin, W.; Zeng, C.; Liu, Z. Increased miR-374b promotes cell proliferation and the production of aberrant glycosylated IgA1 in B cells of IgA nephropathy. FEBS Lett. 2015, 589, 4019–4025. [Google Scholar] [CrossRef]
- Liu, Z.; Yang, S.; Chen, X.; Dong, S.; Zhou, S.; Xu, S. LncRNA LINC00467 acted as an oncogene in esophageal squamous cell carcinoma by accelerating cell proliferation and preventing cell apoptosis via the miR-485-5p/DPAGT1 axis. J. Gastroenterol. Hepatol. 2021, 36, 721–730. [Google Scholar] [CrossRef]
- Cong, J.; Gong, J.; Yang, C.; Xia, Z.; Zhang, H. MiR-200c/FUT4 axis prevents the proliferation of colon cancer cells by downregulating the Wnt/beta-catenin pathway. BMC Cancer 2021, 21, 2. [Google Scholar] [CrossRef]
- Li, Y.; Sun, Z.; Liu, B.; Shan, Y.; Zhao, L.; Jia, L. Tumor-suppressive miR-26a and miR-26b inhibit cell aggressiveness by regulating FUT4 in colorectal cancer. Cell Death. Dis. 2017, 8, e2892. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Xiao, Y.; Liu, B.; Pan, S.; Liu, Q.; Shan, Y.; Li, S.; Qi, Y.; Huang, Y.; Jia, L. Exosomal MALAT1 sponges miR-26a/26b to promote the invasion and metastasis of colorectal cancer via FUT4 enhanced fucosylation and PI3K/Akt pathway. J. Exp. Clin. Cancer Res. 2020, 39, 54. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Zhao, L.; Gao, S.; Song, X.; Dong, W.; Zhao, Y.; Zhou, H.; Cheng, L.; Miao, X.; Jia, L. Increased fucosylation has a pivotal role in multidrug resistance of breast cancer cells through miR-224-3p targeting FUT4. Gene 2016, 578, 232–241. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Feng, X.; Song, X.; Zhou, H.; Zhao, Y.; Cheng, L.; Jia, L. miR-493-5p attenuates the invasiveness and tumorigenicity in human breast cancer by targeting FUT4. Oncol. Rep. 2016, 36, 1007–1015. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Jin, X.; Zhao, Y.; Dai, J.; Cai, Y. Long noncoding RNA GAS6-AS2 sponges microRNA-493, thereby enhancing the malignant characteristics of breast cancer cells via upregulation of FUT4. Pathol. Res. Pract. 2020, 216, 152772. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Liu, J.; Ye, X. Effect of miR-200c on the proliferation, migration and invasion of breast cancer cells and relevant mechanisms. J. Buon. 2019, 24, 61–67. [Google Scholar]
- Zheng, Q.; Cui, X.; Zhang, D.; Yang, Y.; Yan, X.; Liu, M.; Niang, B.; Aziz, F.; Liu, S.; Yan, Q.; et al. miR-200b inhibits proliferation and metastasis of breast cancer by targeting fucosyltransferase IV and alpha1,3-fucosylated glycans. Oncogenesis 2017, 6, e358. [Google Scholar] [CrossRef]
- Li, W.; Li, Y.; Ma, W.; Zhou, J.; Sun, Z.; Yan, X. Long noncoding RNA AC114812.8 promotes the progression of bladder cancer through miR-371b-5p/FUT4 axis. Biomed. Pharmacother. 2020, 121, 109605. [Google Scholar] [CrossRef]
- Liu, B.; Ma, H.; Liu, Q.; Xiao, Y.; Pan, S.; Zhou, H.; Jia, L. MiR-29b/Sp1/FUT4 axis modulates the malignancy of leukemia stem cells by regulating fucosylation via Wnt/β-catenin pathway in acute myeloid leukemia. J. Exp. Clin. Cancer Res. 2019, 38, 200. [Google Scholar] [CrossRef]
- Chen, R.; Zhang, X.; Wang, C. LncRNA HOXB-AS1 promotes cell growth in multiple myeloma via FUT4 mRNA stability by ELAVL1. J. Cell Biochem. 2020, 121, 4043–4051. [Google Scholar] [CrossRef]
- Andolfo, I.; Liguori, L.; De Antonellis, P.; Cusanelli, E.; Marinaro, F.; Pistollato, F.; Garzia, L.; De Vita, G.; Petrosino, G.; Accordi, B.; et al. The micro-RNA 199b-5p regulatory circuit involves Hes1, CD15, and epigenetic modifications in medulloblastoma. Neuro. Oncol. 2012, 14, 596–612. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Wang, Z.; Pan, Y.; Ma, J.; Miao, X.; Qi, X.; Zhou, H.; Jia, L. MiR-26a and miR-26b mediate osteoarthritis progression by targeting FUT4 via NF-kB signaling pathway. Int. J. Biochem. Cell Biol. 2018, 94, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wei, S.; Zhang, Z. MicroRNA-200b relieves LPS-induced inflammatory injury by targeting FUT4 in knee articular chondrocytes in vitro. Exp. Ther. Med. 2021, 21, 407. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Zhang, D.; Yang, Y.U.; Cui, X.; Sun, J.; Liang, C.; Qin, H.; Yang, X.; Liu, S.; Yan, Q. MicroRNA-200c impairs uterine receptivity formation by targeting FUT4 and alpha1,3-fucosylation. Cell Death. Differ. 2017, 24, 2161–2172. [Google Scholar] [CrossRef]
- Liang, L.; Gao, C.; Li, Y.; Sun, M.; Xu, J.; Li, H.; Jia, L.; Zhao, Y. miR-125a-3p/FUT5-FUT6 axis mediates colorectal cancer cell proliferation, migration, invasion and pathological angiogenesis via PI3K-Akt pathway. Cell Death. Dis. 2017, 8, e2968. [Google Scholar] [CrossRef]
- Pan, S.; Liu, Y.; Liu, Q.; Xiao, Y.; Liu, B.; Ren, X.; Qi, X.; Zhou, H.; Zeng, C.; Jia, L. HOTAIR/miR-326/FUT6 axis facilitates colorectal cancer progression through regulating fucosylation of CD44 via PI3K/AKT/mTOR pathway. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 750–760. [Google Scholar] [CrossRef]
- Li, N.; Liu, Y.; Miao, Y.; Zhao, L.; Zhou, H.; Jia, L. MicroRNA-106b targets FUT6 to promote cell migration, invasion, and proliferation in human breast cancer. IUBMB Life 2016, 68, 764–775. [Google Scholar] [CrossRef]
- Bernardi, C.; Soffientini, U.; Piacente, F.; Tonetti, M.G. Effects of microRNAs on fucosyltransferase 8 (FUT8) expression in hepatocarcinoma cells. PLoS ONE 2013, 8, e76540. [Google Scholar] [CrossRef]
- Guo, Y.; Liu, B.; Huang, T.; Qi, X.; Li, S. HOTAIR modulates hepatocellular carcinoma progression by activating FUT8/core-fucosylated Hsp90/MUC1/STAT3 feedback loop via JAK1/STAT3 cascade. Dig. Liver Dis. 2022; Online ahead of print. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, X.; Yang, C.; Xu, S. MicroRNA-198-5p inhibits the migration and invasion of non-small lung cancer cells by targeting fucosyltransferase 8. Clin. Exp. Pharmacol. Physiol. 2019, 46, 955–967. [Google Scholar] [CrossRef]
- Zhao, Y.; Shi, J.; Zhao, Y.; Lu, Z. SNHG1/miR-186/FUT8 regulates cell migration and invasion in oral squamous cell carcinoma. Oral Dis. 2021; Online ahead of print. [Google Scholar] [CrossRef]
- Hu, X.; Shen, N.; Liu, A.; Wang, W.; Zhang, L.; Sui, Z.; Tang, Q.; Du, X.; Yang, N.; Ying, W.; et al. Bone marrow mesenchymal stem cell-derived exosomal miR-34c-5p ameliorates RIF by inhibiting the core fucosylation of multiple proteins. Mol. Ther. 2022, 30, 763–781. [Google Scholar] [CrossRef]
- Dyrskjot, L.; Ostenfeld, M.S.; Bramsen, J.B.; Silahtaroglu, A.N.; Lamy, P.; Ramanathan, R.; Fristrup, N.; Jensen, J.L.; Andersen, C.L.; Zieger, K.; et al. Genomic profiling of microRNAs in bladder cancer: miR-129 is associated with poor outcome and promotes cell death in vitro. Cancer Res. 2009, 69, 4851–4860. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.M.; Zhang, W.Y.; Wang, S.Y.; Qian, G.; Pei, D.L.; Zhang, G.M. microRNA let-7i-5p aggravates kidney fibrosis via targeting GALNT1. Gen. Physiol. Biophys. 2021, 40, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Pan, L.; Yan, X.; Duan, X. The long noncoding RNA DLGAP1-AS2 facilitates cholangiocarcinoma progression via miR-505 and GALNT10. FEBS Open Bio 2021, 11, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Liu, H.O.; Liu, Y.D.; Liu, W.S.; Pan, D.; Zhang, W.J.; Yang, L.; Fu, Q.; Xu, J.J.; Gu, J.X. Decreased Expression of Hepatocyte Nuclear Factor 4α (Hnf4α)/MicroRNA-122 (miR-122) Axis in Hepatitis B Virus-associated Hepatocellular Carcinoma Enhances Potential Oncogenic GALNT10 Protein Activity. J. Biol. Chem. 2015, 290, 1170–1185. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, G.; Zhang, K. MiR-125a regulates ovarian cancer proliferation and invasion by repressing GALNT14 expression. Biomed. Pharmacother. 2016, 80, 381–387. [Google Scholar] [CrossRef]
- Xu, Z.; Jin, H.; Duan, X.; Liu, H.; Zhao, X.; Fan, S.; Wang, Y.; Yao, T. LncRNA PSMA3-AS1 promotes cell proliferation, migration, and invasion in ovarian cancer by activating the PI3K/Akt pathway via the miR-378a-3p/GALNT3 axis. Environ. Toxicol. 2021, 36, 2562–2577. [Google Scholar] [CrossRef]
- Hu, C.Y.; You, P.; Zhang, J.; Zhang, H.; Jiang, N. MiR-506-3p acts as a novel tumor suppressor in prostate cancer through targeting GALNT4. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 5133–5138. [Google Scholar] [CrossRef]
- Xing, L.; Hong, X.; Chang, L.; Ren, P.; Zhang, H. miR-365b regulates the development of non-small cell lung cancer via GALNT4. Exp. Ther. Med. 2020, 20, 1637–1643. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, H.; Yang, L.; Wu, Q.; Liu, W.; Fu, Q.; Zhang, W.; Zhang, H.; Xu, J.; Gu, J. Loss of N-acetylgalactosaminyltransferase-4 orchestrate oncogenic microRNA-9 in hepatocellular carcinoma. J. Biol. Chem. 2017, 292, 3186–3200. [Google Scholar] [CrossRef]
- Gaziel-Sovran, A.; Segura, M.F.; Di Micco, R.; Collins, M.K.; Hanniford, D.; Vega-Saenz, D.M.; Rakus, J.F.; Dankert, J.F.; Shang, S.; Kerbel, R.S.; et al. miR-30b/30d regulation of GalNAc transferases enhances invasion and immunosuppression during metastasis. Cancer Cell 2011, 20, 104–118. [Google Scholar] [CrossRef]
- Li, Y.; Zeng, C.; Hu, J.; Pan, Y.; Shan, Y.; Liu, B.; Jia, L. Long non-coding RNA-SNHG7 acts as a target of miR-34a to increase GALNT7 level and regulate PI3K/Akt/mTOR pathway in colorectal cancer progression. J. Hematol. Oncol. 2018, 11, 89. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Chen, J.; Li, D.; Liu, X.; Li, L.; Wang, K. MicroRNA-30e Functions as a Tumor Suppressor in Cervical Carcinoma Cells through Targeting GALNT7. Transl. Oncol. 2017, 10, 876–885. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, M.E.; Snieckute, G.; Kagias, K.; Nehammer, C.; Multhaupt, H.A.; Couchman, J.R.; Pocock, R. An epidermal microRNA regulates neuronal migration through control of the cellular glycosylation state. Science 2013, 341, 1404–1408. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Ma, H.; Sun, J. MicroRNA34a/c function as tumor suppressors in Hep2 laryngeal carcinoma cells and may reduce GALNT7 expression. Mol. Med. Rep. 2014, 9, 1293–1298. [Google Scholar] [CrossRef]
- Lu, Q.; Xu, L.; Li, C.; Yuan, Y.; Huang, S.; Chen, H. miR-214 inhibits invasion and migration via downregulating GALNT7 in esophageal squamous cell cancer. Tumour. Biol. 2016, 37, 14605–14614. [Google Scholar] [CrossRef]
- Shan, S.W.; Fang, L.; Shatseva, T.; Rutnam, Z.J.; Yang, X.; Du, W.; Lu, W.Y.; Xuan, J.W.; Deng, Z.; Yang, B.B. Mature miR-17-5p and passenger miR-17-3p induce hepatocellular carcinoma by targeting PTEN, GalNT7 and vimentin in different signal pathways. J. Cell Sci. 2013, 126, 1517–1530. [Google Scholar] [CrossRef]
- Gao, F.; Han, J.; Jia, L.; He, J.; Wang, Y.; Chen, M.; Liu, X.; He, X. MiR-30c facilitates natural killer cell cytotoxicity to lung cancer through targeting GALNT7. Genes Genom. 2022; Online ahead of print. [Google Scholar] [CrossRef]
- Kahai, S.; Lee, S.C.; Lee, D.Y.; Yang, J.; Li, M.; Wang, C.H.; Jiang, Z.; Zhang, Y.; Peng, C.; Yang, B.B. MicroRNA miR-378 regulates nephronectin expression modulating osteoblast differentiation by targeting GalNT-7. PLoS ONE 2009, 4, e7535. [Google Scholar] [CrossRef]
- Chao, C.C.; Wu, P.H.; Huang, H.C.; Chung, H.Y.; Chou, Y.C.; Cai, B.H.; Kannagi, R. Downregulation of miR-199a/b-5p is associated with GCNT2 induction upon epithelial-mesenchymal transition in colon cancer. FEBS Lett. 2017, 591, 1902–1917. [Google Scholar] [CrossRef]
- Gonzalez-Vallinas, M.; Molina, S.; Vicente, G.; Zarza, V.; Martin-Hernandez, R.; Garcia-Risco, M.R.; Fornari, T.; Reglero, G.; Ramirez de Molina, A. Expression of MicroRNA-15b and the Glycosyltransferase GCNT3 Correlates with Antitumor Efficacy of Rosemary Diterpenes in Colon and Pancreatic Cancer. PLoS ONE 2014, 9, e98556. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, Y.; Liu, W.; Zhang, Q.; Xiao, H.; Song, H.; Luo, B. MiR-BART1-5p targets core 2 β-1,6-acetylglucosaminyltransferase GCNT3 to inhibit cell proliferation and migration in EBV-associated gastric cancer. Virology 2020, 541, 63–74. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiao, P.; Hu, X. LINC00511 enhances LUAD malignancy by upregulating GCNT3 via miR-195-5p. BMC Cancer 2022, 22, 389. [Google Scholar] [CrossRef] [PubMed]
- Bieg, D.; Sypniewski, D.; Nowak, E.; Bednarek, I. MiR-424-3p suppresses galectin-3 expression and sensitizes ovarian cancer cells to cisplatin. Arch. Gynecol. Obstet. 2019, 299, 1077–1087. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Wang, J.; Yang, G.; Yu, N.; Huang, Z.; Xu, H.; Li, J.; Qiu, J.; Zeng, X.; Chen, S.; et al. Posttranscriptional regulation of Galectin-3 by miR-128 contributes to colorectal cancer progression. Oncotarget 2017, 8, 15242–15251. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Ji, J.; Chen, X.; Xu, W.; Chen, H.; Zhu, S.; Wu, J.; Wu, Y.; Sun, Y.; Sai, W.; et al. Human umbilical cord mesenchymal stem cell-derived exosomes carrying hsa-miRNA-128-3p suppress pancreatic ductal cell carcinoma by inhibiting Galectin-3. Clin. Transl. Oncol. 2021, 24, 517–531. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhou, X.; Luo, G.; Zhang, J.; Yang, M.; Song, C. CircRNA RERE Promotes the Oxidative Stress-Induced Apoptosis and Autophagy of Nucleus Pulposus Cells through the miR-299-5p/Galectin-3 Axis. J. Healthc. Eng 2021, 2021, 2771712. [Google Scholar] [CrossRef]
- Yang, Q.; Hou, C.; Huang, D.; Zhuang, C.; Jiang, W.; Geng, Z.; Wang, X.; Hu, L. miR-455-5p functions as a potential oncogene by targeting galectin-9 in colon cancer. Oncol. Lett. 2017, 13, 1958–1964. [Google Scholar] [CrossRef]
- Yang, Q.; Jiang, W.; Zhuang, C.; Geng, Z.; Hou, C.; Huang, D.; Hu, L.; Wang, X. microRNA-22 downregulation of galectin-9 influences lymphocyte apoptosis and tumor cell proliferation in liver cancer. Oncol. Rep. 2015, 34, 1771–1778. [Google Scholar] [CrossRef]
- Zhu, Q.; Zhan, D.; Yang, Y.; Chong, Y.; Xue, H.; Zhu, P. LINC00173 Promotes Wilms’ Tumor Progression Through MGAT1-mediated MUC3A N-glycosylation. Cell Cycle 2022, 21, 1795–1810. [Google Scholar] [CrossRef]
- Pan, K.; Chen, S.; Wang, Y.; Yao, W.; Gao, X. MicroRNA-23b attenuates tau pathology and inhibits oxidative stress by targeting GnT-III in Alzheimer’s disease. Neuropharmacology 2021, 196, 108671. [Google Scholar] [CrossRef]
- Vaiana, C.A.; Kurcon, T.; Mahal, L.K. MicroRNA-424 Predicts a Role for β1,4 Branched Glycosylation in Cell Cycle Progression. J. Biol. Chem. 2016, 291, 1529–1537. [Google Scholar] [CrossRef]
- Yan, G.; Li, Y.; Zhan, L.; Sun, S.; Yuan, J.; Wang, T.; Yin, Y.; Dai, Z.; Zhu, Y.; Jiang, Z.; et al. Decreased miR-124-3p promoted breast cancer proliferation and metastasis by targeting MGAT5. Am. J. Cancer Res. 2019, 9, 585–596. [Google Scholar] [PubMed]
- Han, D.L.; Wang, L.L.; Zhang, G.F.; Yang, W.F.; Chai, J.; Lin, H.M.; Fu, Z.; Yu, J.M. MiRNA-485-5p, inhibits esophageal cancer cells proliferation and invasion by down-regulating O-linked N-acetylglucosamine transferase. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 2809–2816. [Google Scholar] [CrossRef] [PubMed]
- Kalantzakos, T.J.; Sullivan, T.B.; Sebel, L.E.; Canes, D.; Burks, E.J.; Moinzadeh, A.; Rieger-Christ, K.M. MicroRNAs MiR-15a and MiR-26a cooperatively regulate O-GlcNAc-transferase to control proliferation in clear cell renal cell carcinoma. Cancer Biomark. 2021, 30, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Ning, D.; Chen, J.; Du, P.; Liu, Q.; Cheng, Q.; Li, X.; Zhang, B.; Chen, X.; Jiang, L. The crosstalk network of XIST/miR-424-5p/OGT mediates RAF1 glycosylation and participates in the progression of liver cancer. Liver Int. 2021, 41, 1933–1944. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Cao, M.; Ruan, X.; Jiang, L.; Lee, S.; Lemanek, A.; Ghassemian, M.; Pizzo, D.P.; Wan, Y.; Qiao, Y.; et al. Cancer-cell-secreted miR-122 suppresses O-GlcNAcylation to promote skeletal muscle proteolysis. Nat. Cell Biol. 2022, 24, 793–804. [Google Scholar] [CrossRef]
- Liu, R.; Ma, X.; Chen, L.; Yang, Y.; Zeng, Y.; Gao, J.; Jiang, W.; Zhang, F.; Li, D.; Han, B.; et al. MicroRNA-15b Suppresses Th17 Differentiation and Is Associated with Pathogenesis of Multiple Sclerosis by Targeting O-GlcNAc Transferase. J. Immunol. 2017, 198, 2626–2639. [Google Scholar] [CrossRef]
- Luo, P.; He, T.; Jiang, R.; Li, G. MicroRNA-423-5p targets O-GlcNAc transferase to induce apoptosis in cardiomyocytes. Mol. Med. Rep. 2015, 12, 1163–1168. [Google Scholar] [CrossRef]
- Liu, Y.; Li, X.; Zhang, C.; Zhang, H.; Huang, Y. LINC00973 is involved in cancer immune suppression through positive regulation of Siglec-15 in clear-cell renal cell carcinoma. Cancer Sci. 2020, 111, 3693–3704. [Google Scholar] [CrossRef]
- Ren, Y.; Lyu, J.; Guo, Y.; Yao, Y.; Hu, L. Long Noncoding RNA TUG1 Inhibits Tumor Progression through Regulating Siglec-15-Related Anti-Immune Activity in Hepatocellular Carcinoma. J. Immunol. Res. 2022, 2022, 9557859. [Google Scholar] [CrossRef]
- Gong, A.; Zhao, X.; Pan, Y.; Qi, Y.; Li, S.; Huang, Y.; Guo, Y.; Qi, X.; Zheng, W.; Jia, L. The lncRNA MEG3 mediates renal cell cancer progression by regulating ST3Gal1 transcription and EGFR sialylation. J. Cell Sci. 2020, 133, jcs244020. [Google Scholar] [CrossRef]
- Xi, D.; Hofmann, L.; Alter, T.; Einspanier, R.; Bereswill, S.; Heimesaat, M.M.; Golz, G.; Sharbati, S. The glycosyltransferase ST3GAL2 is regulated by miR-615-3p in the intestinal tract of Campylobacter jejuni infected mice. Gut Pathog. 2021, 13, 42. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Hu, J.; Ma, J.; Qi, X.; Zhou, H.; Miao, X.; Zheng, W.; Jia, L. MiR-193a-3p and miR-224 mediate renal cell carcinoma progression by targeting α2,3-sialyltransferase IV and the phosphatidylinositol 3 kinase/Akt pathway. Mol. Carcinog. 2018, 57, 1067–1077. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Li, Y.; Liu, B.; Shan, Y.; Li, Y.; Zhao, L.; Su, Z.; Jia, L. Downregulation of miR-224 and let-7i contribute to cell survival and chemoresistance in chronic myeloid leukemia cells by regulating ST3GAL IV expression. Gene 2017, 626, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Shao, J.; Wang, Y.; Shen, H.; Yu, S.; Zhang, J.; Yin, L. Hsa-miR-370 inhibited P-selectin-induced cell adhesion in human colon adenocarcinoma cells. Mol. Cell Biochem. 2019, 450, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Hao, Z.; Liu, C.; Yuan, L.; Li, L.; Yin, M.; Li, Q.; Qi, Z.; Wang, Z. MiR-193b modulates osteoarthritis progression through targeting ST3GAL4 via sialylation of CD44 and NF-kB pathway. Cell Signal. 2020, 76, 109814. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Zhou, H.; Miao, Y.; Li, N.; Zhao, L.; Jia, L. MiRNA expression profiles reveal the involvement of miR-26a, miR-548l and miR-34a in hepatocellular carcinoma progression through regulation of ST3GAL5. Lab. Investig. 2017, 97, 530–542. [Google Scholar] [CrossRef]
- Sun, M.; Zhao, X.; Liang, L.; Pan, X.; Lv, H.; Zhao, Y. Sialyltransferase ST3GAL6 mediates the effect of microRNA-26a on cell growth, migration, and invasion in hepatocellular carcinoma through the protein kinase B/mammalian target of rapamycin pathway. Cancer Sci. 2017, 108, 267–276. [Google Scholar] [CrossRef]
- Li, J.; Long, Y.; Sun, J.; Wu, J.; He, X.; Wang, S.; Wang, X.; Miao, X.; Huang, R.; Yan, J. Comprehensive landscape of the ST3GAL family reveals the significance of ST3GAL6-AS1/ST3GAL6 axis on EGFR signaling in lung adenocarcinoma cell invasion. Front. Cell Dev. Biol. 2022, 10, 931132. [Google Scholar] [CrossRef]
- Hu, J.; Shan, Y.; Ma, J.; Pan, Y.; Zhou, H.; Jiang, L.; Jia, L. LncRNA ST3Gal6-AS1/ST3Gal6 axis mediates colorectal cancer progression by regulating α2,3 sialylation via PI3K/Akt signaling. Int. J. Cancer 2019, 145, 450–460. [Google Scholar] [CrossRef]
- Shen, Y.; Feng, Y.; Li, F.; Jia, Y.; Peng, Y.; Zhao, W.; Hu, J.; He, A. lncRNA ST3GAL6AS1 promotes invasion by inhibiting hnRNPA2B1mediated ST3GAL6 expression in multiple myeloma. Int. J. Oncol. 2021, 58, 5. [Google Scholar] [CrossRef]
- Han, Y.; Liu, Y.; Fu, X.; Zhang, Q.; Huang, H.; Zhang, C.; Li, W.; Zhang, J. miR-9 inhibits the metastatic ability of hepatocellular carcinoma via targeting β-galactoside α-2,6-sialyltransferase 1. J. Physiol. Biochem. 2018, 74, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Mei, J.; Lin, W.; Li, S.; Tang, Y.; Ye, Z.; Lu, L.; Wen, Y.; Kan, A.; Zou, J.; Yu, C.; et al. Long noncoding RNA TINCR facilitates hepatocellular carcinoma progression and dampens chemosensitivity to oxaliplatin by regulating the miR-195-3p/ST6GAL1/NF-kB pathway. J. Exp. Clin. Cancer Res. 2022, 41, 5. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Zhao, Z.; Ma, J.; Dong, L.; Liang, Y.; Li, S.; Mao, Y.; Li, Y.; Zhang, Y. MiR-214-3p regulates the viability, invasion, migration and EMT of TNBC cells by targeting ST6GAL1. Cytotechnology 2019, 71, 1155–1165. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Ma, H.; Sun, X.; Liu, B.; Xiao, Y.; Pan, S.; Zhou, H.; Dong, W.; Jia, L. The regulatory ZFAS1/miR-150/ST6GAL1 crosstalk modulates sialylation of EGFR via PI3K/Akt pathway in T-cell acute lymphoblastic leukemia. J. Exp. Clin. Cancer Res. 2019, 38, 199. [Google Scholar] [CrossRef] [PubMed]
- Minami, A.; Shimono, Y.; Mizutani, K.; Nobutani, K.; Momose, K.; Azuma, T.; Takai, Y. Reduction of the ST6 β-Galactosamide α-2,6-Sialyltransferase 1 (ST6GAL1)-catalyzed Sialylation of Nectin-like Molecule 2/Cell Adhesion Molecule 1 and Enhancement of ErbB2/ErbB3 Signaling by MicroRNA-199a. J. Biol. Chem. 2013, 288, 11845–11853. [Google Scholar] [CrossRef]
- Jia, L.; Luo, S.; Ren, X.; Li, Y.; Hu, J.; Liu, B.; Zhao, L.; Shan, Y.; Zhou, H. miR-182 and miR-135b Mediate the Tumorigenesis and Invasiveness of Colorectal Cancer Cells via Targeting ST6GALNAC2 and PI3K/AKT Pathway. Dig. Dis. Sci. 2017, 62, 3447–3459. [Google Scholar] [CrossRef]
- Liu, B.; Liu, Y.; Zhao, L.; Pan, Y.; Shan, Y.; Li, Y.; Jia, L. Upregulation of microRNA-135b and microRNA-182 promotes chemoresistance of colorectal cancer by targeting ST6GALNAC2 via PI3K/AKT pathway. Mol. Carcinog. 2017, 56, 2669–2680. [Google Scholar] [CrossRef]
- Miao, X.; Jia, L.; Zhou, H.; Song, X.; Zhou, M.; Xu, J.; Zhao, L.; Feng, X.; Zhao, Y. miR-4299 mediates the invasive properties and tumorigenicity of human follicular thyroid carcinoma by targeting ST6GALNAC4. IUBMB Life 2016, 68, 136–144. [Google Scholar] [CrossRef]
- Bai, L.; Luo, L.; Gao, W.; Bu, C.; Huang, J. miR-182 modulates cell proliferation and invasion in prostate cancer via targeting ST6GALNAC5. Braz. J. Med. Biol. Res. 2021, 54, e9695. [Google Scholar] [CrossRef]
- Shan, Y.; Liu, Y.; Zhao, L.; Liu, B.; Li, Y.; Jia, L. MicroRNA-33a and let-7e inhibit human colorectal cancer progression by targeting ST8SIA1. Int. J. Biochem. Cell Biol. 2017, 90, 48–58. [Google Scholar] [CrossRef]
- Xing, P.; Wang, Y.; Zhang, L.; Ma, C.; Lu, J. Knockdown of lncRNA MIR44352HG and ST8SIA1 expression inhibits the proliferation, invasion and migration of prostate cancer cells in vitro and in vivo by blocking the activation of the FAK/AKT/β-catenin signaling pathway. Int. J. Mol. Med. 2021, 47, 93. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wang, F.; Fan, L.; Wang, H.; Gu, S. Long Noncoding RNA TUG1 Aggravates Cerebral Ischemia/Reperfusion Injury by Acting as a ceRNA for miR-3072-3p to Target St8sia2. Oxid. Med. Cell. Longev. 2022, 2022, 9381203. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Dong, W.; Su, Z.; Zhao, L.; Miao, Y.; Li, N.; Zhou, H.; Jia, L. Functional roles of sialylation in breast cancer progression through miR-26a/26b targeting ST8SIA4. Cell Death. Dis. 2016, 7, e2561. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Cao, S.; Wang, X.; Zhang, L.; Yuan, H.; Ma, X. lncRNA MALAT1/miR26a/26b/ST8SIA4 axis mediates cell invasion and migration in breast cancer cell lines. Oncol. Rep. 2021, 46, 181. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Zhao, X.; Liang, L.; Wang, G.; Li, Y.; Miao, X.; Zhao, Y. miR-146a and miR-146b promote proliferation, migration and invasion of follicular thyroid carcinoma via inhibition of ST8SIA4. Oncotarget 2017, 8, 28028–28041. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Yu, G.; Liang, J.; Fan, P.; Dong, K.; Zhang, B.; Chen, X.; Zhu, H.; Chu, L. miR-144-5p and miR-451a Inhibit the Growth of Cholangiocarcinoma Cells Through Decreasing the Expression of ST8SIA4. Front. Oncol. 2020, 10, 563486. [Google Scholar] [CrossRef]
- Pepe, F.; Pagotto, S.; Soliman, S.; Rossi, C.; Lanuti, P.; Braconi, C.; Mariani-Costantini, R.; Visone, R.; Veronese, A. Regulation of miR-483-3p by the O-linked N-acetylglucosamine transferase links chemosensitivity to glucose metabolism in liver cancer cells. Oncogenesis 2017, 6, e328. [Google Scholar] [CrossRef]
- Herzog, K.; Bandiera, S.; Pernot, S.; Fauvelle, C.; Juhling, F.; Weiss, A.; Bull, A.; Durand, S.C.; Chane-Woon-Ming, B.; Pfeffer, S.; et al. Functional microRNA screen uncovers O-linked N-acetylglucosamine transferase as a host factor modulating hepatitis C virus morphogenesis and infectivity. Gut 2020, 69, 380–392. [Google Scholar] [CrossRef]
- Ma, M.; Guo, D.; Tan, Z.; Du, J.; Guan, F.; Li, X. Fucosyltransferase 8 regulation and breast cancer suppression by transcription factor activator protein 2γ. Cancer Sci. 2021, 112, 3190–3204. [Google Scholar] [CrossRef]
- Qi, Y.; Shan, Y.; Li, S.; Huang, Y.; Guo, Y.; Huang, T.; Zhao, X.; Jia, L. LncRNA LEF1-AS1/LEF1/FUT8 Axis Mediates Colorectal Cancer Progression by Regulating α1, 6-Fucosylation via Wnt/β-Catenin Pathway. Dig. Dis. Sci. 2022, 67, 2182–2194. [Google Scholar] [CrossRef]
- Dennis, J.W.; Laferte, S.; Waghorne, C.; Breitman, M.L.; Kerbel, R.S. β 1-6 branching of Asn-linked oligosaccharides is directly associated with metastasis. Science 1987, 236, 582–585. [Google Scholar] [CrossRef]
- Ye, J.; Wei, X.; Shang, Y.; Pan, Q.; Yang, M.; Tian, Y.; He, Y.; Peng, Z.; Chen, L.; Chen, W.; et al. Core 3 mucin-type O-glycan restoration in colorectal cancer cells promotes MUC1/p53/miR-200c-dependent epithelial identity. Oncogene 2017, 36, 6391–6407. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; He, R.Q.; Wu, H.Y.; Zhang, T.T.; Liang, H.W.; Ye, Z.H.; Li, Z.Y.; Xie, T.T.; Shi, Q.; Ma, J.; et al. Expression Signature and Role of miR-30d-5p in Non-Small Cell Lung Cancer: A Comprehensive Study Based on in Silico Analysis of Public Databases and in Vitro Experiments. Cell Physiol. Biochem. 2018, 50, 1964–1987. [Google Scholar] [CrossRef] [PubMed]
- Pucci, M.; Gomes, F.I.; Malagolini, N.; Ferracin, M.; Dall’Olio, F. The Sda Synthase B4GALNT2 Reduces Malignancy and Stemness in Colon Cancer Cell Lines Independently of Sialyl Lewis X Inhibition. Int. J. Mol. Sci. 2020, 21, 6558. [Google Scholar] [CrossRef] [PubMed]
- Terraneo, L.; Avagliano, L.; Caretti, A.; Bianciardi, P.; Tosi, D.; Bulfamante, G.P.; Samaja, M.; Trinchera, M. Expression of carbohydrate-antigen sialyl-Lewis a on colon cancer cells promotes xenograft growth and angiogenesis in nude mice. Int. J. Biochem. Cell Biol. 2013, 45, 2796–2800. [Google Scholar] [CrossRef] [PubMed]
- Trinchera, M.; Aronica, A.; Dall’Olio, F. Selectin Ligands Sialyl-Lewis a and Sialyl-Lewis x in Gastrointestinal Cancers. Biology 2017, 6, 16. [Google Scholar] [CrossRef]
- Trinchera, M.; Malagolini, N.; Chiricolo, M.; Santini, D.; Minni, F.; Caretti, A.; Dall’Olio, F. The biosynthesis of the selectin-ligand sialyl Lewis x in colorectal cancer tissues is regulated by fucosyltransferase VI and can be inhibited by an RNA interference-based approach. Int. J. Biochem. Cell Biol. 2011, 43, 130–139. [Google Scholar] [CrossRef]
- Ronchetti, D.; Todoerti, K.; Vinci, C.; Favasuli, V.; Agnelli, L.; Manzoni, M.; Pelizzoni, F.; Chiaramonte, R.; Platonova, N.; Giuliani, N.; et al. Expression Pattern and Biological Significance of the lncRNA ST3GAL6-AS1 in Multiple Myeloma. Cancers 2020, 12, 782. [Google Scholar] [CrossRef]
- Wang, L.; Chen, X.; Meng, F.; Huang, T.; Wang, S.; Zheng, Z.; Zheng, G.; Li, W.; Zhang, J.; Liu, Y. α2,6-Sialylation promotes hepatocellular carcinoma cells migration and invasion via enhancement of nSmase2-mediated exosomal miRNA sorting. J. Physiol Biochem. 2022; Online ahead of print. [Google Scholar] [CrossRef]
- Du, Q.; Xiao, R.D.; Luo, R.G.; Xie, J.B.; Su, Z.D.; Wang, Y. Construction of long non-coding RNA- and microRNA-mediated competing endogenous RNA networks in alcohol-related esophageal cancer. PLoS ONE 2022, 17, e0269742. [Google Scholar] [CrossRef]
- Dall’Olio, F.; Malagolini, N.; Trinchera, M.; Chiricolo, M. Sialosignaling: Sialyltransferases as engines of self-fueling loops in cancer progression. Biochim. Biophys. Acta 2014, 1840, 2752–2764. [Google Scholar] [CrossRef]
- Gadwal, A.; Modi, A.; Khokhar, M.; Vishnoi, J.R.; Choudhary, R.; Elhence, P.; Banerjee, M.; Purohit, P. Critical appraisal of epigenetic regulation of galectins in cancer. Int. J. Clin. Oncol. 2021, 27, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Compagno, D.; Jaworski, F.M.; Gentilini, L.; Contrufo, G.; Gonzalez, P.I.; Elola, M.T.; Pregi, N.; Rabinovich, G.A.; Laderach, D.J. Galectins: Major signaling modulators inside and outside the cell. Curr. Mol. Med. 2014, 14, 630–651. [Google Scholar] [CrossRef] [PubMed]
- Adams, O.J.; Stanczak, M.A.; von Gunten, G.S.; Laubli, H. Targeting sialic acid-Siglec interactions to reverse immune suppression in cancer. Glycobiology 2018, 28, 640–647. [Google Scholar] [CrossRef] [PubMed]
- Nagarajah, S.; Xia, S.; Rasmussen, M.; Tepel, M. Endogenous intronic antisense long non-coding RNA, MGAT3-AS1, and kidney transplantation. Sci. Rep. 2019, 9, 14743. [Google Scholar] [CrossRef]
- Nagarajah, S.; Rasmussen, M.; Hoegh, S.V.; Tepel, M. Prospective Study of Long Noncoding RNA, MGAT3-AS1, and Viremia of BK Polyomavirus and Cytomegalovirus in Living Donor Renal Transplant Recipients. Kidney Int. Rep. 2020, 5, 2218–2227. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Sun, G.B.; Dou, G.X.; Wang, B.Q. Long non-coding RNA B3GALT5-AS1 contributes to the progression of gastric cancer via interacting with CSNK2A1. Exp. Ther. Med. 2021, 22, 927. [Google Scholar] [CrossRef]
- Luo, M.L.; Li, J.; Shen, L.; Chu, J.; Guo, Q.; Liang, G.; Wu, W.; Chen, J.; Chen, R.; Song, E. The Role of APAL/ST8SIA6-AS1 lncRNA in PLK1 Activation and Mitotic Catastrophe of Tumor Cells. J. Natl. Cancer Inst. 2020, 112, 356–368. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, Y.; Zhang, Y.; Liu, Z. lncRNA ST8SIA6-AS1 facilitates proliferation and invasion in liver cancer by regulating miR-142-3p. Exp. Ther. Med. 2021, 22, 1348. [Google Scholar] [CrossRef]
- Kuai, J.; Zheng, L.; Yi, X.; Liu, Z.; Qiu, B.; Lu, Z.; Jiang, Y. ST8SIA6-AS1 promotes the development of hepatocellular carcinoma cells through miR-338-3p/NONO Axis. Dig. Liver Dis. 2021, 53, 1192–1200. [Google Scholar] [CrossRef]
- Mou, Y.; Ding, X. LncRNA ST8SIA6-AS1 facilitates hepatocellular carcinoma progression by governing miR-651-5p/TM4SF4 axis. Anticancer Drugs 2022, 33, 741–751. [Google Scholar] [CrossRef]
- Qiao, Y.; Wang, B.; Yan, Y.; Niu, L. Long noncoding RNA ST8SIA6-AS1 promotes cell proliferation and metastasis in triple-negative breast cancer by targeting miR-145-5p/CDCA3 to inactivate the p53/p21 signaling pathway. Environ. Toxicol. 2022, 37, 2398–2411. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Zhang, X.; Cao, J.; Yang, J.; Chen, Z.; Wang, W.; Wang, S.; Zhang, L.; Xie, L.; Fang, L.; et al. The novel role of circular RNA ST3GAL6 on blocking gastric cancer malignant behaviours through autophagy regulated by the FOXP2/MET/mTOR axis. Clin. Transl. Med. 2022, 12, e707. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wu, S.; He, H.; Ai, K.; Xu, R.; Zhang, L.; Zhu, X. CircRNA-ST6GALNAC6 increases the sensitivity of bladder cancer cells to erastin-induced ferroptosis by regulating the HSPB1/P38 axis. Lab. Investig. 2022, 102, 1323–1334. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Lian, C.; Chen, G.; Zou, P.; Qin, B.G. CircRNA FUT10 regulates the regenerative potential of aged skeletal muscle stem cells by targeting HOXA9. Aging 2021, 13, 17428–17441. [Google Scholar] [CrossRef] [PubMed]
- Joshi, H.J.; Hansen, L.; Narimatsu, Y.; Freeze, H.H.; Henrissat, B.; Bennett, E.; Wandall, H.H.; Clausen, H.; Schjoldager, K.T. Glycosyltransferase genes that cause monogenic congenital disorders of glycosylation are distinct from glycosyltransferase genes associated with complex diseases. Glycobiology 2018, 28, 284–294. [Google Scholar] [CrossRef]


| Target Glycogene | Upstream ncRNA | Downstream ncRNA | Downstream Target | Tissue and Reference |
|---|---|---|---|---|
| A/B glycosyltransferases | miR-331-3p miR-1908-5p | Biosynthesis of AB0 antigens [18] | ||
| ALG3 | miR-98-5p | Lung cancer [19] | ||
| B3GNT3 | miR-149-5p | Lung cancer [20] | ||
| B3GNT5 | miR1365-5p | MIR44352HG | Liver cancer [21] | |
| B4GALT3 | miR-27 | β1 integrins | Cervical cancer [22] | |
| miR-338-3p | DANCR | Neuroblastoma [23] | ||
| B4GALT5 | miR-491-5p | circ_0009910 | miR-491-5p | Acute myeloid leukemia [24] |
| C1GALT1 | miR-181d-5p | RAC1 | Lung cancer [25] | |
| miR-1-3p | circ HP1BP3 | Bladder cancer [26] | ||
| miR-124-3p | Aging colon [27] | |||
| COSMC | miR-374b | IgA nephropathy [28] | ||
| DPAGT1 | miR-485-5p | LINC00467 | Esophageal cancer [29] | |
| FUT4 | miR-200c | Colorectal cancer [30] | ||
| miR-26a/b | MALAT1 | PI3/AKT | Colorectal cancer [31,32] | |
| miR-224-3p | Breast cancer [33] | |||
| miR-493 | GAS6-AS2 | Breast cancer [34,35] | ||
| miR-200c | Breast cancer [36] | |||
| miR-493-5p | Breast cancer [34] | |||
| miR-200b | Breast cancer [37] | |||
| miR-371b-5p | AC114812.8 | Bladder cancer [38] | ||
| miR-29b | Sp1 | CD44 | Leukemia stem cells [39] | |
| HOXB-AS1 | Multiple myeloma [40] | |||
| miR-199b-5p | Medulloblastoma [41] | |||
| miR26a/b | NFkB | Osteoarthritis [42] | ||
| miR200b | Arthritis [43] | |||
| miR200c | Uterine receptivity [44] | |||
| FUT5 | miR-125a-3p | PI3K/AKT | Colorectal cancer [45] | |
| FUT6 | miR-125a-3p | PI3K/AKT | Colorectal cancer [45] | |
| miR-326 | HOTAIR | CD44/PI3K/AKT | Colorectal cancer [46] | |
| miR-106b | Breast cancer [47] | |||
| FUT8 | miR-34a, miR-122 | Liver cancer [48] | ||
| HOTAIR | JAK/STAT3 | Liver cancer [49] | ||
| miR-198-5p | Lung cancer [50] | |||
| miR-186 | SNHG1 | MMP2/MMP9 | Oral cancer [51] | |
| miR-34c-5p | Renal interstitial fibrosis [52] | |||
| GALNT1 | miR-129 | Bladder cancer [53] | ||
| let-7i-5p | Kidney fibrosis [54] | |||
| GALNT10 | mir-505 | DLGAP1-AS2 | Cholangiocarcinoma [55] | |
| miR-122 | Liver cancer [56] | |||
| GALNT14 | miR-125a | MMP2 and MMP9 | Ovarian cancer [57] | |
| GALNT3 | miR-378a-3p | PSMA3-AS1 | PI3K/Akt | Ovarian cancer [58] |
| GALNT4 | miR-506-3p | Lung cancer [59] | ||
| miR-365b | Lung cancer [60] | |||
| miR-9 | Liver cancer [61] | |||
| GALNT7 | miR-30b/30d | Melanoma [62] | ||
| miR-34a | SNHG7 | PI3K/Akt/mTOR | Colorectal cancer [63] | |
| miR-30e | Cervical cancer [64] | |||
| miR-214 | Cervical cancer [65] | |||
| miR-34a/c | Laryngeal cancer [66] | |||
| miR-214 | Esophageal cancer [67] | |||
| miR-17-5p/miR-17-3p | Liver cancer [68] | |||
| miR-30c | PI3K/AKT | Natural killer activity [69] in lung cancer | ||
| miR-378 | Osteoblast differentiation [70] | |||
| GCNT2 | miR-199a/b-5p | Colorectal cancer [71] | ||
| GCNT3 | miR-15b | Pancreatic and colorectal cancer [72] | ||
| miR-BART1-5p | EBV-induced gastric cancer [73] | |||
| miR-195-5p | LINC00511 | Lung cancer [74] | ||
| LGALS3 | miR-424-3p | Ovarian cancer [75] | ||
| miR-128 | Colorectal cancer [76] | |||
| miR-128-3p | Pancreatic cancer [77] | |||
| miR-299-5p | circRERE | Apoptosis of nucleus polposum cells [78] | ||
| LGALS9 | miR-455-5p | Colorectal cancer [79] | ||
| miR-22 | Liver cancer [80] | |||
| MGAT1 | LINC00173 | mucin 3A | Wilms’ tumor [81] | |
| MGAT3 | miR-23b | Tau protein | Alzheimer’s disease [82] | |
| MGAT4A | miR-424 | cyclin D1 | Breast cancer [83] | |
| MGAT5 | miR-124 | Breast cancer [84] | ||
| OGT | miR-485-5p | Esophageal cancer [85] | ||
| miR-15a/miR-26a | Kidney cancer [86] | |||
| miR-424-5p | XIST | Raf1 | Liver cancer [87] | |
| miR-122 | RYR1 | Breast cancer [88] | ||
| miR-15b | RORγt | Th17 differentiation [89] | ||
| miR-423-5p | Apoptosis of cardiomyocytes [90] | |||
| SIGLEC15 | miR-7109-3p | LINC00973 | Kidney cancer [91] | |
| miR-582-5p | TUG1 | Liver cancer [92] | ||
| ST3GAL1 | MEG3 | EGFR/PI3K/AKT | Kidney cancer [93] | |
| ST3GAL2 | Gut infection [94] | |||
| ST3GAL4 | miR-193a-3p miR-224 | PI3K/AKT | Kidney cancer [95] | |
| miR-224, let-7i | Chronic myeloid leukemia [96] | |||
| miR-370 | Colorectal cancer [97] | |||
| miR193-b | CD44/NF-kB | Arthritis [98] | ||
| ST3GAL5 | miR-26a, miR-548l, miR-34a | Liver cancer [99] | ||
| ST3GAL6 | miR-26a | AKT/mTOR | Liver cancer [100] | |
| ST3GAL6-AS1 | EGFR | Lung cancer [101] | ||
| ST3GAL6-AS1 | MLL1 | ST3GAL6 | Colorectal cancer [102] | |
| ST3GAL6-AS1 | HNRNPA2B1 | ST3GAL6 | Multiple myeloma [103] | |
| ST6GAL1 | miR-9 | β1-integrins/FAK | Liver cancer [104] | |
| miR-195-3p | TINCR | NFkB | Liver cancer [105] | |
| miR-214-3p | Breast cancer [106] | |||
| miR-150 | ZF-AS1 | EGFR/PI3K/Akt | Acute lymphoblastic leukemia [107] | |
| miR-199a | ErbB2/ErbB3 | Various cancers [108] | ||
| ST6GALNAC2 | miR-182/miR-135b | PI3K/AKT | Colorectal cancer [109,110] | |
| ST6GALNAC4 | miR-4299 | Thyroid cancer [111] | ||
| ST6GALNAC5 | miR-182 | Prostate cancer [112] | ||
| ST8SIA1 | miR-33a/let-7e | Colorectal cancer [113] | ||
| MIR44352HG | FAK/AKT/β-catenin | Prostate cancer [114] | ||
| ST8SIA2 | miR-3072-3p | TUG1 | Brain ischemia [115] | |
| ST8SIA4 | miR-26a/b | MALAT1 | Breast cancer [116,117] | |
| miR-146a/b | PI3K-AKT-mTOR | Thyroid cancer [118] | ||
| miR-144-5p/miR-451a | Cholangiocarcinoma [119] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duca, M.; Malagolini, N.; Dall’Olio, F. The Mutual Relationship between Glycosylation and Non-Coding RNAs in Cancer and Other Physio-Pathological Conditions. Int. J. Mol. Sci. 2022, 23, 15804. https://doi.org/10.3390/ijms232415804
Duca M, Malagolini N, Dall’Olio F. The Mutual Relationship between Glycosylation and Non-Coding RNAs in Cancer and Other Physio-Pathological Conditions. International Journal of Molecular Sciences. 2022; 23(24):15804. https://doi.org/10.3390/ijms232415804
Chicago/Turabian StyleDuca, Martina, Nadia Malagolini, and Fabio Dall’Olio. 2022. "The Mutual Relationship between Glycosylation and Non-Coding RNAs in Cancer and Other Physio-Pathological Conditions" International Journal of Molecular Sciences 23, no. 24: 15804. https://doi.org/10.3390/ijms232415804
APA StyleDuca, M., Malagolini, N., & Dall’Olio, F. (2022). The Mutual Relationship between Glycosylation and Non-Coding RNAs in Cancer and Other Physio-Pathological Conditions. International Journal of Molecular Sciences, 23(24), 15804. https://doi.org/10.3390/ijms232415804









