Structural, Thermodynamic and Enzymatic Characterization of N,N-Diacetylchitobiose Deacetylase from Pyrococcus chitonophagus
Abstract
1. Introduction
2. Results and Discussion
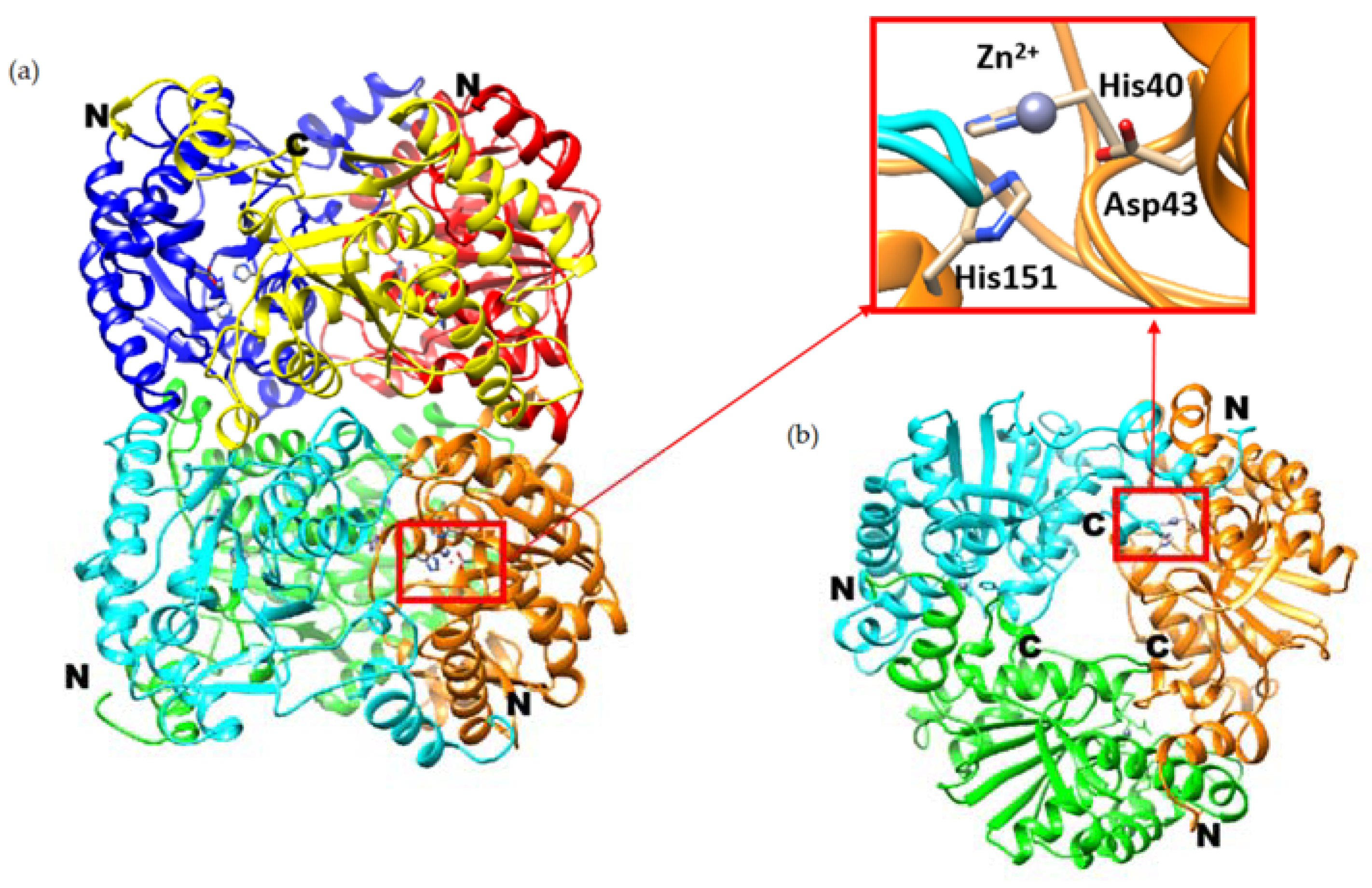
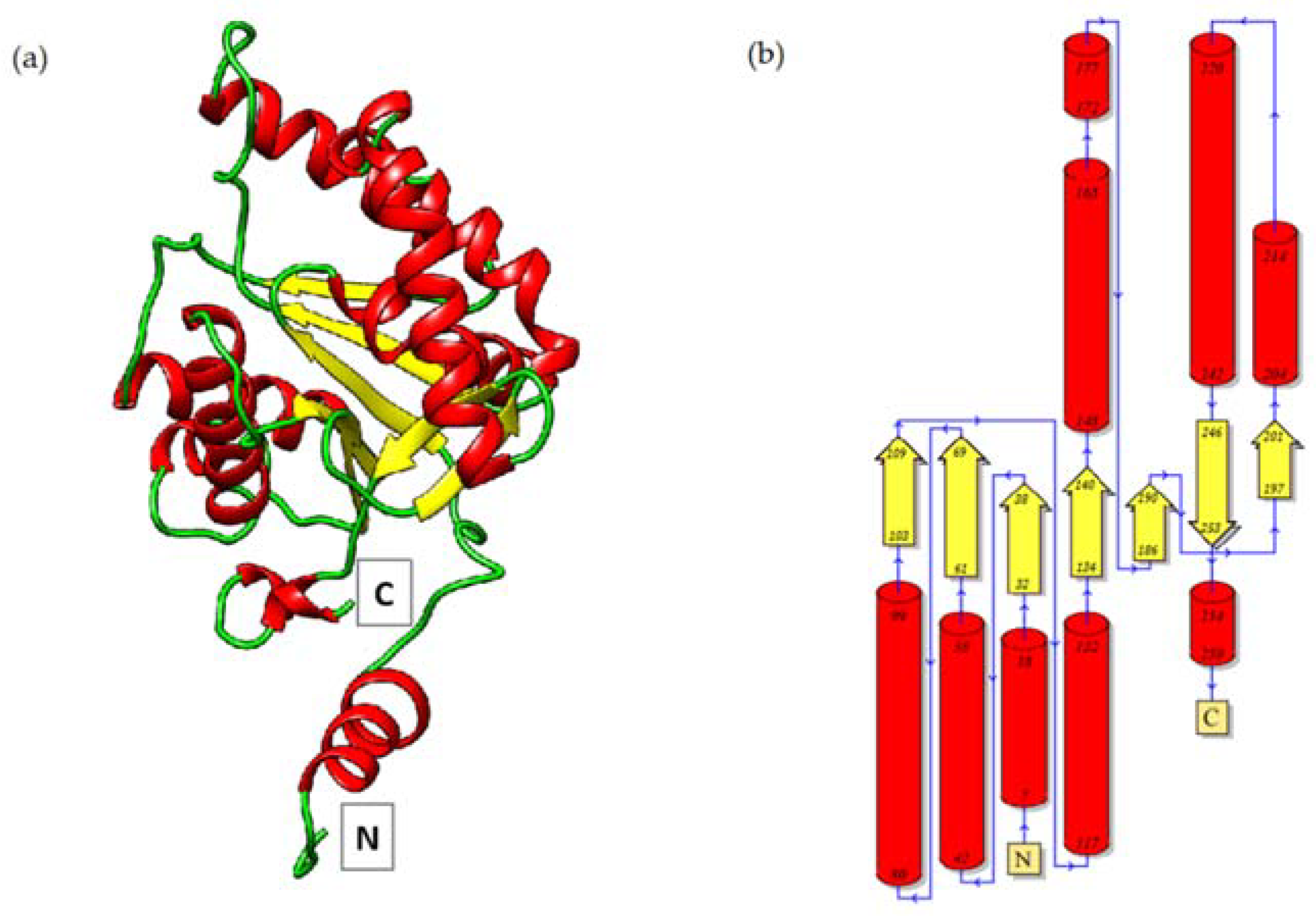
2.1. Oligomeric State and the Overall Structure
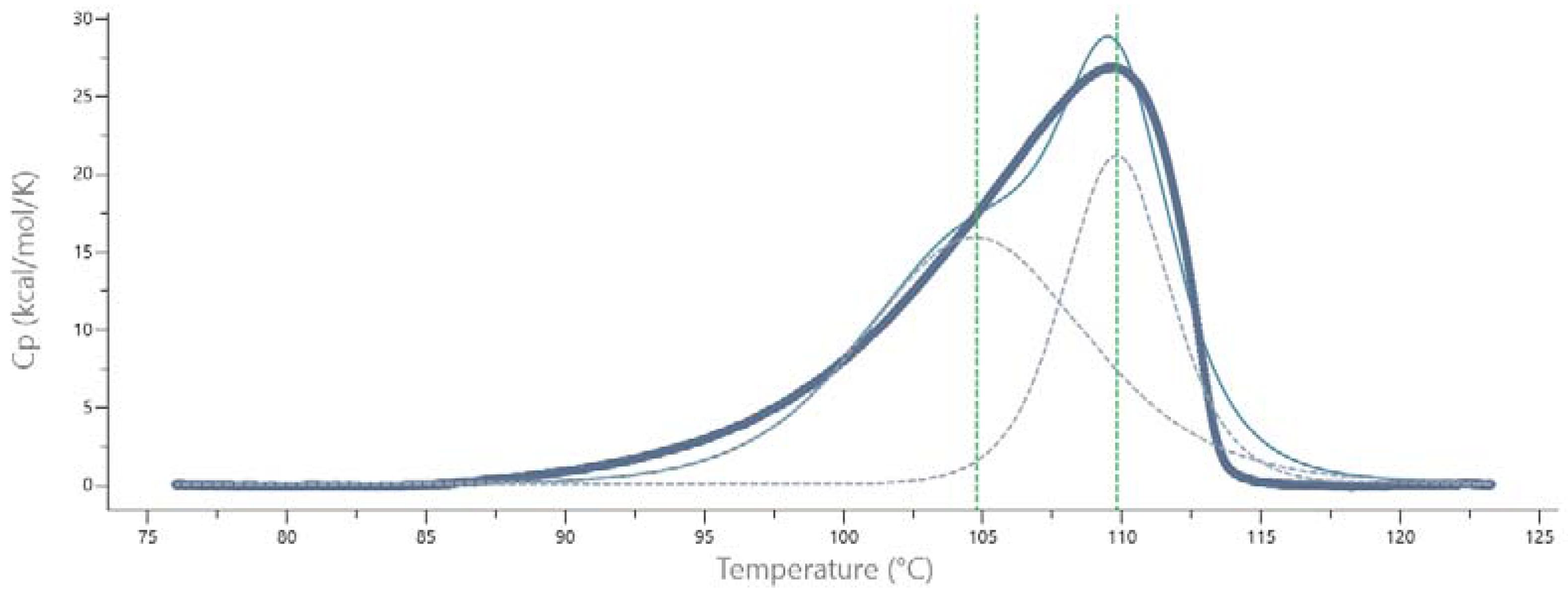
2.2. Thermal Stability and Melting Profile of Pch-Dac
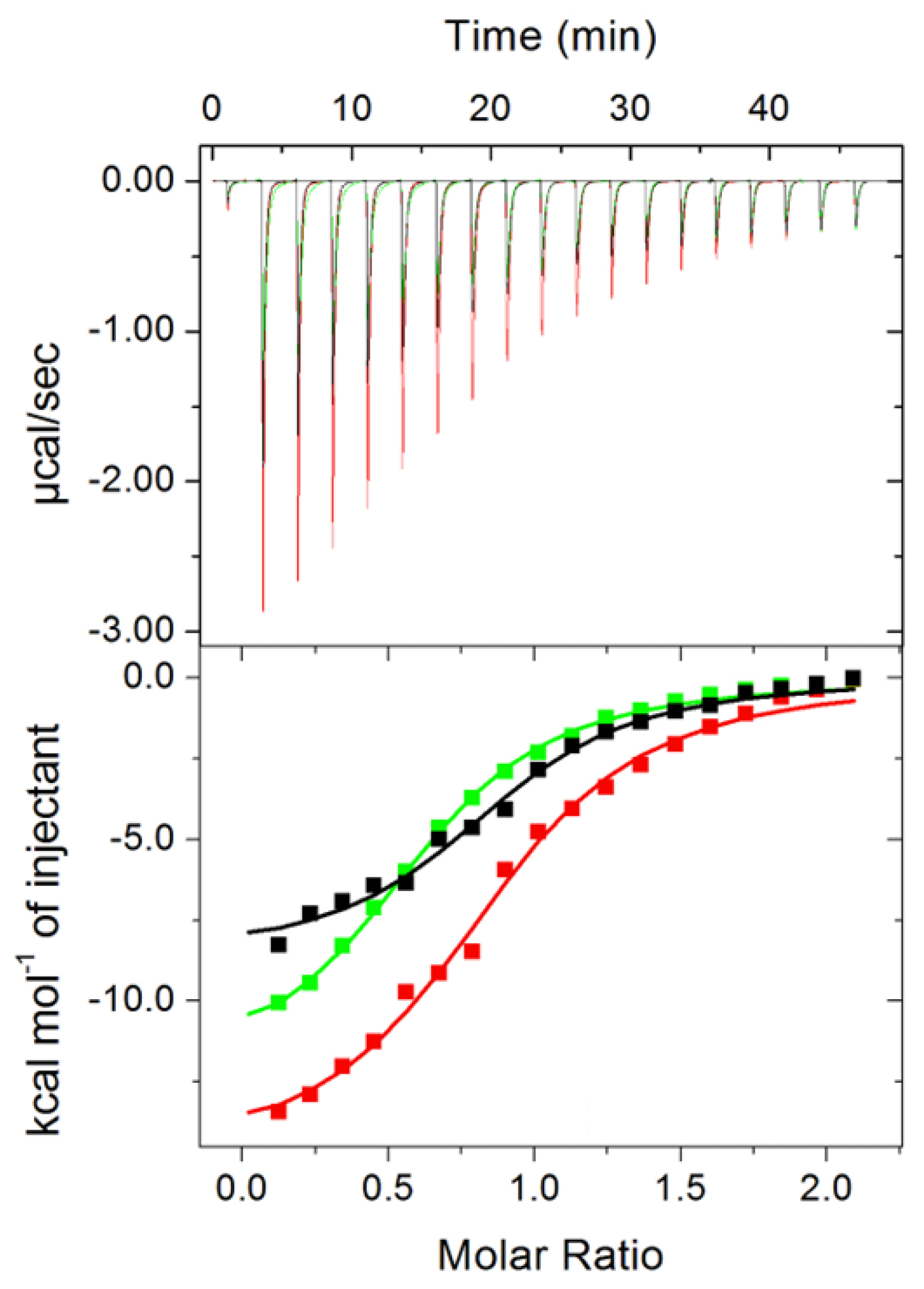
2.3. Identifying the Prosthetic Group by Microcalorimetry and Anomalous X-ray Scattering
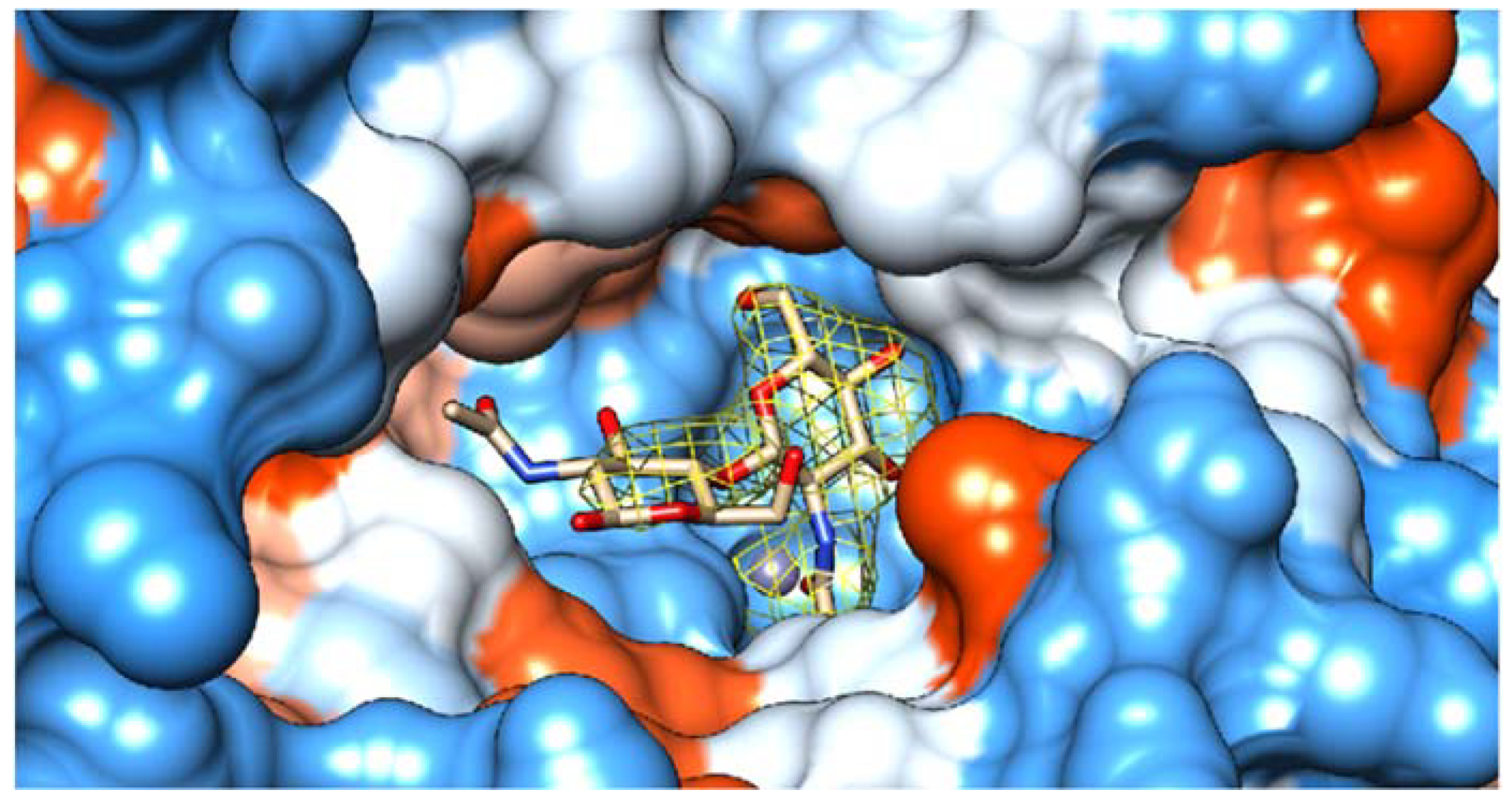
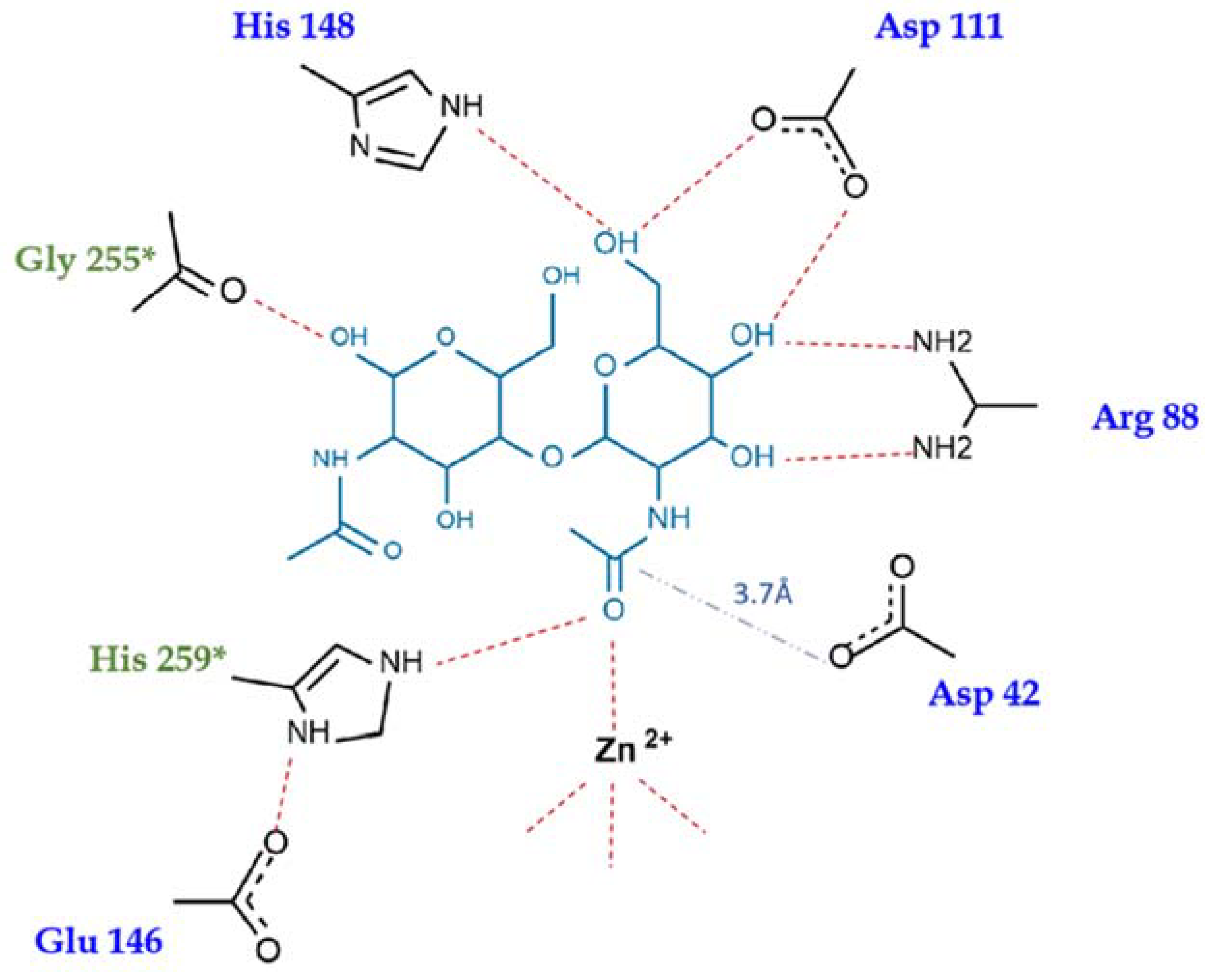
2.4. Substrate Binding
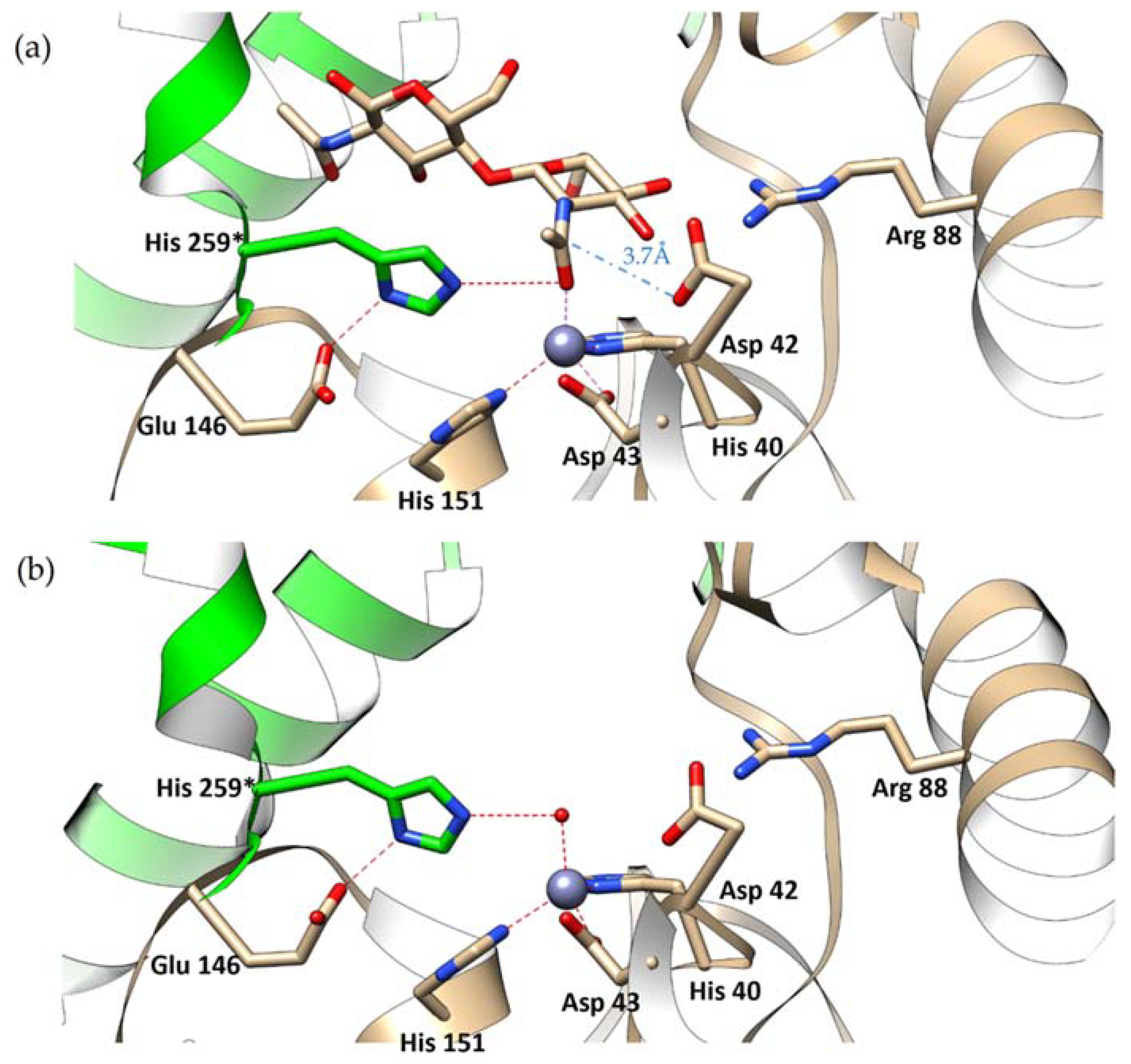
- The unliganded lipoglycopeptide antibiotic deacetylase (Orf2*) has two clear water molecules bound to the Zn2+ cation, thus making the Zn2+ pentacoordinated (PDB ID 3DFF) [10]. This is consistent with the model according to which one water molecules is displaced by the substrate while the second is the ‘hydrolytic water’ acting as the nucleophile. This enzyme, however, has a different architecture of the binding site from Pch-Dac. The gap between the Asp and His residues, flanking the active site, is wider in Orf2* by approximately 2 Å (ca 9 Å), compared to Pch-Dac (7 Å), probably due to the different nature of its substrate. The wider substrate binding site in Orf2* can more easily accommodate the two water molecules. An amino acid sequence comparison of the two enzymes shows that whereas the Asp residues come from corresponding places, the His residues are unrelated. The monomeric Orf2* is self-contained, whereas in the oligomeric Pch-Dac the catalytic His259* comes from the neighboring subunit.
- The two deacetylases from Pyrococcus horikoshii and Pyrococcus furiosus that are closely related to Pch-Dac have been described with Zn2+ or Cd2+ in the metal-binding site and different ligands [5,6,7]. In the complex with the reaction intermediate analog (MPG), two oxygen atoms of the ligand approach the active site but they interact asymmetrically: one makes a direct interaction with Zn2+ (the average distance is 1.9 Å), while the other is more distant (2.8 Å). In these complexes, Zn2+ appears tetrahedrally coordinated [6]. Asymmetric interactions with Zn2+ are also observed in complexes with the phosphate: one of the oxygen atoms is closer to the cation (1.9 Å) than the other (3.1 Å) (PDB ID 3WL3) and in a complex with an acetate ion (1.9 and 2.4 Å) (PDB ID 3WE7) [5]. In the absence of ligands, the six crystallographically independent subunits contain water molecules that can be interpreted as either partially disordered or they can be modeled as a single site near Zn2+ or as two water molecules but also at different distances to the Zn2+ cation (PDB ID 4XM0, 4XM2) [7]. Overall, Zn2+ shows a clear tendency to be tetrahedrally coordinated, taking one ligand in addition to the conserved His-Asp-His triad. This is different from the Cd2+-substituted proteins, in which the Cd2+ cations show a tendency to be octahedrally coordinated, with some distortions to this geometry or one ligand missing in the geometrically octahedral six-ligand coordination shell (PDB ID 3Wl4, 4XM1, 4XLZ) [5,7].
2.5. Enzyme Kinetics
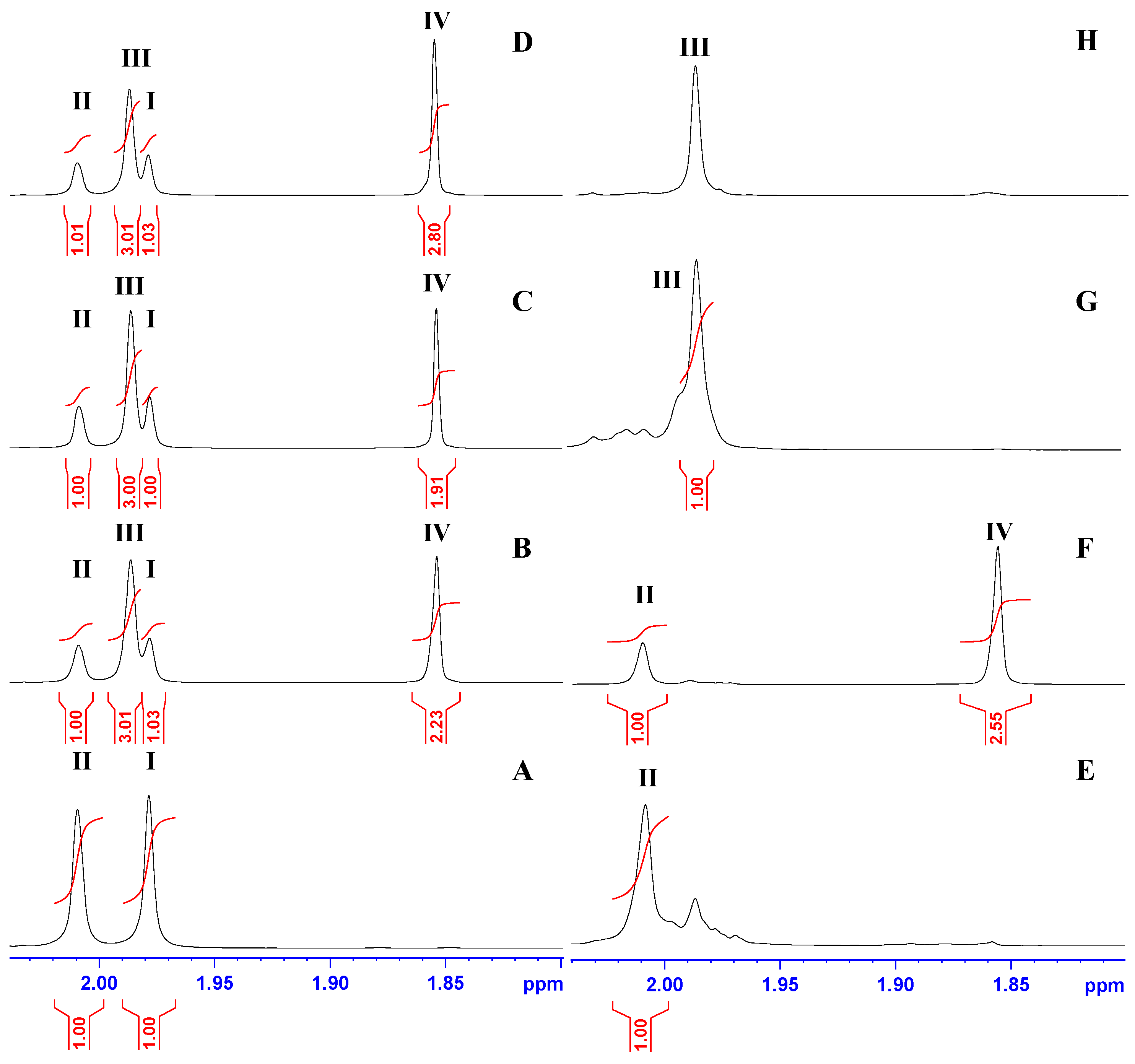
2.6. Identifying the Reaction Products by NMR Spectroscopy
3. Materials and Methods
3.1. Gene Cloning, Expression and Protein Purification
3.2. Protein Crystallization, Data Collection, Processing, Structure Solution and Refinement
3.3. Small-Angle X-ray Scattering (SAXS and SEC-SAXS)
3.4. Differential Scanning Calorimetry (DSC)
3.5. Microcalorimetric Measurements of Pch-Dac Interactions with Divalent Cations
3.6. Microcalorimetric Measurements of Pch-Dac Enzyme kinetics
3.7. NMR Spectroscopy
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Pch-Dac | N:N-diacetylchitobiose deacetylase from Pyrococcus chitonophagus |
| Pch-Dac-lig | Crystal structure of Pch-Dac with bound substrate |
| Pch-Dac-anom | Crystal structure of Pch-Dac used for identifying the prosthetic group |
| GlcNAc | N-acetyl-d-glucosamine |
| (GlcNAc)2 | 1,4-β-d-N-acetyl glucosaminyl-d-N-acetyl glucosamine |
| GlcN-GlcNAc | 1,4-β-d-glucosaminyl-d-N-acetylglucosamine |
| GlcNAc-GlcN | 1,4-β-d-acetyl glucosaminyl-d-N-glucosamine |
| r.m.s.d. | root-mean-square deviation |
| DSC | Differential Scanning Calorimetry |
| ITC | Isothermal Titration Calorimetry |
| TPEN | N,N,N′,N′-tetrakis(2-pyridinylmethyl)-1,2-ethanediamine; ion chelator |
| DSS | 4,4-dimethyl-4-silapentane-1-sulfonic acid; standard used in NMR |
References
- Cauchie, H.-M. Chitin production by arthropods in the hydrosphere. Hydrobiologia 2002, 470, 63–95. [Google Scholar] [CrossRef]
- Huber, R.; Stöhr, J.; Hohenhaus, S.; Rachel, R.; Burggraf, S.; Jannasch, H.W.; Stetter, K.O. Thermococcus chitonophagus sp. nov., a novel, chitin-degrading, hyperthermophilic archaeum from a deep-sea hydrothermal vent environment. Arch. Microbiol. 1995, 164, 255–264. [Google Scholar] [CrossRef]
- Papadimitriou, K.; Baharidis, P.K.; Georgoulis, A.; Engel, M.; Louka, M.; Karamolegkou, G.; Tsoka, A.; Blom, J.; Pot, B.; Malecki, P.; et al. Analysis of the complete genome sequence of the archaeon Pyrococcus chitonophagus DSM 10152 (formerly Thermococcus chitonophagus). Extremophiles 2016, 20, 351–361. [Google Scholar] [CrossRef]
- Blair, D.E.; Hekmat, O.; Schüttelkopf, A.W.; Shrestha, B.; Tokuyasu, K.; Withers, S.G.; van Aalten, D.M.F. Structure and Mechanism of Chitin Deacetylase from the Fungal Pathogen Colletotrichum lindemuthianum, Biochemistry 2006, 45, 9416–9426. [Google Scholar] [CrossRef]
- Mine, S.; Niiyama, M.; Hashimoto, W.; Ikegami, T.; Koma, D.; Ohmoto, T.; Fukuda, Y.; Inoue, T.; Abe, Y.; Ueda, T.; et al. Expression from engineered Escherichia coli chromosome and crystallographic study of archaeal N,N′-diacetylchitobiose deacetylase. FEBS J. 2014, 281, 2584–2596. [Google Scholar] [CrossRef]
- Nakamura, T.; Yonezawa, Y.; Tsuchiya, Y.; Niiyama, M.; Ida, K.; Oshima, M.; Morita, J.; Uegaki, K. Substrate recognition of N,N′-diacetylchitobiose deacetylase from Pyrococcus horikoshii. J. Struct. Biol. 2016, 195, 286–293. [Google Scholar] [CrossRef]
- Nakamura, T.; Niiyama, M.; Hashimoto, W.; Ida, K.; Abe, M.; Morita, J.; Uegaki, K. Multiple crystal forms of N,N′-diacetylchitobiose deacetylase from Pyrococcus furiosus. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2015, 71, 657–662. [Google Scholar] [CrossRef]
- Mondal, S.; Bobbili, K.B.; Paul, S.; Swamy, M.J. DSC and FCS Studies Reveal the Mechanism of Thermal and Chemical Unfolding of CIA17, a Polydisperse Oligomeric Protein from Coccinia Indica. J. Phys. Chem. B 2021, 125, 7117–7127. [Google Scholar] [CrossRef]
- Feliciano, G.T.; da Silva, A.J.R. Unravelling the reaction mechanism of matrix metalloproteinase 3 using QM/MM calculations. J. Mol. Struct. 2015, 1091, 125–132. [Google Scholar] [CrossRef]
- Zou, Y.; Brunzelle, J.S.; Nair, S.K. Crystal Structures of Lipoglycopeptide Antibiotic Deacetylases: Implications for the Biosynthesis of A40926 and Teicoplanin. Chem. Biol. 2008, 15, 533–545. [Google Scholar] [CrossRef]
- Mine, S.; Ikegami, T.; Kawasaki, K.; Nakamura, T.; Uegaki, K. Expression, refolding, and purification of active diacetylchitobiose deacetylase from Pyrococcus horikoshii. Protein Expr. Purif. 2012, 84, 265–269. [Google Scholar] [CrossRef]
- Cianci, M.; Bourenkov, G.; Pompidor, G.; Karpics, I.; Kallio, J.; Bento, I.; Roessle, M.; Cipriani, F.; Fiedler, S.; Schneider, T.R. P13, the EMBL macromolecular crystallography beamline at the low-emittance PETRA III ring for high- and low-energy phasing with variable beam focusing. J. Synchrotron Radiat. 2017, 24, 323–332. [Google Scholar] [CrossRef]
- Kabsch, W. XDS. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.W.; McCoy, A.; et al. Overview of theCCP4 suite and current developments. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Murshudov, G.N.; Skubák, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 355–367. [Google Scholar] [CrossRef]
- Holm, L. Using Dali for Protein Structure Comparison. In Structural Bioinformatics; Methods in Molecular Biology; Humana: New York, NY, USA, 2020; Volume 2112, pp. 29–42. [Google Scholar] [CrossRef]
- Krissinel, E.; Henrick, K. Detection of protein assemblies in crystals. Lect. Notes Comput. Sci. 2005, 3695, 163–174. [Google Scholar] [CrossRef]
- STARANISO Anisotropy & Bayesian Estimation Server. Available online: https://staraniso.globalphasing.org/cgi-bin/staraniso.cgi (accessed on 28 September 2022).
- Jeffries, C.M.; Graewert, M.A.; Blanchet, C.E.; Langley, D.B.; Whitten, A.E.; Svergun, D.I. Preparing monodisperse macromolecular samples for successful biological small-angle X-ray and neutron-scattering experiments. Nat. Protoc. 2016, 11, 2122–2153. [Google Scholar] [CrossRef]
- Blanchet, C.E.; Spilotros, A.; Schwemmer, F.; Graewert, M.A.; Kikhney, A.; Jeffries, C.M.; Franke, D.; Mark, D.; Zengerle, R.; Cipriani, F.; et al. Versatile sample environments and automation for biological solution X-ray scattering experiments at the P12 beamline (PETRA III, DESY). J. Appl. Crystallogr. 2015, 48, 431–443. [Google Scholar] [CrossRef]
- Round, A.; Felisaz, F.; Fodinger, L.; Gobbo, A.; Huet, J.; Villard, C.; Blanchet, C.E.; Pernot, P.; McSweeney, S.; Rössle, M.; et al. BioSAXS Sample Changer: A robotic sample changer for rapid and reliable high-throughput X-ray solution scattering experiments. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015, 71, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Graewert, M.A.; Franke, D.; Jeffries, C.M.; Blanchet, C.E.; Ruskule, D.; Kuhle, K.; Flieger, A.; Schäfer, B.; Tartsch, B.; Meijers, R.; et al. Automated Pipeline for Purification, Biophysical and X-ray Analysis of Biomacromolecular Solutions. Sci. Rep. 2015, 5, 10734. [Google Scholar] [CrossRef] [PubMed]
- Hajizadeh, N.R.; Franke, D.; Jeffries, C.M.; Svergun, D.I. Consensus Bayesian assessment of protein molecular mass from solution X-ray scattering data. Sci. Rep. 2018, 8, 7204. [Google Scholar] [CrossRef] [PubMed]
- Molodenskiy, D.; Mertens, H.; Svergun, D.I. An automated data processing and analysis pipeline for transmembrane proteins in detergent solutions. Sci. Rep. 2020, 10, 8081. [Google Scholar] [CrossRef]
- Panjkovich, A.; Svergun, D.I. CHROMIXS: Automatic and interactive analysis of chromatography-coupled small-angle X-ray scattering data. Bioinformatics 2017, 34, 1944–1946. [Google Scholar] [CrossRef]
- Manalastas-Cantos, K.; Konarev, P.V.; Hajizadeh, N.R.; Kikhney, A.G.; Petoukhov, M.V.; Molodenskiy, D.S.; Panjkovich, A.; Mertens, H.D.T.; Gruzinov, A.; Borges, C.; et al. ATSAS 3.0: Expanded functionality and new tools for small-angle scattering data analysis. J. Appl. Crystallogr. 2021, 54, 343–355. [Google Scholar] [CrossRef]
- Svergun, D.I.; Barberato, C.; Koch, M.H.J. CRYSOL—A Program to Evaluate X-ray Solution Scattering of Biological Macromolecules from Atomic Coordinates. J. Appl. Crystallogr. 1995, 28, 768–773. [Google Scholar] [CrossRef]
- Konarev, P.V.; Volkov, V.V.; Sokolova, A.V.; Koch, M.H.J.; Svergun, D.I. PRIMUS: A Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003, 36, 1277–1282. [Google Scholar] [CrossRef]
- Guinier, A. La diffraction des rayons X aux très petits angles: Application à l’étude de phénomènes ultramicroscopiques. Ann. Phys. 1939, 11, 161–237. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, G.; Moitessier, N.; Mittermaier, A.K. Enzyme Kinetics by Isothermal Titration Calorimetry: Allostery, Inhibition, and Dynamics. Front. Mol. Biosci. 2020, 7, 285. [Google Scholar] [CrossRef]
- Hwang, T.-L.; Shaka, A. Water Suppression That Works. Excitation Sculpting Using Arbitrary Wave-Forms and Pulsed-Field Gradients. J. Magn. Reson. Ser. A 1995, 112, 275–279. [Google Scholar] [CrossRef]

| Zn2+ | Cd2+ | Ni2+ | |
|---|---|---|---|
| Stoichiometry | 0.86 ± 0.02 | 0.87 ± 0.02 | 0.62 ± 0.01 |
| Kd [µM] | 9.5 ± 1.3 | 11.0 ± 1.3 | 12.7 ± 1.0 |
| ΔH [cal/mol] | −8666 ± 314 | −15,040 ± 498 | −12,400 ± 332 |
| ΔS [cal/mol/deg] | −6.1 | −27.8 | −19.2 |
| Substrate | Km [mM] | kcat [s−1] | kcat/Km |
|---|---|---|---|
| GlcNAc | 22.4 ± 1.5 | 226 ± 6 | 10 ± 1 |
| (GlcNAc)2 | 2.6 ± 0.2 | 178 ± 2 | 68 ± 6 |
| (GlcNAc)3 | 11.2 ± 1.1 | 71 ± 4 | 6 ± 1 |
| pH | KM [mM] | kcat [s−1] | kcat/KM |
|---|---|---|---|
| 6.5 | 2.6 ± 0.2 | 178 ± 2 | 68 ± 6 |
| 7.5 | 3.7 ± 0.1 | 263 ± 1 | 71 ± 2 |
| 8.5 | 4.8 ± 0.4 | 169 ± 4 | 35 ± 4 |
| Substrate | Temp. (°C) | 25 | 35 | 45 | 55 | 65 | 75 |
|---|---|---|---|---|---|---|---|
| GlcNAc | KM [mM] | - | 9.2 ± 0.2 | 19.5 ± 1.1 | 14.8 ± 0.4 | 13.3 ± 0.4 | 22.4 ± 1.5 |
| kcat [s−1] | - | 32.6 ± 0.2 | 75.1 ± 1.6 | 117 ± 1 | 102 ± 1 | 226 ± 6 | |
| kcat/KM [s−1·mM−1] | - | 3.5 ± 0.1 | 3.8 ± 0.3 | 7.9 ± 0.3 | 7.7 ± 0.3 | 10.2 ± 0.9 | |
| (GlcNAc)2 | KM | 3.8 ± 0.2 | 6.3 ± 0.2 | 4.3 ± 0.1 | 4.5 ± 0.2 | 5.1 ± 0.2 | 3.7 ± 0.1 |
| kcat | 23.4 ± 0.4 | 53.1 ± 0.5 | 84.3 ± 0.3 | 153 ± 2 | 222 ± 3 | 263 ± 2 | |
| kcat/KM | 6.2 ± 0.4 | 8.4 ± 0.3 | 19.6 ± 0.5 | 34 ± 2 | 44 ± 2 | 71 ± 2 | |
| (GlcNAc)3 | KM | - | - | - | 14.2 ± 1.3 | 11.1 ± 1.1 | 11.2 ± 1.1 |
| kcat | - | - | - | 31.4 ± 1.8 | 48.1 ± 2.8 | 70.6 ± 4.2 | |
| kcat/KM | - | - | - | 2.2 ± 0.3 | 4.3 ± 0.7 | 6.3 ± 1.0 |
| Protein | Pch-Dac-lig | Pch-Dac | Pch-Dac-anom ‡ |
|---|---|---|---|
| PDB code | 8BGO | 8BGN | 8BGP |
| Space group | P21 | P3221 | P212121 |
| Unit cell parameters: | |||
| a (Å) | 79.2 | 150.6 | 78.6 |
| b (Å) | 151.7 | 150.6 | 158.7 |
| c (Å) | 167.1 | 72.2 | 158.6 |
| α (°) | 90 | 90 | 90 |
| β (°) | 93.3 | 90 | 90 |
| γ (°) | 90 | 120 | 90 |
| Wavelength (Å) | 0.9797 | 0.9797 | 1.2815 |
| Resolution (Å) | 3.08 | 2.76 | 2.51 |
| Rmerge *,# | 0.077 (0.657) | 0.114 (0.966) | 0.199 (1.090) |
| Rpim † | 0.057 (0.303) | ||
| Completeness (%) | 99.3 | 99.08 | 94.2 |
| Observed reflections | 488,298 | 470,509 | 565,577 |
| Unique reflections | 6943 | 24,234 | 43,577 |
| <I/σ(I)> | 14.6 (2.0) | 18.4 (2.4) | 11.1 (3.03) |
| Protein subunits/asymmetric unit | 12 | 3 | 6 |
| R §/Rfree $ | 0.1687/0.2147 | 0.1763/0.2271 | 0.2183/0.2614 |
| Sample Concentration | 0.70 mg/mL (22 µM), 1.24 mg/mL(40 µM), 1.39 mg/mL (45 µM), 1.79 mg/mL (58 µM) |
| Sample Solution Components | Buffer A: 20 mM TRIS pH 7.4, 200 mM NaCl Buffer B: 25 mM HEPES pH 7.5, 200 mM NaCl, 10 µM ZnCl2 |
| Scan Rates | 45 °C/h, 60 °C/h, 90 °C/h, 240 °C/h. |
| Feedback Modes | high, medium |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Biniek-Antosiak, K.; Bejger, M.; Śliwiak, J.; Baranowski, D.; Mohammed, A.S.A.; Svergun, D.I.; Rypniewski, W. Structural, Thermodynamic and Enzymatic Characterization of N,N-Diacetylchitobiose Deacetylase from Pyrococcus chitonophagus. Int. J. Mol. Sci. 2022, 23, 15736. https://doi.org/10.3390/ijms232415736
Biniek-Antosiak K, Bejger M, Śliwiak J, Baranowski D, Mohammed ASA, Svergun DI, Rypniewski W. Structural, Thermodynamic and Enzymatic Characterization of N,N-Diacetylchitobiose Deacetylase from Pyrococcus chitonophagus. International Journal of Molecular Sciences. 2022; 23(24):15736. https://doi.org/10.3390/ijms232415736
Chicago/Turabian StyleBiniek-Antosiak, Katarzyna, Magdalena Bejger, Joanna Śliwiak, Daniel Baranowski, Ahmed S. A. Mohammed, Dmitri I. Svergun, and Wojciech Rypniewski. 2022. "Structural, Thermodynamic and Enzymatic Characterization of N,N-Diacetylchitobiose Deacetylase from Pyrococcus chitonophagus" International Journal of Molecular Sciences 23, no. 24: 15736. https://doi.org/10.3390/ijms232415736
APA StyleBiniek-Antosiak, K., Bejger, M., Śliwiak, J., Baranowski, D., Mohammed, A. S. A., Svergun, D. I., & Rypniewski, W. (2022). Structural, Thermodynamic and Enzymatic Characterization of N,N-Diacetylchitobiose Deacetylase from Pyrococcus chitonophagus. International Journal of Molecular Sciences, 23(24), 15736. https://doi.org/10.3390/ijms232415736






