The New General Biological Property of Stem-like Tumor Cells (Part II: Surface Molecules, Which Belongs to Distinctive Groups with Particular Functions, Form a Unique Pattern Characteristic of a Certain Type of Tumor Stem-like Cells)
Abstract
1. Introduction
Historical Background and the General Logic of the Study
2. Results
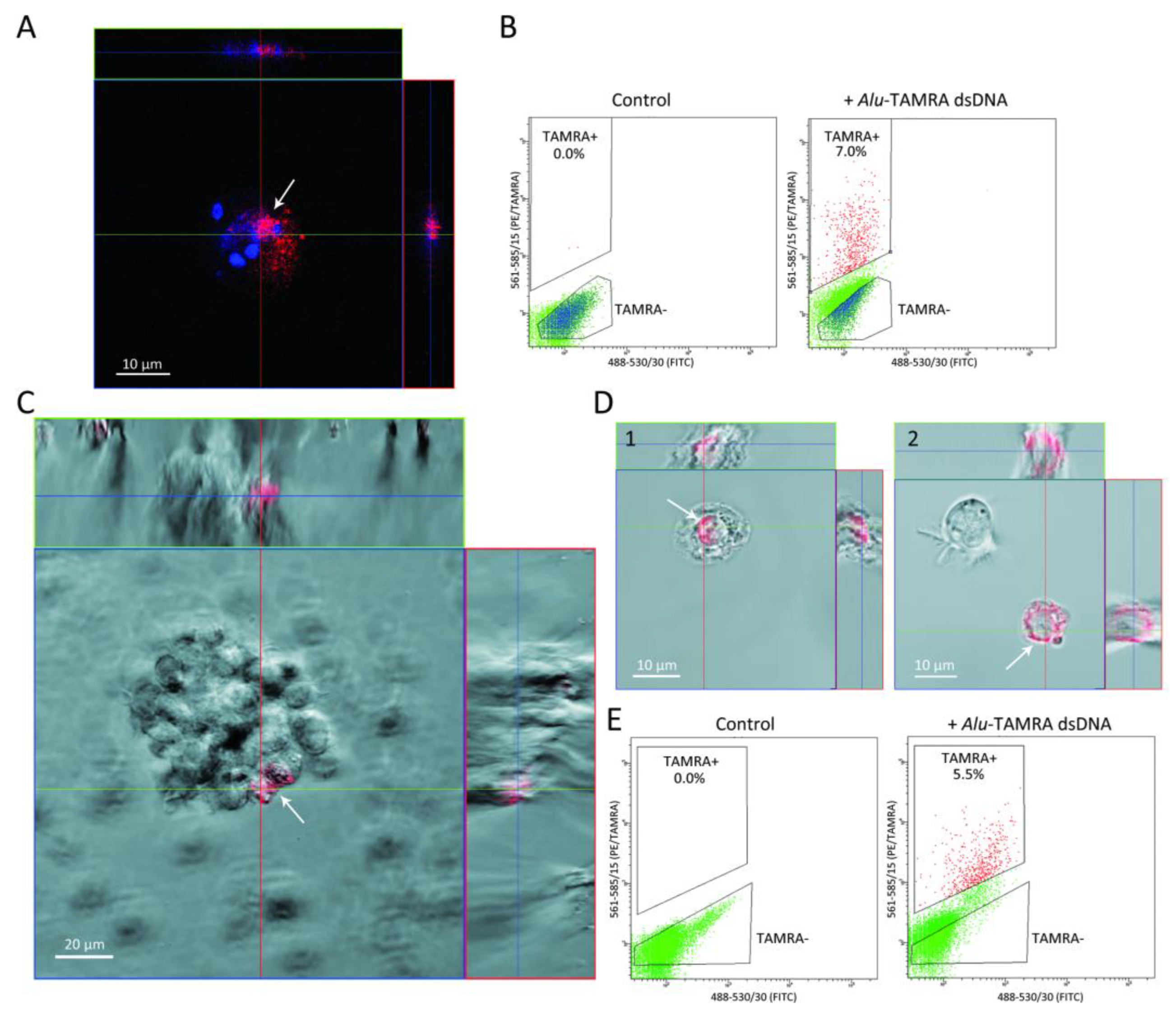
2.1. Searching for the Putative Cell Surface Proteins Determining the Interaction of the TAMRA-Labeled dsDNA Probe with Krebs-2 and HH47 Tumor Stem-like Cells
2.1.1. Search-Based Analysis of the Bioinformatic and Published Data on Functions of Putative Surface Proteins That Are Present on Krebs-2 and HH47 Cells and Are Responsible for Binding and Internalizing the dsDNA Probe
| Functional Group (Binding Site) | Amino Acid Sequence | Brief Description | Reference |
|---|---|---|---|
| SRs (collagen-like domain) | GRGNPGAPGKPGRSGSPGPKGQKGEKGSV | Heparin-binding sites are shown in blue | [25] |
| C1q protein (C1q-like domain) | VFTVTRQTHQPPAONSLIRFNAVLTNPQGDYDTSTGKFTCKVPGLYYFVYHASHTANLCVLLYRSGVKVVTFCGHTSKTNQVNSGGVLLRLQVGEEVWLAV | Heparin-binding sites are shown in blue; the DNA-binding site is shown in red | [26] |
| PGs/GPs and GPI-APs (cluster of positively charged amino acids) | IFLLVTLVTVCACWKPSKRKQKKLEKQNSLEYMDQNDD | The cluster of positively charged amino acids is shown in green | [20] |
2.1.2. Identification of Group-Specific Genes for PGs, SRs, and GPI-APs Overexpressed in TAMRA+ Krebs-2 and HH47 Cells
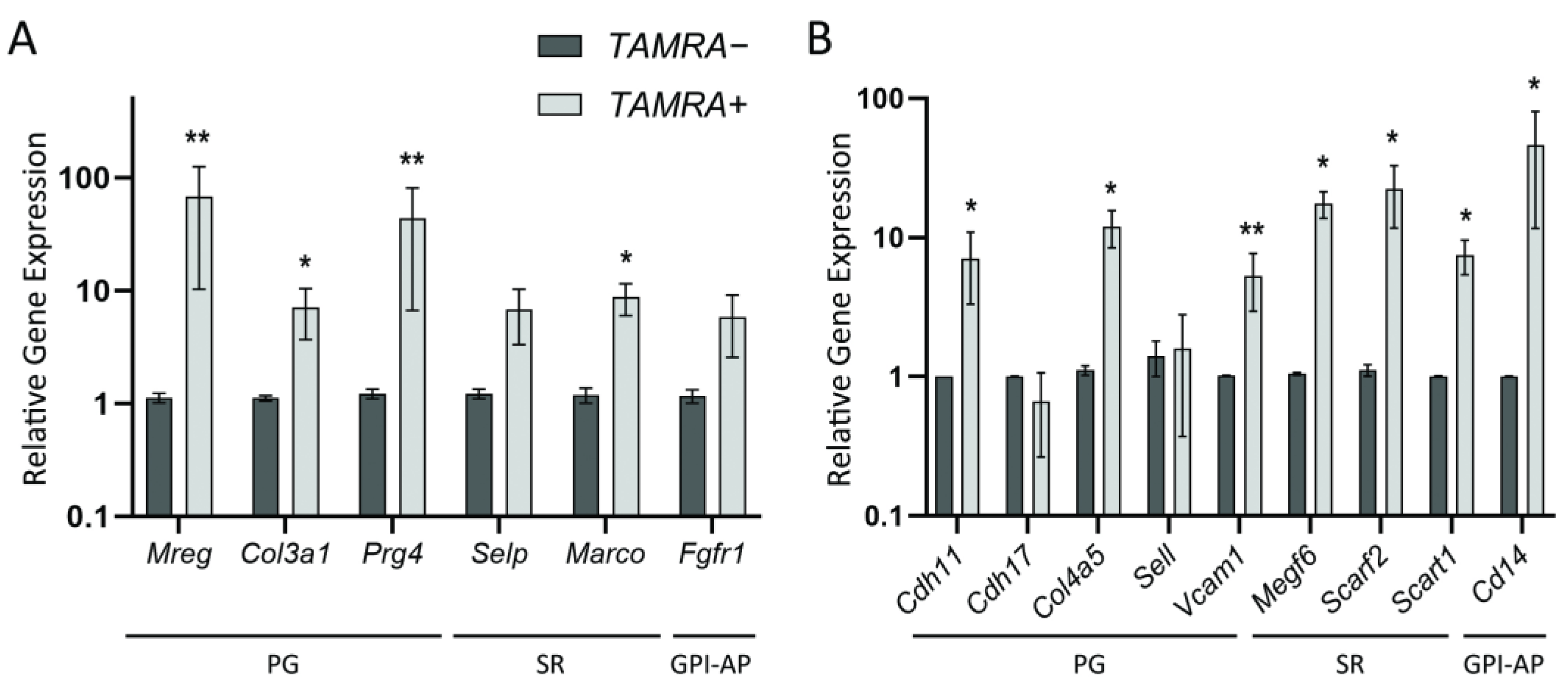
2.2. Quantitative Comparison of the Expression Levels of Several Group-Specific Factors by Real-Time PCR
3. Discussion
- What can provide the general positive charge of tumor stem-like cells? and
- What inherent functions of tumor stem-like cells are ensured by such an exceedingly plastic mechanism?
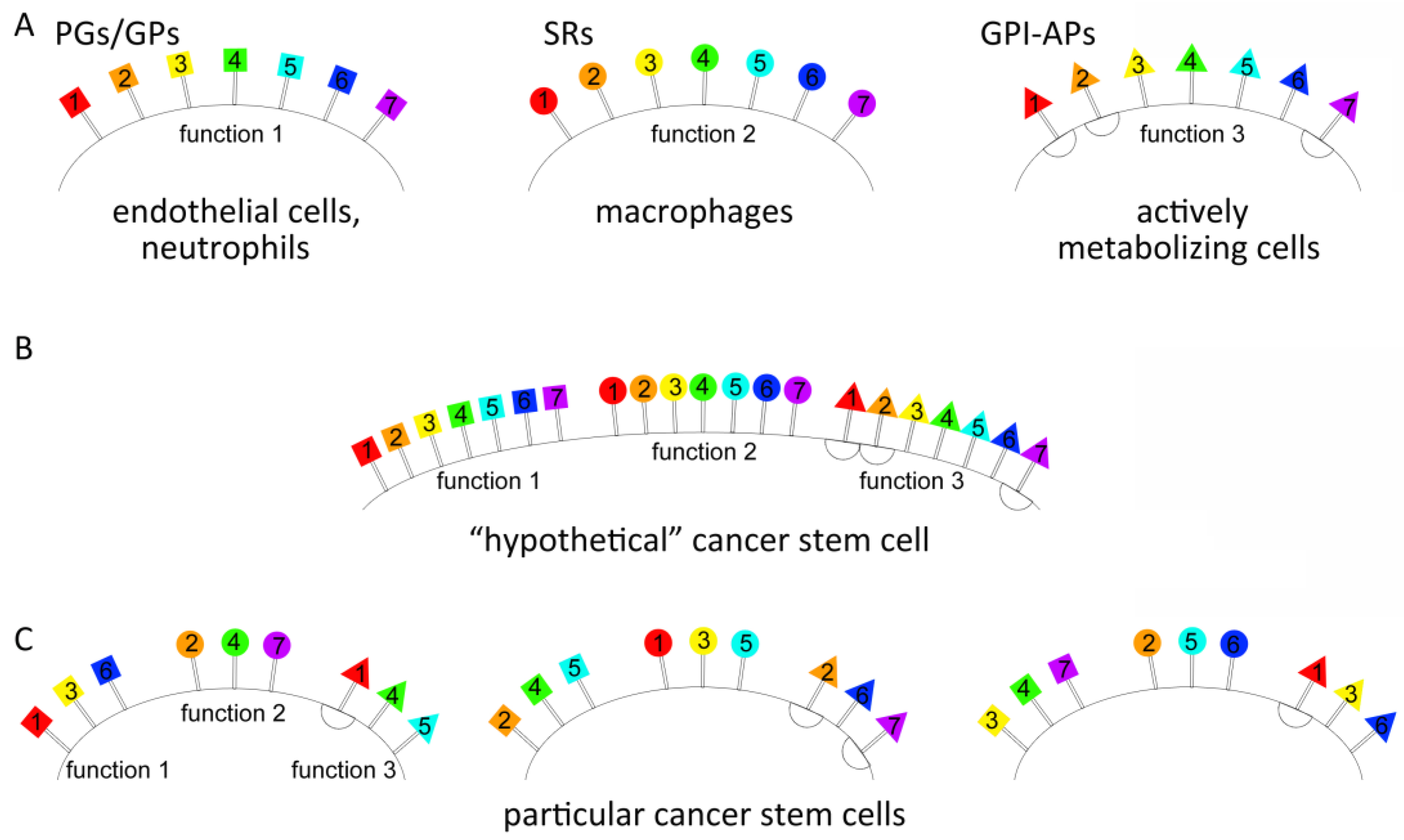
3.1. What Can Provide the General Positive Charge of Tumor Stem-like Cells?
3.2. What Inherent Functions of Tumor Stem-like Cells Are Ensured by Such an Exceedingly Plastic Mechanism?
3.2.1. Proteoglycans/Glycoproteins
3.2.2. Scavenger Receptors
3.2.3. Glycosylphosphatidylinositol-Anchored Proteins
3.2.4. Caveolae-Dependent Transport Is Typical of Cells with Intensive Metabolism
4. Materials and Methods
4.1. Experimental Design and Study Logic
4.2. Tumor Model
4.3. Isolation of HH47-Internalizing, TAMRA-Labeled dsDNA
4.4. RNA Isolation and RNA-seq
4.5. Transcriptome Analysis
4.6. Real-Time PCR and Primer Selection
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ritter, G.S.; Dolgova, E.V.; Petrova, D.D.; Efremov, Y.R.; Proskurina, A.S.; Potter, E.A.; Ruzanova, V.S.; Kirikovich, S.S.; Levites, E.V.; Taranov, O.S.; et al. The New General Biological Property of Stem-like Tumor Cells. Part I. Peculiarities of the Process of Double-Stranded DNA Fragments Internalization into Stem-like Tumor Cells. Front. Genet. 2022, 13, 954395. [Google Scholar] [CrossRef] [PubMed]
- Dolgova, E.V.; Alyamkina, E.A.; Efremov, Y.R.; Nikolin, V.P.; Popova, N.A.; Tyrinova, T.V.; Kozel, A.V.; Minkevich, A.M.; Andrushkevich, O.M.; Zavyalov, E.L.; et al. Identification of Cancer Stem Cells and a Strategy for Their Elimination. Cancer Biol. Ther. 2014, 15, 1378–1394. [Google Scholar] [CrossRef] [PubMed]
- Dolgova, E.V.; Efremov, Y.R.; Orishchenko, K.E.; Andrushkevich, O.M.; Alyamkina, E.A.; Proskurina, A.S.; Bayborodin, S.I.; Nikolin, V.P.; Popova, N.A.; Chernykh, E.R.; et al. Delivery and Processing of Exogenous Double-Stranded DNA in Mouse CD34 + Hematopoietic Progenitor Cells and Their Cell Cycle Changes upon Combined Treatment with Cyclophosphamide and Double-Stranded DNA. Gene 2013, 528, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Dolgova, E.V.; Nikolin, V.P.; Popova, N.A.; Proskurina, A.S.; Orishenko, K.E.; Alyamkina, E.A.; Efremov, Y.R.; Chernykh, E.R.; Ostanin, A.A.; Malkova, E.M.; et al. Internalization of Exogenous DNA into Internal Compartments of Murine Bone Marrow Cells. Russ. J. Genet. Appl. Res. 2012, 2, 440–452. [Google Scholar] [CrossRef]
- Potter, E.A.; Dolgova, E.V.; Proskurina, A.S.; Zavyalov, E.L.; Taranov, O.S.; Baiborodin, S.I.; Alexander, A. Gene Expression Profiling of Tumor-Initiating Stem Cells from Mouse Krebs-2 Carcinoma Using a Novel Marker of Poorly Differentiated Cells. Oncotarget 2017, 8, 9425–9441. [Google Scholar] [CrossRef][Green Version]
- Dolgova, E.V.; Shevela, E.Y.; Tyrinova, T.V.; Minkevich, A.M.; Proskurina, A.S.; Potter, E.A.; Orishchenko, K.E.; Zavjalov, E.L.; Bayborodin, S.I.; Nikolin, V.P.; et al. Nonadherent Spheres With Multiple Myeloma Surface Markers Contain Cells That Contribute to Sphere Formation and Are Capable of Internalizing Extracellular Double-Stranded DNA. Clin. Lymphoma Myeloma Leuk. 2016, 16, 563–576. [Google Scholar] [CrossRef]
- Dolgova, E.V.; Petrova, D.D.; Proskurina, A.S.; Ritter, G.S.; Kisaretova, P.E.; Potter, E.A.; Efremov, Y.R.; Bayborodin, S.I.; Karamysheva, T.V.; Romanenko, M.V.; et al. Identification of the Xenograft and Its Ascendant Sphere-Forming Cell Line as Belonging to EBV-Induced Lymphoma, and Characterization of the Status of Sphere-Forming Cells. Cancer Cell Int. 2019, 19, 120. [Google Scholar] [CrossRef]
- Proskurina, A.S.; Kupina, V.V.; Efremov, Y.R.; Dolgova, E.V.; Ruzanova, V.S.; Ritter, G.S.; Potter, E.A.; Kirikovich, S.S.; Levites, E.V.; Ostanin, A.A.; et al. Karanahan: A Potential New Treatment Option for Human Breast Cancer and Its Validation in a Clinical Setting. Breast Cancer Basic Clin. Res. 2022, 16. [Google Scholar] [CrossRef]
- Kisaretova, P.E.; Kirikovich, S.S.; Ritter, G.S.; Efremov, Y.R.; Taranov, O.S.; Dubatolova, T.D.; Proskurina, A.S.; Potter, E.A.; Dolgova, E.V.; Sidorov, S.V.; et al. Approbation of the Cancer Treatment Approach Based on the Eradication of TAMRA+ Cancer Stem Cells in a Model of Murine Cyclophosphamide Resistant Lymphosarcoma. Anticancer Res. 2020, 40, 795–805. [Google Scholar] [CrossRef]
- Ruzanova, V.; Proskurina, A.; Efremov, Y.; Kirikovich, S.; Ritter, G.; Levites, E.; Dolgova, E.; Potter, E.; Babaeva, O.; Sidorov, S.; et al. Chronometric Administration of Cyclophosphamide and a Double-Stranded DNA-Mix at Interstrand Crosslinks Repair Timing, Called “Karanahan” Therapy, Is Highly Efficient in a Weakly Immunogenic Lewis Carcinoma Model. Pathol. Oncol. Res. 2022, 28, 1610180. [Google Scholar] [CrossRef]
- Potter, E.A.; Dolgova, E.V.; Proskurina, A.S.; Minkevich, A.M.; Efremov, Y.R.; Taranov, O.S.; Omigov, V.V.; Nikolin, V.P.; Popova, N.A.; Bayborodin, S.I.; et al. A Strategy to Eradicate Well-Developed Krebs-2 Ascites in Mice. Oncotarget 2016, 7, 11580–11594. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ruzanova, V.S.; Proskurina, A.S.; Ritter, G.S.; Efremov, Y.R.; Nikolin, V.P.; POPOVA, N.A.; Naprimerov, V.A.; Dolgova, E.V.; POTTER, E.A.; KIRIKOVICH, S.S.; et al. Experimental Comparison of the In Vivo Efficacy of Two Novel Anticancer Therapies. Anticancer Res. 2021, 41, 3371–3387. [Google Scholar] [CrossRef] [PubMed]
- Dolgova, E.V.; Andrushkevich, O.M.; Kisaretova, P.E.; Proskurina, A.S.; Ritter, G.S.; Dubatolova, T.D.; Romanenko, M.V.; Taranov, O.S.; Efremov, Y.R.; Zavyalov, E.L.; et al. Efficacy of the New Therapeutic Approach in Curing Malignant Neoplasms on the Model of Human Glioblastoma. Cancer Biol. Med. 2021, 18, 910–930. [Google Scholar] [CrossRef]
- Dolgova, E.V.; Potter, E.A.; Proskurina, A.S.; Minkevich, A.M.; Chernych, E.R.; Ostanin, A.A.; Efremov, Y.R.; Bayborodin, S.I.; Nikolin, V.P.; Popova, N.A.; et al. Properties of Internalization Factors Contributing to the Uptake of Extracellular DNA into Tumor-Initiating Stem Cells of Mouse Krebs-2 Cell Line. Stem Cell Res. Ther. 2016, 7, 76. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Outschoorn, U.E.; Sotgia, F.; Lisanti, M.P. Caveolae and Signalling in Cancer. Nat. Rev. Cancer 2015, 15, 225–237. [Google Scholar] [CrossRef] [PubMed]
- Dyer, A.P.; Banfield, B.W.; Martindale, D.; Spannier, D.M.; Tufaro, F. Dextran Sulfate Can Act as an Artificial Receptor to Mediate a Type-Specific Herpes Simplex Virus Infection via Glycoprotein B. J. Virol. 1997, 71, 191–198. [Google Scholar] [CrossRef]
- Narazaki, M.; Segarra, M.; Tosato, G. Sulfated Polysaccharides Identified as Inducers of Neuropilin-1 Internalization and Functional Inhibition of VEGF 165 and Semaphorin3A. Blood 2008, 111, 4126–4136. [Google Scholar] [CrossRef]
- Zhu, X.D.; Zhuang, Y.; Ben, J.J.; Qian, L.L.; Huang, H.P.; Bai, H.; Sha, J.H.; He, Z.G.; Chen, Q. Caveolae-Dependent Endocytosis Is Required for Class a Macrophage Scavenger Receptor-Mediated Apoptosis in Macrophages. J. Biol. Chem. 2011, 286, 8231–8239. [Google Scholar] [CrossRef]
- Sarrazin, S.; Lamanna, W.C.; Esko, J.D. Heparan Sulfate Proteoglycans. Cold Spring Harb. Perspect. Biol. 2011, 3, a004952. [Google Scholar] [CrossRef]
- Ori, A.; Wilkinson, M.C.; Fernig, D.G. The Heparanome and Regulation of Cell Function: Structures, Functions and Challenges. Front. Biosci. 2008, 13, 4309–4338. [Google Scholar] [CrossRef]
- Ori, A.; Free, P.; Wilkinson, M.C.; Fernig, D.G. Identification of Heparin-Binding Sites in Proteins by Selective Labeling *. Mol. Cell. Proteom. 2009, 8, 2256–2265. [Google Scholar] [CrossRef] [PubMed]
- Gómez Toledo, A.; Sorrentino, J.T.; Sandoval, D.R.; Malmström, J.; Lewis, N.E.; Esko, J.D. A Systems View of the Heparan Sulfate Interactome. J. Histochem. Cytochem. 2021, 69, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Sathyapriya, R.; Vishveshwara, S. Interaction of DNA with Clusters of Amino Acids in Proteins. Nucleic Acids Res. 2004, 32, 4109–4118. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jureti, D.; Zoranic, L.; Zucic, D. Basic Charge Clusters and Predictions of Membrane Protein Topology. J. Chem. Inf. Comput. Sci 2002, 42, 620–632. [Google Scholar] [CrossRef] [PubMed]
- Doi, T.; Higashino, K.I.; Kurihara, Y.; Wada, Y.; Miyazaki, T.; Nakamura, H.; Uesugi, S.; Imanishi, T.; Kawabe, Y.; Itakura, H.; et al. Charged Collagen Structure Mediates the Recognition of Negatively Charged Macromolecules by Macrophage Scavenger Receptors. J. Biol. Chem. 1993, 268, 2126–2133. [Google Scholar] [CrossRef] [PubMed]
- Garlatti, V.; Chouquet, A.; Lunardi, T.; Vivès, R.; Païdassi, H.; Lortat-Jacob, H.; Thielens, N.M.; Arlaud, G.J.; Gaboriaud, C. Cutting Edge: C1q Binds Deoxyribose and Heparan Sulfate through Neighboring Sites of Its Recognition Domain. J. Immunol. 2010, 185, 808–812. [Google Scholar] [CrossRef]
- Davies, N.A.; Watkeys, L.; Butcher, L.; Potter, S.; Hughes, M.G.; Moir, H.; Morris, K.; Thomas, A.W.; Webb, R. The Contributions of Oxidative Stress, Oxidised Lipoproteins and AMPK towards Exercise-Associated PPARγ Signalling within Human Monocytic Cells. Free Radic. Res. 2015, 49, 45–56. [Google Scholar] [CrossRef]
- Kinoshita, T. Biosynthesis and Biology of Mammalian GPI-Anchored Proteins. Open Biol. 2020, 10, 190290. [Google Scholar] [CrossRef]
- Hussein, N.H.; Amin, N.S.; El Tayebi, H.M. GPI-AP: Unraveling a New Class of Malignancy Mediators and Potential Immunotherapy Targets. Front. Oncol. 2020, 10, 537311. [Google Scholar] [CrossRef]
- Sharonov, G.V.; Balatskaya, M.N.; Tkachuk, V.A. Glycosylphosphatidylinositol-Anchored Proteins as Regulators of Cortical Cytoskeleton. Biochemistry 2016, 81, 636–650. [Google Scholar] [CrossRef]
- Ikezawa, H. Glycosylphosphatidylinositol (GPI)-Anchored Proteins. Biol. Pharm. Bull. 2002, 25, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Le, W.; Chen, B.; Cui, Z.; Liu, Z.; Shi, D. Detection of Cancer Cells Based on Glycolytic-Regulated Surface Electrical Charges. Biophys. Rep. 2019, 5, 10–18. [Google Scholar] [CrossRef]
- Li, Z.; Gao, Y.; Li, W.; Li, Y.; Lv, H.; Zhang, D.; Peng, J.; Cheng, W.; Mei, L.; Chen, H.; et al. Charge-Reversal Nanomedicines as a Smart Bullet for Deep Tumor Penetration. Smart Mater. Med. 2022, 3, 243–253. [Google Scholar] [CrossRef]
- Zhang, P.; Chen, D.; Li, L.; Sun, K. Charge Reversal Nano-Systems for Tumor Therapy. J. Nanobiotechnol. 2022, 20, 31. [Google Scholar] [CrossRef]
- Peixoto, A.; Relvas-Santos, M.; Azevedo, R.; Lara Santos, L.; Ferreira, J.A. Protein Glycosylation and Tumor Microenvironment Alterations Driving Cancer Hallmarks. Front. Oncol. 2019, 9, 380. [Google Scholar] [CrossRef]
- De Pasquale, V.; Pavone, L.M. Heparan Sulfate Proteoglycan Signaling in Tumor Microenvironment. Int. J. Mol. Sci. 2020, 21, 6588. [Google Scholar] [CrossRef]
- Furini, G.; Verderio, E. Spotlight on the Transglutaminase 2-Heparan Sulfate Interaction. Med. Sci. 2019, 7, 5. [Google Scholar] [CrossRef]
- Singh, P.; Carraher, C.; Schwarzbauer, J.E. Assembly of Fibronectin Extracellular Matrix. Annu. Rev. Cell Dev. Biol. 2010, 26, 397–419. [Google Scholar] [CrossRef]
- Cheng, J.P.X.; Nichols, B.J. Caveolae: One Function or Many? Trends Cell Biol. 2016, 26, 177–189. [Google Scholar] [CrossRef]
- Ahrens, T.D.; Bang-Christensen, S.R.; Jørgensen, A.M.; Løppke, C.; Spliid, C.B.; Sand, N.T.; Clausen, T.M.; Salanti, A.; Agerbæk, M.O. The Role of Proteoglycans in Cancer Metastasis and Circulating Tumor Cell Analysis. Front. Cell Dev. Biol. 2020, 8, 749. [Google Scholar] [CrossRef]
- Iannuzzi, C.; Irace, G.; Sirangelo, I. The Effect of Glycosaminoglycans (GAGs) on Amyloid Aggregation and Toxicity. Molecules 2015, 20, 2510–2528. [Google Scholar] [CrossRef] [PubMed]
- Maksimenko, A.V.; Turashev, A.D. Endothelial Glycocalyx of Blood Circulation System. I. Detection, Components, and Structural Organization. Russ. J. Bioorganic Chem. 2014, 40, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Elfenbein, A.; Simons, M. Auxiliary and Autonomous Proteoglycan Signaling Networks. In Methods in Enzymology; Academic Press Inc.: Cambridge, MA, USA, 2010; Volume 480, pp. 3–31. [Google Scholar]
- Pries, A.R.; Secomb, T.W.; Gaehtgens, P. The Endothelial Surface Layer. Pflug. Arch. Eur. J. Physiol. 2000, 440, 653–666. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Le, W.; Wang, Y.; Li, Z.; Wang, D.; Ren, L.; Lin, L.; Cui, S.; Hu, J.J.; Hu, Y.; et al. Targeting Negative Surface Charges of Cancer Cells by Multifunctional Nanoprobes. Theranostics 2016, 6, 1887–1898. [Google Scholar] [CrossRef]
- Adamson, R.; Clough, G. Plasma Proteins Modify the Endothelial Cell Glycocalyx of Frog Mesenteric Microvessels. J. Physiol. 1992, 445, 473–486. [Google Scholar] [CrossRef]
- Yarilin, A.A. Role of Adhesion Molecules in the Pathogenesis of Rheumatoid Arthritis. Rheumatol. Sci. Pract. 2000, 38, 61–68. [Google Scholar] [CrossRef]
- Sokologorskiy, S.V. Glycocalyx—Birth of a New Clinical Paradigm. Russ. J. Anaesthesiol. Reanimatol. 2018, 4, 22–29. [Google Scholar] [CrossRef]
- Chen, Y.; Guan, Q.; Han, X.; Bai, D.; Li, D.; Tian, Y. Proteoglycans in the Periodontium: A Review with Emphasis on Specific Distributions, Functions, and Potential Applications. J. Periodontal Res. 2021, 56, 617–632. [Google Scholar] [CrossRef]
- Taylor, K.R.; Gallo, R.L. Glycosaminoglycans and Their Proteoglycans: Host-associated Molecular Patterns for Initiation and Modulation of Inflammation. FASEB J. 2006, 20, 9–22. [Google Scholar] [CrossRef]
- Gandhi, N.S.; Mancera, R.L. The Structure of Glycosaminoglycans and Their Interactions with Proteins. Chem. Biol. Drug Des. 2008, 72, 455–482. [Google Scholar] [CrossRef]
- Heinegård, D. Fell-Muir Lecture: Proteoglycans and More—From Molecules to Biology. Int. J. Exp. Pathol. 2009, 90, 575–586. [Google Scholar] [CrossRef] [PubMed]
- Iozzo, R.V.; Schaefer, L. Proteoglycan Form and Function: A Comprehensive Nomenclature of Proteoglycans. Matrix Biol. 2015, 42, 11–55. [Google Scholar] [CrossRef] [PubMed]
- Dane, M.; van den Berg, B.; Lee, D.; Boels, M.; Tiemeier, G.; Avramut, M.; van Zonneveld, A.; van der Vlag, J.; Vink, H.; Rabelink, T. A Microscopic View on the Renal Endothelial Glycocalyx. Am. J. Physiol. Ren. Physiol. 2015, 308, F956–F966. [Google Scholar] [CrossRef] [PubMed]
- Esko, J.; Kimata, K.; Lindahl, U. Proteoglycans and Sulfated Glycosaminoglycans. In Essentials of Glycobiology, 2nd ed.; NCBI Bookshelf: Montreal, QC, Canada, 2009; pp. 1–14. [Google Scholar]
- Tanaka, K.A.; Key, N.S.; Levy, J.H. Blood Coagulation: Hemostasis and Thrombin Regulation. Anesth. Analg. 2009, 108, 1433–1446. [Google Scholar] [CrossRef]
- Levi, M.; Van Der Poll, T.; Büller, H.R. Bidirectional Relation between Inflammation and Coagulation. Circulation 2004, 109, 2698–2704. [Google Scholar] [CrossRef]
- Mallard, B.W.; Tiralongo, J. Cancer Stem Cell Marker Glycosylation: Nature, Function and Significance. Glycoconj. J. 2017, 34, 441–452. [Google Scholar] [CrossRef]
- Kang, H.; Wu, Q.; Sun, A.; Liu, X.; Fan, Y.; Deng, X. Cancer Cell Glycocalyx and Its Significance in Cancer Progression. Int. J. Mol. Sci. 2018, 19, 2484. [Google Scholar] [CrossRef]
- Cherfils-Vicini, J.; Iltis, C.; Cervera, L.; Pisano, S.; Croce, O.; Sadouni, N.; Győrffy, B.; Collet, R.; Renault, V.M.; Rey-Millet, M.; et al. Cancer Cells Induce Immune Escape via Glycocalyx Changes Controlled by the Telomeric Protein TRF2. EMBO J. 2019, 38, e100012. [Google Scholar] [CrossRef]
- Girigoswami, K.; Saini, D.; Girigoswami, A. Extracellular Matrix Remodeling and Development of Cancer. Stem Cell Rev. Rep. 2021, 17, 739–747. [Google Scholar] [CrossRef]
- Canton, J.; Neculai, D.; Grinstein, S. Scavenger Receptors in Homeostasis and Immunity. Nat. Rev. Immunol. 2013, 13, 621–634. [Google Scholar] [CrossRef]
- PrabhuDas, M.R.; Baldwin, C.L.; Bollyky, P.L.; Bowdish, D.M.E.; Drickamer, K.; Febbraio, M.; Herz, J.; Kobzik, L.; Krieger, M.; Loike, J.; et al. A Consensus Definitive Classification of Scavenger Receptors and Their Roles in Health and Disease. J. Immunol. 2017, 198, 3775–3789. [Google Scholar] [CrossRef] [PubMed]
- Zani, I.; Stephen, S.; Mughal, N.; Russell, D.; Homer-Vanniasinkam, S.; Wheatcroft, S.; Ponnambalam, S. Scavenger Receptor Structure and Function in Health and Disease. Cells 2015, 4, 178–201. [Google Scholar] [CrossRef] [PubMed]
- Gusev, E.Y.; Zotova, N.V.; Zhuravleva, Y.A.; Chereshnev, V.A. Physiological and Pathogenic Role of Scavenger Receptors in Humans. Med. Immunol. 2020, 22, 7–48. [Google Scholar] [CrossRef]
- Alquraini, A.; El Khoury, J. Scavenger Receptors. Curr. Biol. 2020, 30, R790–R795. [Google Scholar] [CrossRef] [PubMed]
- Todt, J.C.; Hu, B.; Curtis, J.L. The Scavenger Receptor SR-A I/II (CD204) Signals via the Receptor Tyrosine Kinase Mertk during Apoptotic Cell Uptake by Murine Macrophages. J. Leukoc. Biol. 2008, 84, 510–518. [Google Scholar] [CrossRef]
- Vasquez, M.; Simões, I.; Consuegra-Fernández, M.; Aranda, F.; Lozano, F.; Berraondo, P. Exploiting Scavenger Receptors in Cancer Immunotherapy: Lessons from CD5 and SR-B1. Eur. J. Immunol. 2017, 47, 1108–1118. [Google Scholar] [CrossRef]
- Pandey, M.S.; Baggenstoss, B.A.; Washburn, J.; Harris, E.N.; Weigel, P.H. The Hyaluronan Receptor for Endocytosis (HARE) Activates NF-ΚB-Mediated Gene Expression in Response to 40-400-KDa, but Not Smaller or Larger, Hyaluronans. J. Biol. Chem. 2013, 288, 14068–14079. [Google Scholar] [CrossRef]
- Khaidakov, M.; Mitra, S.; Kang, B.Y.; Wang, X.; Kadlubar, S.; Novelli, G.; Raj, V.; Winters, M.; Carter, W.C.; Mehta, J.L. Oxidized LDL Receptor 1 (OLR1) as a Possible Link between Obesity, Dyslipidemia and Cancer. PLoS ONE 2011, 6, e20277. [Google Scholar] [CrossRef]
- Zurzolo, C.; Simons, K. Glycosylphosphatidylinositol-Anchored Proteins: Membrane Organization and Transport. Biochim. Biophys. Acta 2016, 1858, 632–639. [Google Scholar] [CrossRef]
- Fujihara, Y.; Ikawa, M. GPI-AP Release in Cellular, Developmental, and Reproductive Biology. J. Lipid Res. 2016, 57, 538–545. [Google Scholar] [CrossRef]
- Lakhan, S.E.; Sabharanjak, S.; De, A. Endocytosis of Glycosylphosphatidylinositol-Anchored Proteins. J. Biomed. Sci. 2009, 16, 93. [Google Scholar] [CrossRef] [PubMed]
- Echarri, A.; Pavón, D.; Sánchez, S.; García-García, M.; Calvo, E.; Huerta-López, C.; Velázquez-Carreras, D.; Viaris de Lesegno, C.; Ariotti, N.; Lázaro-Carrillo, A.; et al. An Abl-FBP17 Mechanosensing System Couples Local Plasma Membrane Curvature and Stress Fiber Remodeling during Mechanoadaptation. Nat. Commun. 2019, 10, 5828. [Google Scholar] [CrossRef] [PubMed]
- Golani, G.; Ariotti, N.; Parton, R.; Kozlov, M. Membrane Curvature and Tension Control the Formation and Collapse of Caveolar Superstructures. Dev. Cell 2019, 48, 523–538.e4. [Google Scholar] [CrossRef]
- Hubert, M.; Larsson, E.; Lundmark, R. Keeping in Touch with the Membrane; Protein- And Lipid-Mediated Confinement of Caveolae to the Cell Surface. Biochem. Soc. Trans. 2020, 48, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Cameron, P.; Ruffin, J.; Bollag, R.; Rasmussen, H.; Cameron, R. Identification of Caveolin and Caveolin-Related Proteins in the Brain. J. Neurosci. 1997, 17, 9520–9535. [Google Scholar] [CrossRef] [PubMed]
- Parton, R. Caveolae--from Ultrastructure to Molecular Mechanisms. Nat. Rev. Mol. Cell Biol. 2003, 4, 162–167. [Google Scholar] [CrossRef]
- Parton, R.; del Pozo, M. Caveolae as Plasma Membrane Sensors, Protectors and Organizers. Nat. Rev. Mol. Cell Biol. 2013, 14, 98–112. [Google Scholar] [CrossRef] [PubMed]
- Parton, R.; Tillu, V.; Collins, B. Caveolae. Curr. Biol. 2018, 28, R402–R405. [Google Scholar] [CrossRef]
- Yan, S.; Wang, Y.; Zhang, Y.; Wang, L.; Zhao, X.; Du, C.; Gao, P.; Yan, F.; Liu, F.; Gong, X.; et al. Synaptotagmin-11 Regulates the Functions of Caveolae and Responds to Mechanical Stimuli in Astrocytes. FASEB J. 2020, 34, 2609–2624. [Google Scholar] [CrossRef]
- Low, J.Y.; Laiho, M. Caveolae-Associated Molecules, Tumor Stroma, and Cancer Drug Resistance: Current Findings and Future Perspectives. Cancers 2022, 14, 589. [Google Scholar] [CrossRef]
- Parton, R.G.; Kozlov, M.M.; Ariotti, N. Caveolae and Lipid Sorting: Shaping the Cellular Response to Stress. J. Cell Biol. 2020, 219, e201905071. [Google Scholar] [CrossRef] [PubMed]
- Graf, G.; Connell, P.; van der Westhuyzen, D.; Smart, E. The Class B, Type I Scavenger Receptor Promotes the Selective Uptake of High Density Lipoprotein Cholesterol Ethers into Caveolae. J. Biol. Chem. 1999, 274, 12043–12048. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Ariotti, N.; Rae, J.; Liang, H.; Tillu, V.; Tee, S.; Bastiani, M.; Bademosi, A.T.; Collins, B.M.; Meunier, F.A.; et al. Dissecting the Nanoscale Lipid Profile of Caveolae. bioRxiv 2020. [Google Scholar] [CrossRef]
- Hubert, M.; Larsson, E.; Vegesna, N.; Ahnlund, M.; Johansson, A.; Moodie, L.; Lundmark, R. Lipid Accumulation Controls the Balance between Surface Connection and Scission of Caveolae. Elife 2020, 9, e55038. [Google Scholar] [CrossRef]
- Pilch, P.; Liu, L. Fat Caves: Caveolae, Lipid Trafficking and Lipid Metabolism in Adipocytes. Trends Endocrinol. Metab. 2011, 22, 318–324. [Google Scholar] [CrossRef]
- Pilch, P.; Meshulam, T.; Ding, S.; Liu, L. Caveolae and Lipid Trafficking in Adipocytes. Clin. Lipidol. 2011, 6, 49–58. [Google Scholar] [CrossRef][Green Version]
- Frank, P.; Pavlides, S.; Lisanti, M. Caveolae and Transcytosis in Endothelial Cells: Role in Atherosclerosis. Cell Tissue Res. 2009, 335, 41–47. [Google Scholar] [CrossRef]
- Potter, E.A.; Ritter, G.S.; Dolgova, E.V.; Proskurina, A.S.; Kisaretova, P.E.; Efremov, Y.R.; Nikolin, V.P.; Popova, N.A.; Taranov, O.S.; Ostanin, A.A.; et al. Evaluating the efficiency of the tumor-initiating stem cells eradication strategy on the example of ascite form of mouse gepatocarcinoma G-29. Oncol. Issue 2018, 64, 818–829. (In Russian) [Google Scholar]
| Groups of the Membrane-Anchored Proteins with the Same Particular Function | “Hypothetical” Tumor Stem-like Cell Based on the Published Data [20,27,28,29,30,31] | Krebs-2 | HH47 |
|---|---|---|---|
| PGs/GPs | Adipoq Caprin2 Cdh13 Clec4 Col12A1 Fcn2 Gldn Gpc3 Hip Itgam Ncam Otol1 Sell Selplg | Apoc1 Catsperg2 Cd200 Col3a1 Col6a2 Cldn1 Dsc2 Dnaja4 Fcna Adgrg3 Hepacam Itm2a Itga9 Itln1 Jsrp1 Lyve1 Mreg Nectin1 Ncam Psd2 Pgr Selplg Rab15 Tnfrsf13c | Acvr1 Cdh11 Cdh17 Chodl Col4a5 Col4a6 Col7a1 Cthrc1 Fcrl3 Gpc 1 Gpc2 Igf1r Itga10 Itga5 Kcnip3 Kel Ncam Sell Podxl Ptprm Tenm4 Tspan15 Treml4 Vcam1 |
| SRs | Marco Megf Scara Scarb | Marco | Megf6 Scarf2 Scart1 |
| GPI-APs | Aamp Ache Car4 Cd14 Cd24a Cd48 Cd55 Cd59a Cdh1 Ceacam5 Cntfr Cpm Fgfr Folr1 Gpc3 Gpihbp1 H2-Q7 Ly6a Lypd1 Mdp1 Msln Nrp1 Parp1 Plaur Robo Thy1 | Cd55 Fgfr1 | Cd14 Gdpd5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petrova, D.D.; Dolgova, E.V.; Proskurina, A.S.; Ritter, G.S.; Ruzanova, V.S.; Efremov, Y.R.; Potter, E.A.; Kirikovich, S.S.; Levites, E.V.; Taranov, O.S.; et al. The New General Biological Property of Stem-like Tumor Cells (Part II: Surface Molecules, Which Belongs to Distinctive Groups with Particular Functions, Form a Unique Pattern Characteristic of a Certain Type of Tumor Stem-like Cells). Int. J. Mol. Sci. 2022, 23, 15800. https://doi.org/10.3390/ijms232415800
Petrova DD, Dolgova EV, Proskurina AS, Ritter GS, Ruzanova VS, Efremov YR, Potter EA, Kirikovich SS, Levites EV, Taranov OS, et al. The New General Biological Property of Stem-like Tumor Cells (Part II: Surface Molecules, Which Belongs to Distinctive Groups with Particular Functions, Form a Unique Pattern Characteristic of a Certain Type of Tumor Stem-like Cells). International Journal of Molecular Sciences. 2022; 23(24):15800. https://doi.org/10.3390/ijms232415800
Chicago/Turabian StylePetrova, Daria D., Evgeniya V. Dolgova, Anastasia S. Proskurina, Genrikh S. Ritter, Vera S. Ruzanova, Yaroslav R. Efremov, Ekaterina A. Potter, Svetlana S. Kirikovich, Evgeniy V. Levites, Oleg S. Taranov, and et al. 2022. "The New General Biological Property of Stem-like Tumor Cells (Part II: Surface Molecules, Which Belongs to Distinctive Groups with Particular Functions, Form a Unique Pattern Characteristic of a Certain Type of Tumor Stem-like Cells)" International Journal of Molecular Sciences 23, no. 24: 15800. https://doi.org/10.3390/ijms232415800
APA StylePetrova, D. D., Dolgova, E. V., Proskurina, A. S., Ritter, G. S., Ruzanova, V. S., Efremov, Y. R., Potter, E. A., Kirikovich, S. S., Levites, E. V., Taranov, O. S., Ostanin, A. A., Chernykh, E. R., Kolchanov, N. A., & Bogachev, S. S. (2022). The New General Biological Property of Stem-like Tumor Cells (Part II: Surface Molecules, Which Belongs to Distinctive Groups with Particular Functions, Form a Unique Pattern Characteristic of a Certain Type of Tumor Stem-like Cells). International Journal of Molecular Sciences, 23(24), 15800. https://doi.org/10.3390/ijms232415800








