MiR-19 Family Impairs Adipogenesis by the Downregulation of the PPARγ Transcriptional Network
Abstract
1. Introduction
2. Results
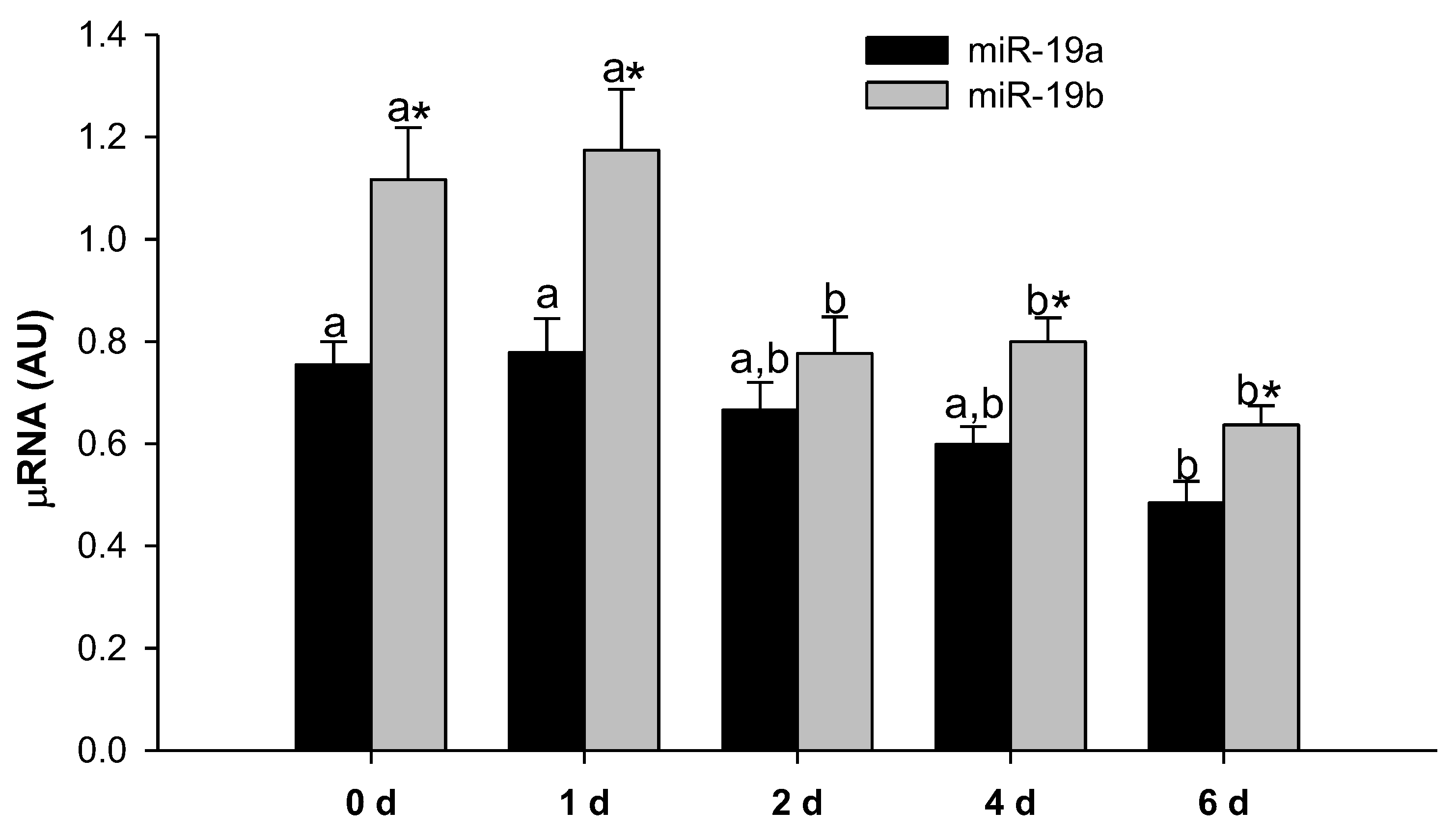
2.1. MiR-19a and miR-19b Expression Levels Decrease during 3T3-L1 Adipocyte Differentiation
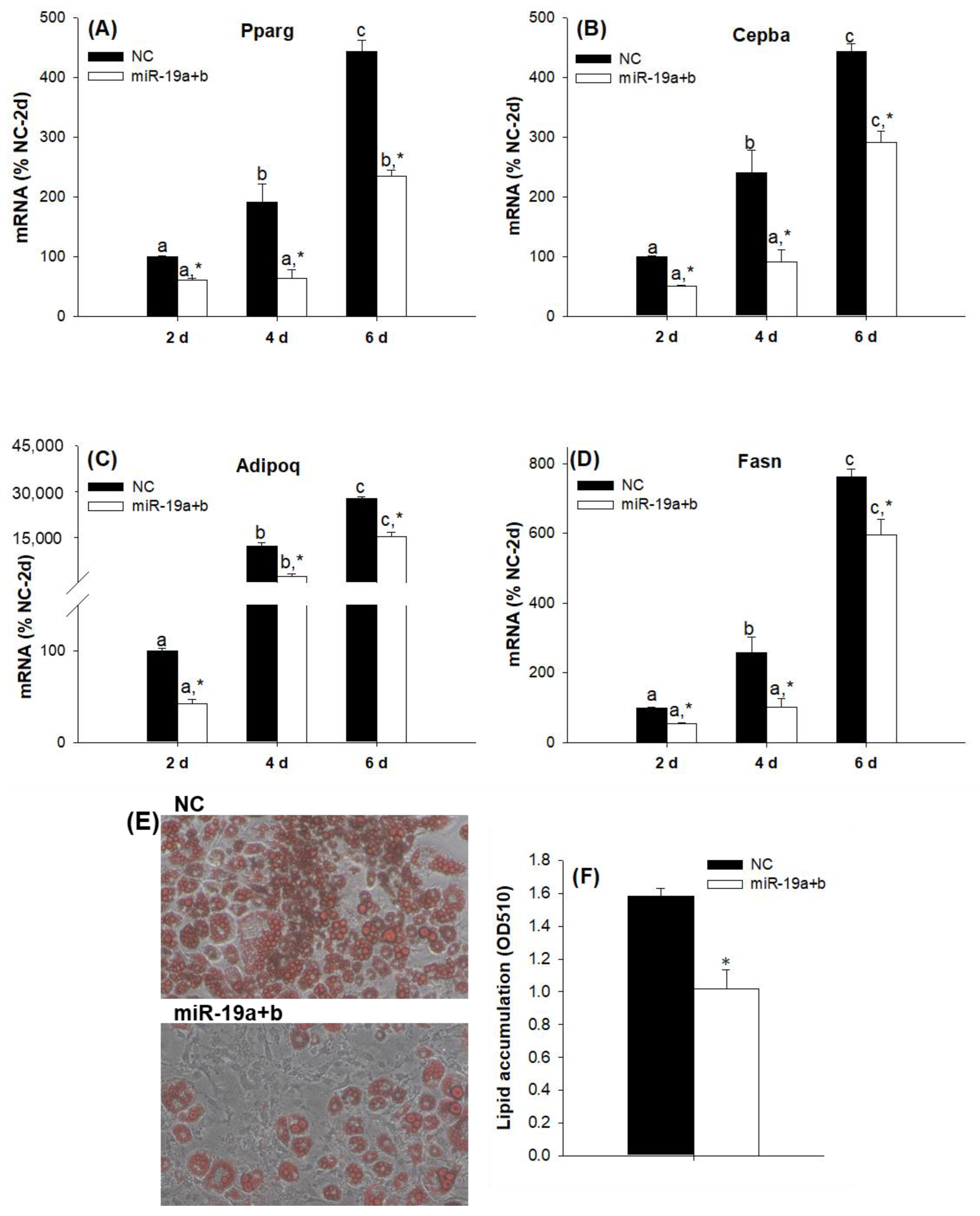
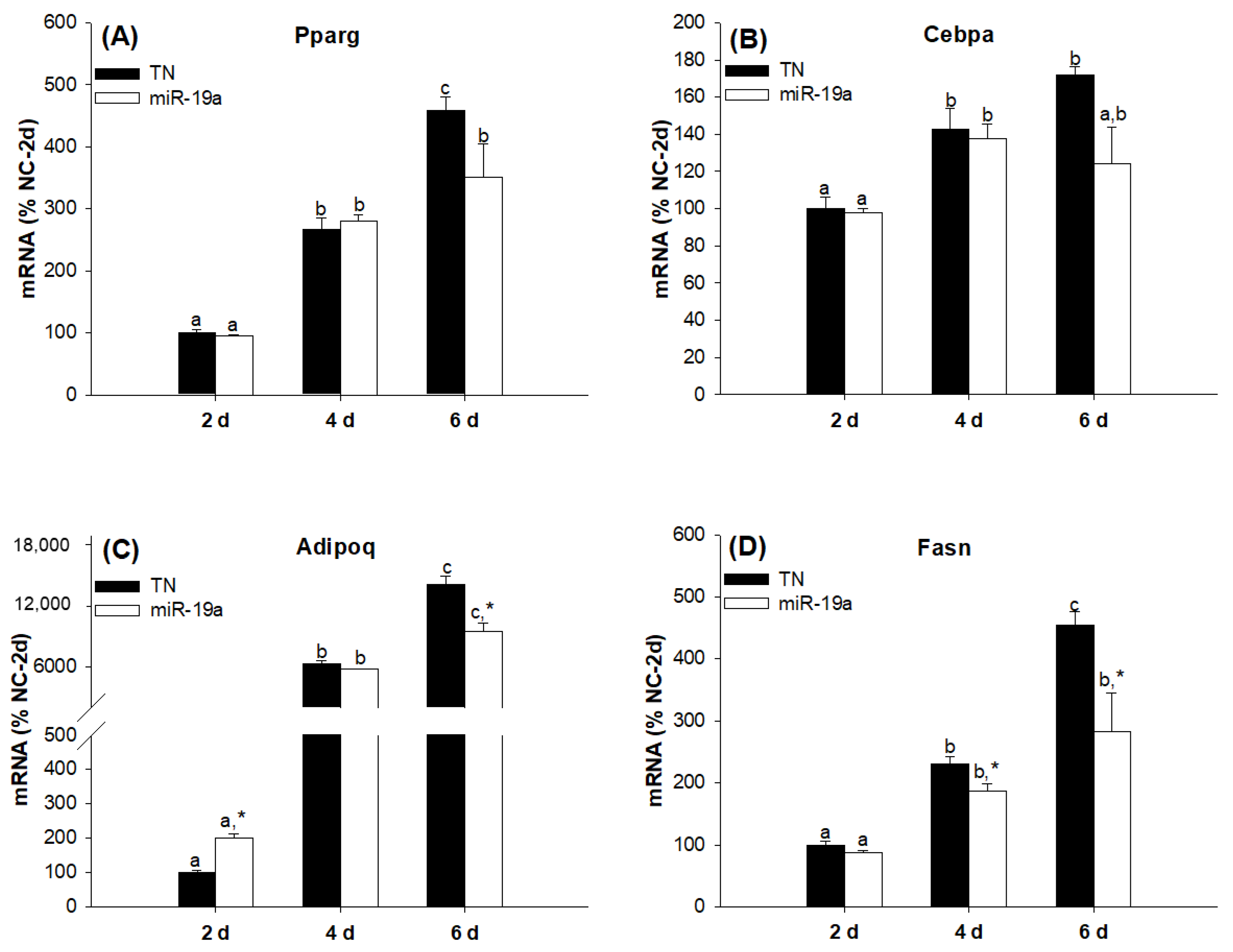
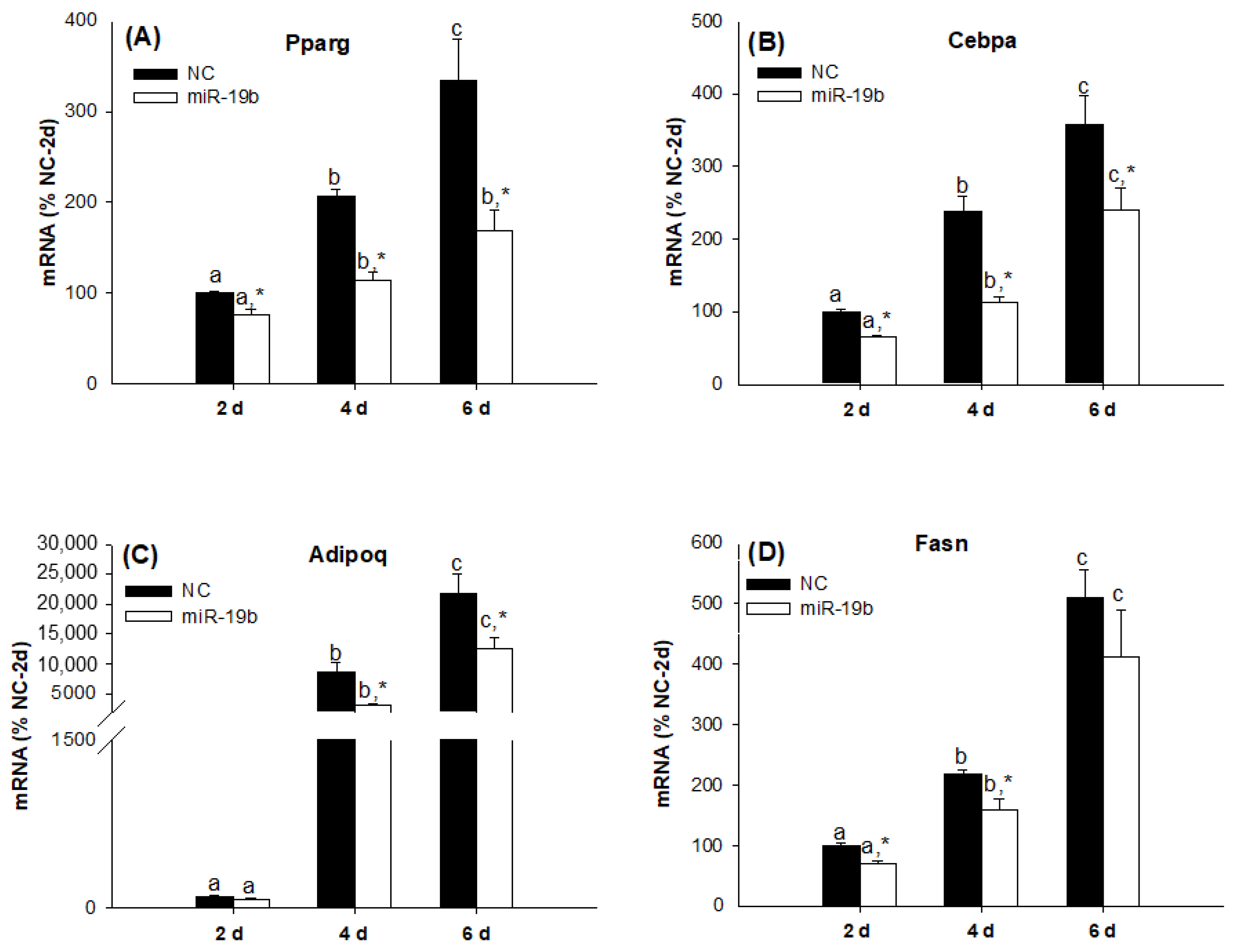
2.2. Overexpression of the miR-19 Family Inhibits 3T3-L1 Cell Differentiation
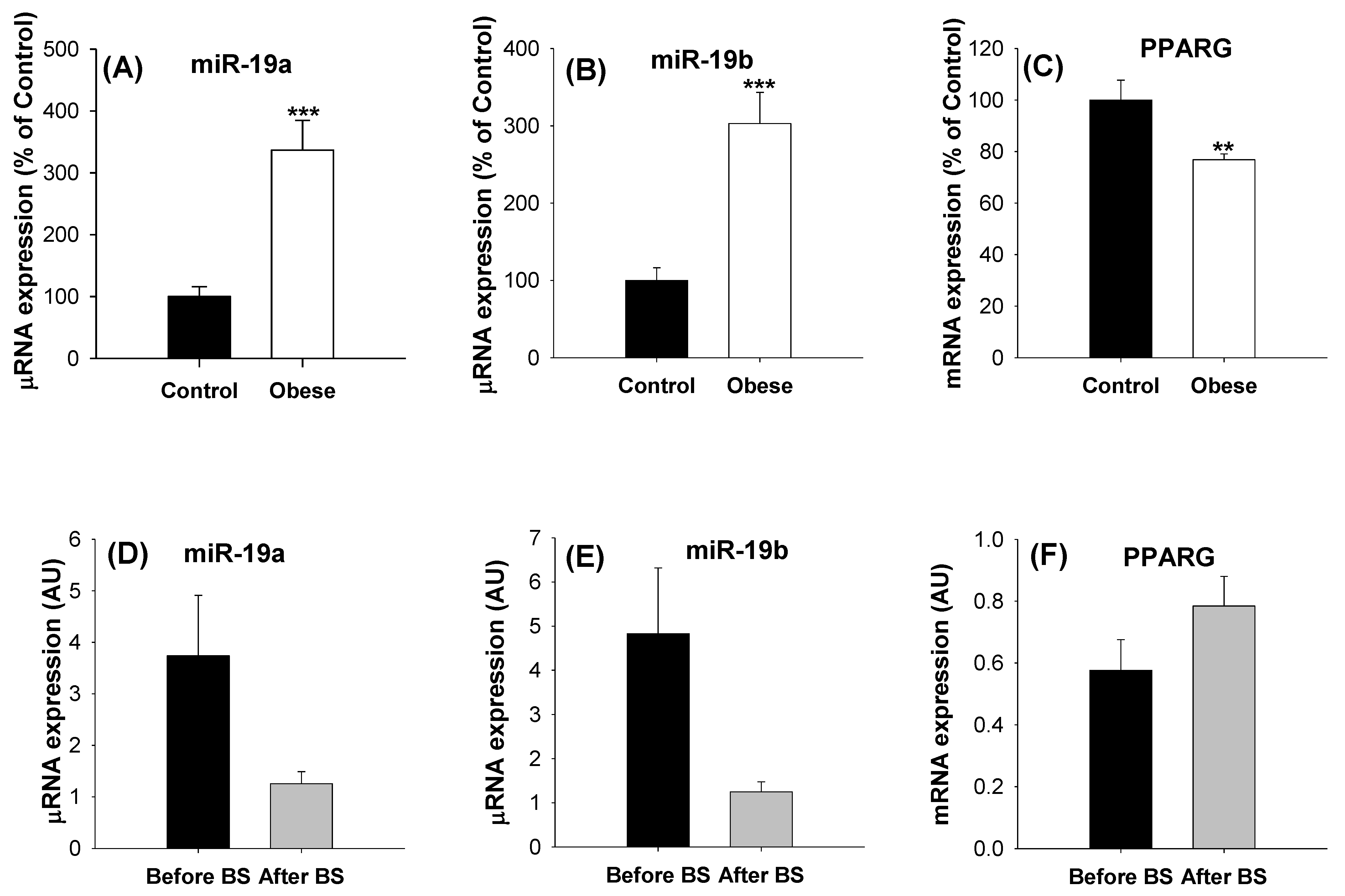
2.3. Levels of miR19s in aSAT Are Modulated by Body Weight
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. 3T3-L1 Transfection
4.3. 3T3-L1 Differentiation
4.4. Patients and Adipose Tissue Sample Collection
4.5. RNA Extraction and Rea-Time (RT) Quantitative (q)PCR
4.6. Oil Red O Staining and Quantification
4.7. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Manrique-Acevedo, C.; Chinnakotla, B.; Padilla, J.; Martinez-Lemus, L.A.; Gozal, D. Obesity and cardiovascular disease in women. Int. J. Obes. 2020, 44, 1210–1226. [Google Scholar] [CrossRef] [PubMed]
- Baron, M.; Maillet, J.; Huyvaert, M.; Dechaume, A.; Boutry, R.; Loiselle, H.; Durand, E.; Toussaint, B.; Vaillant, E.; Philippe, J.; et al. Loss-of-function mutations in MRAP2 are pathogenic in hyperphagic obesity with hyperglycemia and hypertension. Nat. Med. 2019, 25, 1733–1738. [Google Scholar] [CrossRef] [PubMed]
- Ormazabal, V.; Nair, S.; Elfeky, O.; Aguayo, C.; Salomon, C.; Zuñiga, F.A. Association between insulin resistance and the development of cardiovascular disease. Cardiovasc. Diabetol. 2018, 17, 122. [Google Scholar] [CrossRef]
- Ambele, M.A.; Dhanraj, P.; Giles, R.; Pepper, M.S. Adipogenesis: A Complex Interplay of Multiple Molecular Determinants and Pathways. Int. J. Mol. Sci. 2020, 21, 4283. [Google Scholar] [CrossRef] [PubMed]
- Vishvanath, L.; Gupta, R.K. Contribution of adipogenesis to healthy adipose tissue expansion in obesity. J. Clin. Investig. 2019, 129, 4022–4031. [Google Scholar] [CrossRef]
- Moseti, D.; Regassa, A.; Kim, W.-K. Molecular Regulation of Adipogenesis and Potential Anti-Adipogenic Bioactive Molecules. Int. J. Mol. Sci. 2016, 17, 124. [Google Scholar] [CrossRef]
- Gharanei, S.; Shabir, K.; Brown, J.; Weickert, M.; Barber, T.; Kyrou, I.; Randeva, H. Regulatory microRNAs in Brown, Brite and White Adipose Tissue. Cells 2020, 9, 2489. [Google Scholar] [CrossRef]
- Zaiou, M.; El Amri, H.; Bakillah, A. The clinical potential of adipogenesis and obesity-related microRNAs. Nutr. Metab. Cardiovasc. Dis. 2018, 28, 91–111. [Google Scholar] [CrossRef]
- Vienberg, S.; Geiger, J.; Madsen, S.; Dalgaard, L.T. MicroRNAs in metabolism. Acta Physiol. 2017, 219, 346–361. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Mogilyansky, E.; Rigoutsos, I. The miR-17/92 cluster: A comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013, 20, 1603–1614. [Google Scholar] [CrossRef]
- Li, X.; Teng, C.; Ma, J.; Fu, N.; Wang, L.; Wen, J.; Wang, T.-Y. miR-19 family: A promising biomarker and therapeutic target in heart, vessels and neurons. Life Sci. 2019, 232, 116651. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Yang, S.; Zhao, Y.; Wu, Q.; Chen, F. Dynamic evolution of mir-17–92 gene cluster and related miRNA gene families in vertebrates. Mol. Biol. Rep. 2013, 40, 3147–3153. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Jia, X.; Zhou, J.; Sun, Q.; Ma, Z. The MiR-17-92 Gene Cluster is a Blood-Based Marker for Cancer Detection in Non-Small-Cell Lung Cancer. Am. J. Med. Sci. 2020, 360, 248–260. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Hua, S.; Zhang, J.; Xu, S. The MicroRNA Family Both in Normal Development and in Different Diseases: The miR-17-92 Cluster. BioMed Res. Int. 2019, 2019, 9450240. [Google Scholar] [CrossRef]
- Wan, S.; Chen, X.; He, Y.; Yu, X. Novel Functions of MicroRNA-17-92 Cluster in the Endocrine System. Curr. Drug Targets 2018, 18, 191–200. [Google Scholar] [CrossRef]
- Wang, Q.; Li, Y.C.; Wang, J.; Kong, J.; Qi, Y.; Quigg, R.J.; Li, X. miR-17-92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130. Proc. Natl. Acad. Sci. USA 2008, 105, 2889–2894. [Google Scholar] [CrossRef]
- Wang, J.; Xu, X.; Li, P.; Zhang, B.; Zhang, J. HDAC3 protects against atherosclerosis through inhibition of inflammation via the mi-croRNA-19b/PPARgamma/NF-kappaB axis. Atherosclerosis 2021, 323, 1–12. [Google Scholar] [CrossRef]
- Vidigal, J.A.; Ventura, A. The biological functions of miRNAs: Lessons from in vivo studies. Trends Cell Biol. 2015, 25, 137–147. [Google Scholar] [CrossRef]
- Massart, J.; Sjögren, R.J.O.; Egan, B.; Garde, C.; Lindgren, M.; Gu, W.; Ferreira, D.M.S.; Katayama, M.; Ruas, J.L.; Barrès, R.; et al. Endurance exercise training-responsive miR-19b-3p improves skeletal muscle glucose metabolism. Nat. Commun. 2021, 12, 5948. [Google Scholar] [CrossRef] [PubMed]
- Dou, L.; Wang, S.; Huang, X.; Sun, X.; Zhang, Y.; Shen, T.; Guo, J.; Man, Y.; Tang, W.; Xuelin, S. MiR-19a mediates gluconeogenesis by targeting PTEN in hepatocytes. Mol. Med. Rep. 2018, 17, 3967–3971. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lin, T.-Y.; Chen, L.; Liu, Y.; Dian, M.-J.; Hao, W.-C.; Lin, X.-L.; Li, X.-Y.; Li, Y.-L.; Lian, M.; et al. miR-19 regulates the expression of interferon-induced genes and MHC class I genes in human cancer cells. Int. J. Med. Sci. 2020, 17, 953–964. [Google Scholar] [CrossRef] [PubMed]
- Gantier, M.P.; Stunden, H.J.; McCoy, C.E.; Behlke, M.A.; Wang, D.; Kaparakis-Liaskos, M.; Sarvestani, S.T.; Yang, Y.H.; Xu, D.; Corr, S.C.; et al. A miR-19 regulon that controls NF-kappaB signaling. Nucleic Acids Res. 2012, 40, 8048–8058. [Google Scholar] [CrossRef] [PubMed]
- Olive, V.; Bennett, M.J.; Walker, J.C.; Ma, C.; Jiang, I.; Cordon-Cardo, C.; Li, Q.J.; Lowe, S.W.; Hannon, G.J.; He, L. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009, 23, 2839–2849. [Google Scholar] [CrossRef]
- Gruszka, R.; Zakrzewska, M. The Oncogenic Relevance of miR-17-92 Cluster and Its Paralogous miR-106b-25 and miR-106a-363 Clusters in Brain Tumors. Int. J. Mol. Sci. 2018, 19, 879. [Google Scholar] [CrossRef]
- Kabekkodu, S.P.; Shukla, V.; Varghese, V.K.; Adiga, D.; Jishnu, P.V.; Chakrabarty, S.; Satyamoorthy, K. Cluster miRNAs and cancer: Diagnostic, prognostic and therapeutic opportunities. Wiley Interdiscip. Rev. RNA 2020, 11, e1563. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, J.; Wu, L.; Hu, X. MicroRNAs of the miR-17~92 family maintain adipose tissue macrophage homeostasis by sustaining IL-10 expression. eLife 2020, 9, e55676. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Lim, L.P.; Lau, N.C.; Garrett-Engele, P.; Grimson, A.; Schelter, J.M.; Castle, J.; Bartel, D.P.; Linsley, P.S.; Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005, 433, 769–773. [Google Scholar] [CrossRef]
- Baek, D.; Villén, J.; Shin, C.; Camargo, F.D.; Gygi, S.P.; Bartel, D.P. The impact of microRNAs on protein output. Nature 2008, 455, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Selbach, M.; Schwanhäusser, B.; Thierfelder, N.; Fang, Z.; Khanin, R.; Rajewsky, N. Widespread changes in protein synthesis induced by microRNAs. Nature 2008, 455, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Clement, T.; Salone, V.; Rederstorff, M. Dual luciferase gene reporter assays to study miRNA function. Methods Mol. Biol. 2015, 1296, 187–198. [Google Scholar]
- Hammarstedt, A.; Gogg, S.; Hedjazifar, S.; Nerstedt, A.; Smith, U. Impaired Adipogenesis and Dysfunctional Adipose Tissue in Human Hypertrophic Obesity. Physiol. Rev. 2018, 98, 1911–1941. [Google Scholar] [CrossRef] [PubMed]
- Koenen, M.; Hill, M.A.; Cohen, P.; Sowers, J.R. Obesity, Adipose Tissue and Vascular Dysfunction. Circ. Res. 2021, 128, 951–968. [Google Scholar] [CrossRef] [PubMed]
- Porro, S.; Genchi, V.A.; Cignarelli, A.; Natalicchio, A.; Laviola, L.; Giorgino, F.; Perrini, S. Dysmetabolic adipose tissue in obesity: Morphological and functional characteristics of adipose stem cells and mature adipocytes in healthy and unhealthy obese subjects. J. Endocrinol. Investig. 2021, 44, 921–941. [Google Scholar] [CrossRef]
- Kwiatkowski, S.; Kajdy, A.; Stefańska, K.; Bednarek-Jędrzejek, M.; Dzidek, S.; Tousty, P.; Sokołowska, M.; Kwiatkowska, E. PPARgamma-A Factor Linking Metabolically Unhealthy Obesity with Placental Pathologies. Int. J. Mol. Sci. 2021, 22, 13167. [Google Scholar] [CrossRef]
- Torres, J.L.; Usategui-Martin, R.; Hernandez-Cosido, L.; Bernardo, E.; Manzanedo-Bueno, L.; Hernandez-Garcia, I.; Mateos-Diaz, A.M.; Rozo, O.; Matesanz, N.; Salete-Granado, D.; et al. PPAR-gamma Gene Expression in Human Adipose Tissue Is Associated with Weight Loss After Sleeve Gastrectomy. J. Gastrointest. Surg. 2022, 26, 286–297. [Google Scholar] [CrossRef]
- Ruschke, K.; Fishbein, L.; Dietrich, A.; Klöting, N.; Tönjes, A.; Oberbach, A.; Fasshauer, M.; Jenkner, J.; Schön, M.R.; Stumvoll, M.; et al. Gene expression of PPARgamma and PGC-1alpha in human omental and subcutaneous adipose tissues is related to insulin resistance markers and mediates beneficial effects of physical training. Eur. J. Endocrinol. 2010, 162, 515–523. [Google Scholar] [CrossRef]
- Zarkesh, M.; Tabaei, K.; Akbarzadeh, M.; Daneshafrooz, A.; Zadeh-Vakili, A. Association of miR-34a and miR-143 levels with PPARgamma gene expression in adipose tissues of non-diabetic adults. J. Physiol. Anthropol. 2022, 41, 13. [Google Scholar] [CrossRef]
- Castellano-Castillo, D.; Denechaud, P.-D.; Fajas, L.; Moreno-Indias, I.; Oliva-Olivera, W.; Tinahones, F.; Queipo-Ortuño, M.I.; Cardona, F. Human adipose tissue H3K4me3 histone mark in adipogenic, lipid metabolism and inflammatory genes is positively associated with BMI and HOMA-IR. PLoS ONE 2019, 14, e0215083. [Google Scholar] [CrossRef]
- Singh, P.B.; Pua, H.H.; Happ, H.C.; Schneider, C.; Von Moltke, J.; Locksley, R.M.; Baumjohann, D.; Ansel, K.M. MicroRNA regulation of type 2 innate lymphoid cell homeostasis and function in allergic inflammation. J. Exp. Med. 2017, 214, 3627–3643. [Google Scholar] [CrossRef]
- Ortega, F.J.; Moreno, M.; Mercader, J.M.; Moreno-Navarrete, J.M.; Fuentes-Batllevell, N.; Sabater, M.; Ricart, W.; Fernández-Real, J.M. Inflammation triggers specific microRNA profiles in human adipocytes and macrophages and in their supernatants. Clin. Epigenetics 2015, 7, 49. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Autieri, M.V.; Scalia, R. Adipose tissue inflammation and metabolic dysfunction in obesity. Am. J. Physiol. Physiol. 2021, 320, C375–C391. [Google Scholar] [CrossRef] [PubMed]
- Varela-Rodriguez, B.M.; Juiz-Valiña, P.; Varela, L.; Outeiriño-Blanco, E.; Bravo, S.B.; García-Brao, M.J.; Mena, E.; Noguera, J.F.; Valero-Gasalla, J.; Cordido, F.; et al. Beneficial Effects of Bariatric Surgery-Induced by Weight Loss on the Proteome of Abdominal Subcu-taneous Adipose Tissue. J. Clin. Med. 2020, 9, 213. [Google Scholar] [CrossRef]
- Unamuno, X.; Gómez-Ambrosi, J.; Rodríguez, A.; Becerril, S.; Frühbeck, G.; Catalán, V. Adipokine dysregulation and adipose tissue inflammation in human obesity. Eur. J. Clin. Investig. 2018, 48, e12997. [Google Scholar] [CrossRef] [PubMed]

| MiR-19 Family Study | PPARG Study | |||
|---|---|---|---|---|
| Control | Obese | Control | Obese | |
| Gender | 17 ♂ and 5 ♀ | 14 ♂ and 28 ♀ | 25 ♂ and 7 ♀ | 25 ♂ and 109 ♀ |
| Age (years) | 47.12 ± 2.26 | 43.49 ± 1.37 | 50.16 ± 2.29 | 46.47 ± 0.84 |
| BMI (kg/m2) | 24.05 ± 0.27 | 48.46 ± 1.23 * | 24.75 ± 0.29 | 49.95 ± 0.74 * |
| Glucose (mg/dL) | 86.64 ± 1.74 | 101.88 ± 4.95 * | 88.81 ± 1.55 | 104.42 ± 2.42 * |
| Diabetic patients (%) | 0% | 40.48% * | 0% | 32.84% * |
| Before Bariatric Surgery | After Bariatric Surgery | |
|---|---|---|
| Gender | 2 ♂ and 4 ♀ | 2 ♂ and 4 ♀ |
| Age (years) | 50.80 ± 5.00 | 53.13 ± 4.98 |
| BMI (kg/m2) | 48.52 ± 3.75 | 29.73 ± 0.98 * |
| Glucose (mg/dL) | 96.83 ± 6.67 | 85.50 ± 2.58 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Juiz-Valiña, P.; Varela-Rodríguez, B.M.; Outeiriño-Blanco, E.; García-Brao, M.J.; Mena, E.; Cordido, F.; Sangiao-Alvarellos, S. MiR-19 Family Impairs Adipogenesis by the Downregulation of the PPARγ Transcriptional Network. Int. J. Mol. Sci. 2022, 23, 15792. https://doi.org/10.3390/ijms232415792
Juiz-Valiña P, Varela-Rodríguez BM, Outeiriño-Blanco E, García-Brao MJ, Mena E, Cordido F, Sangiao-Alvarellos S. MiR-19 Family Impairs Adipogenesis by the Downregulation of the PPARγ Transcriptional Network. International Journal of Molecular Sciences. 2022; 23(24):15792. https://doi.org/10.3390/ijms232415792
Chicago/Turabian StyleJuiz-Valiña, Paula, Bárbara María Varela-Rodríguez, Elena Outeiriño-Blanco, María Jesús García-Brao, Enrique Mena, Fernando Cordido, and Susana Sangiao-Alvarellos. 2022. "MiR-19 Family Impairs Adipogenesis by the Downregulation of the PPARγ Transcriptional Network" International Journal of Molecular Sciences 23, no. 24: 15792. https://doi.org/10.3390/ijms232415792
APA StyleJuiz-Valiña, P., Varela-Rodríguez, B. M., Outeiriño-Blanco, E., García-Brao, M. J., Mena, E., Cordido, F., & Sangiao-Alvarellos, S. (2022). MiR-19 Family Impairs Adipogenesis by the Downregulation of the PPARγ Transcriptional Network. International Journal of Molecular Sciences, 23(24), 15792. https://doi.org/10.3390/ijms232415792









