Deciphering the Role of miR-200c-3p in Type 1 Diabetes (Subclinical Cardiovascular Disease) and Its Correlation with Inflammation and Vascular Health
Abstract
1. Introduction
2. Results
2.1. Subjects’ Characteristics
2.2. Cytokine Profiles
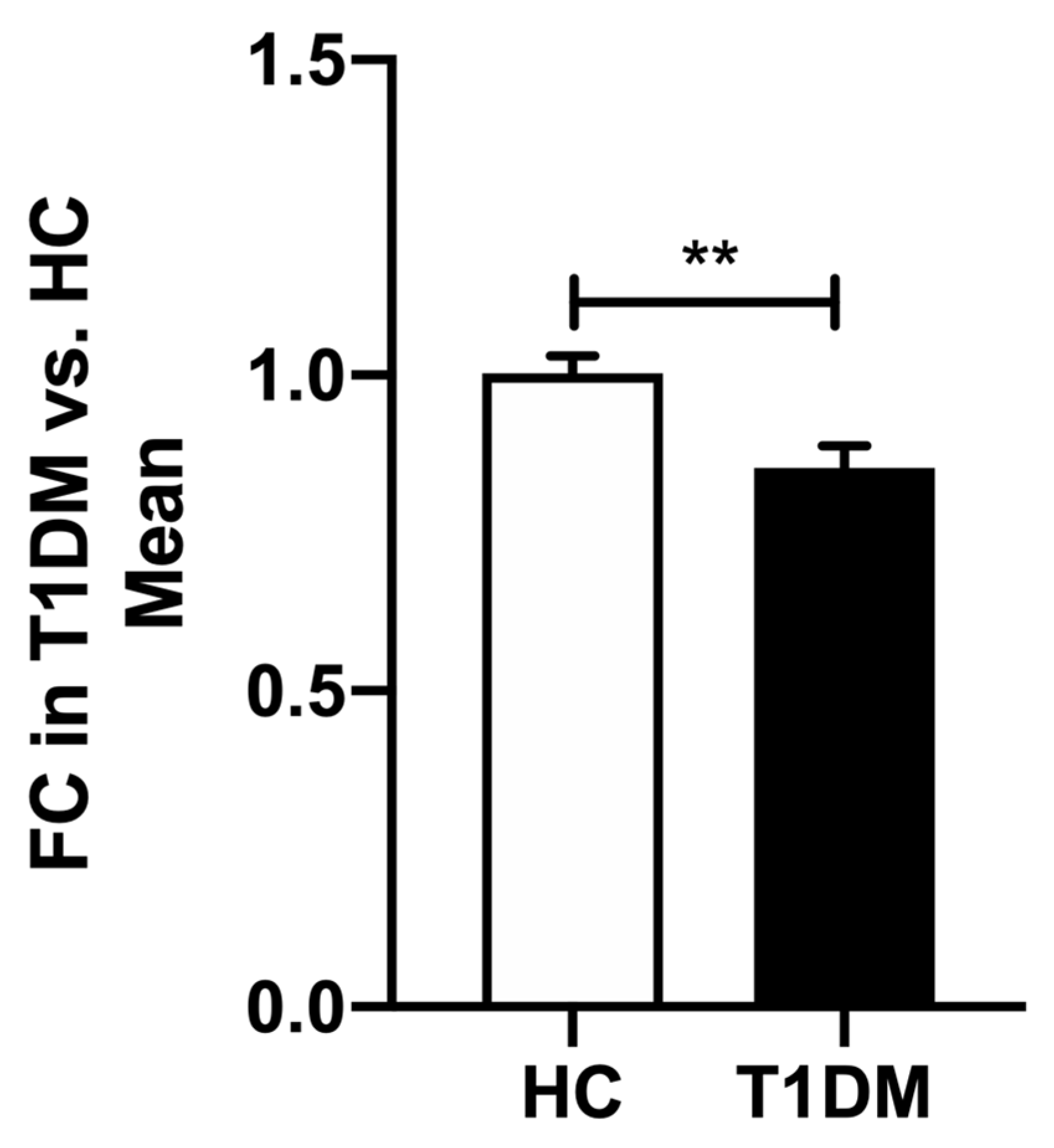
2.3. Expression of hsa-miR-200c-3p
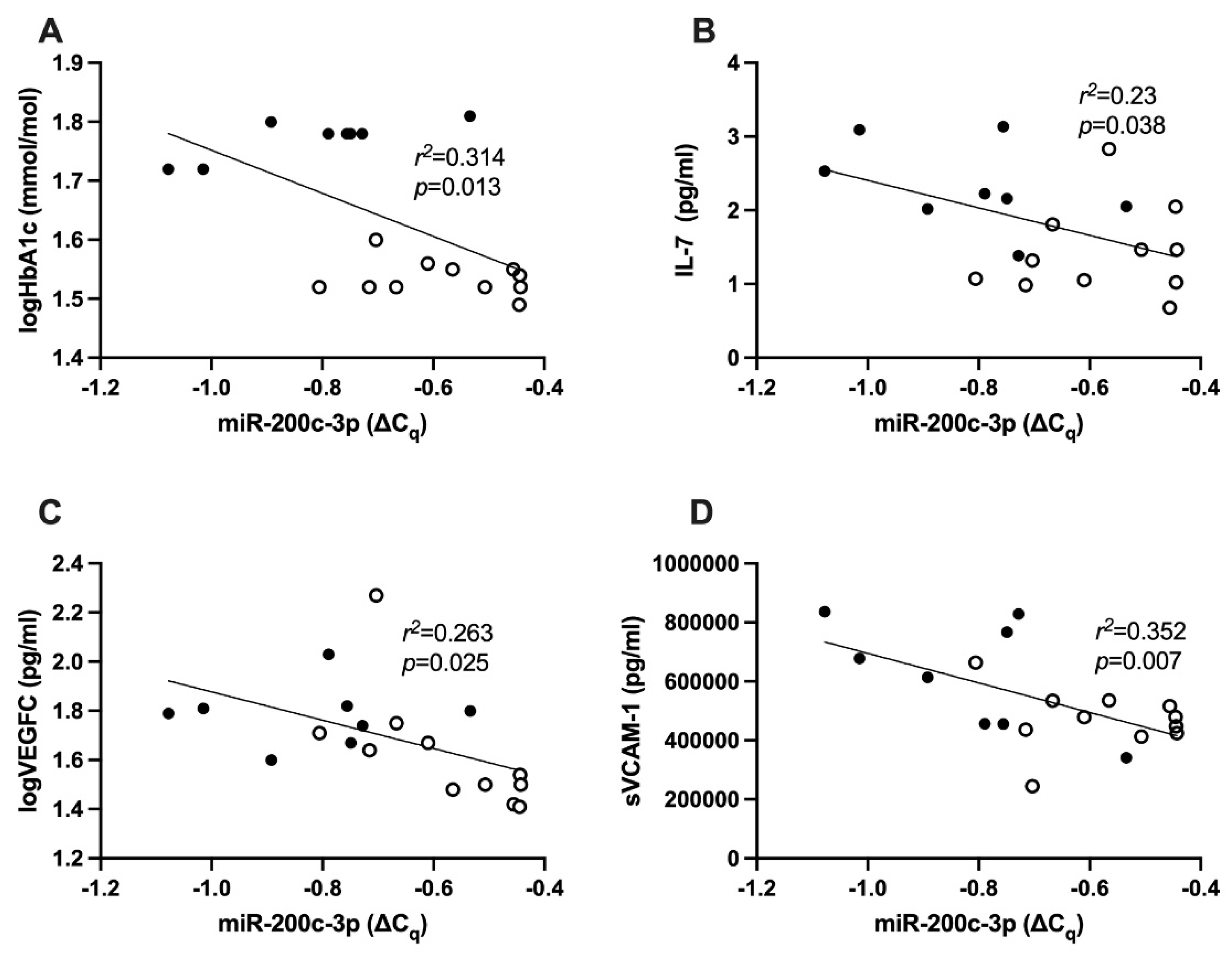
2.4. Correlation between HbA1c and hsa-miR-200c-3p
2.5. Correlation between hsa-miR-200c-3p and Cytokines
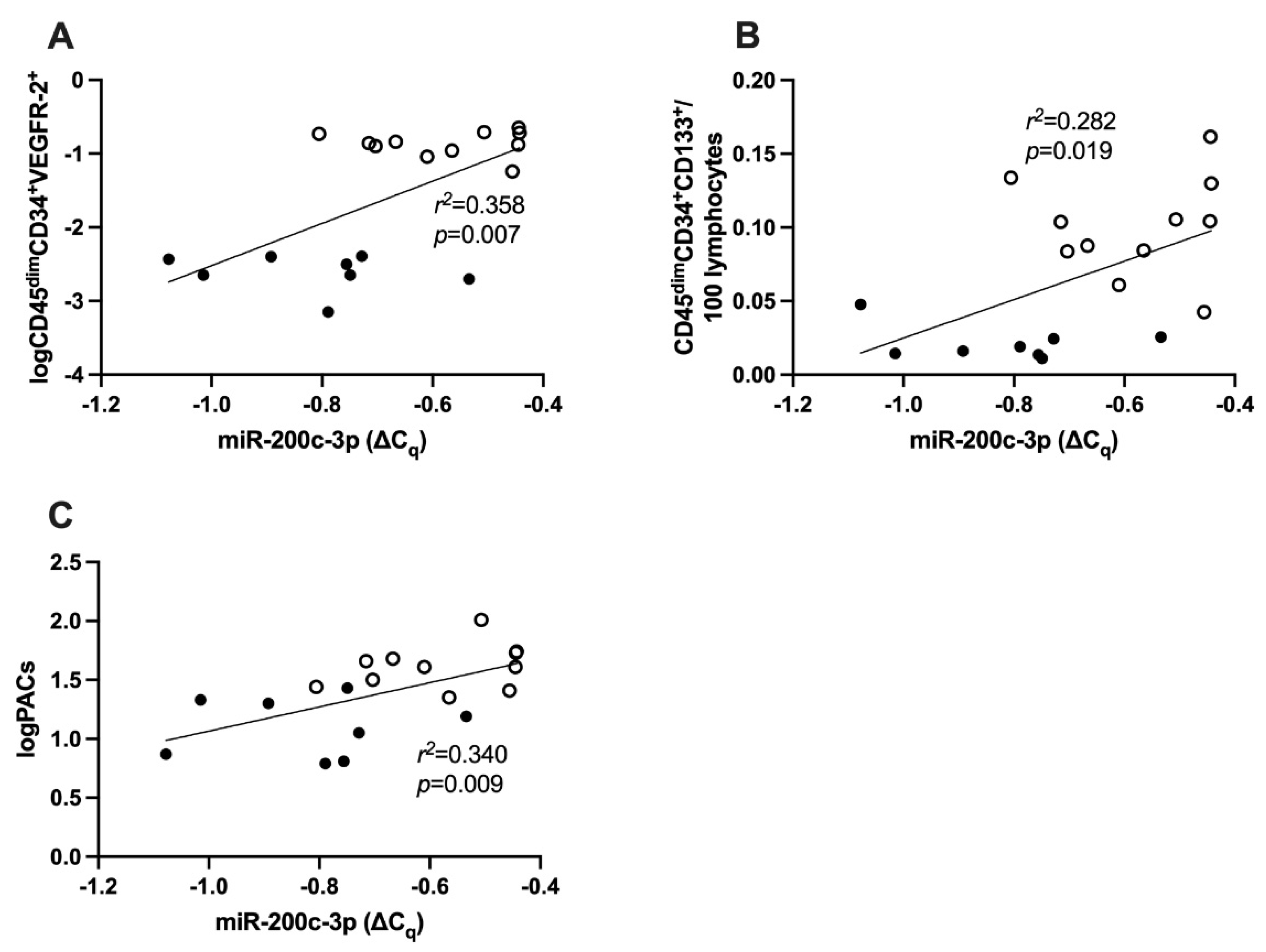
2.6. Correlation between miR-200c-3p and cEPCs
2.7. Correlation between miR-200c-3p and PACs
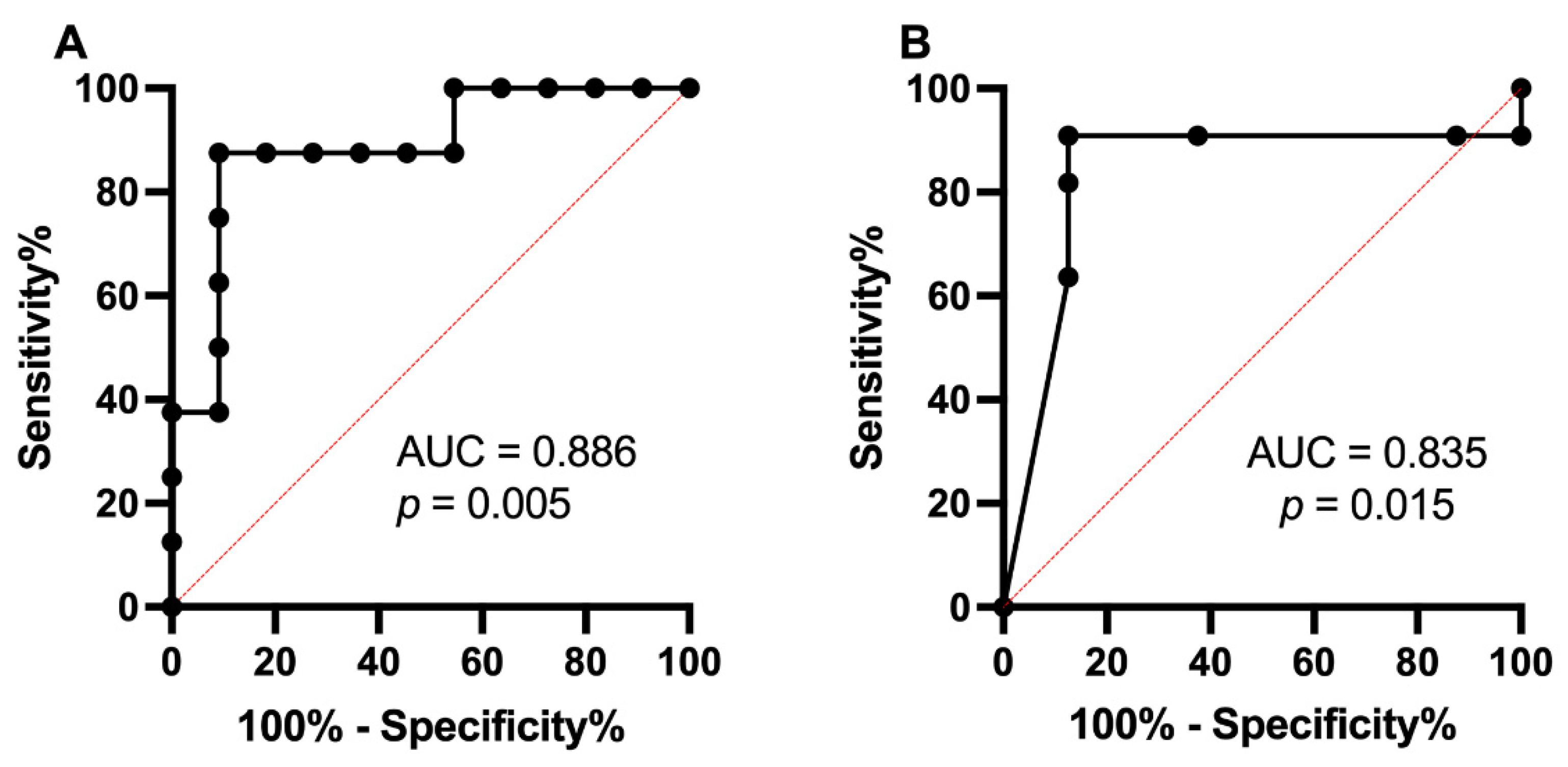
2.8. Receiver Operating Characteristic Curve Analysis of miR-200c-3p and T1DM
3. Discussion
3.1. Inflammation in T1DM
3.2. miR-200c-3p Expression in T1DM
3.3. miR-200c-3p and Inflammation
3.4. miR-200c-3p and Vascular Health
3.5. Causation/Contribution
4. Materials and Methods
4.1. Study
4.2. Cytokine Analysis
4.3. Plasma miRNA Extraction
4.4. Peripheral Blood Mononuclear Cell Total RNA Extraction
4.5. Quantitative Real-Time Polymerase Chain Reaction for miRNA
4.6. In Vitro Assays for Vascular Health
4.6.1. Evaluation of Circulating Endothelial Progenitor Cells (cEPCs) and Circulating Endothelial Cells (cECs) via Flow Cytometry
4.6.2. Proangiogenic Cells (PACs) In Vitro Assay
4.7. Ingenuity Pathway Analysis (IPA)
4.8. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- de Ferranti, S.D.; de Boer, I.H.; Fonseca, V.; Fox, C.S.; Golden, S.H.; Lavie, C.J.; Magge, S.N.; Marx, N.; McGuire, D.K.; Orchard, T.J.; et al. Type 1 diabetes mellitus and cardiovascular disease: A scientific statement from the American Heart Association and American Diabetes Association. Circulation 2014, 130, 1110–1130. [Google Scholar] [CrossRef] [PubMed]
- Soedamah-Muthu, S.S.; Fuller, J.H.; Mulnier, H.E.; Raleigh, V.S.; Lawrenson, R.A.; Colhoun, H.M. High risk of cardiovascular disease in patients with type 1 diabetes in the U.K.: A cohort study using the general practice research database. Diabetes Care 2006, 29, 798–804. [Google Scholar] [CrossRef] [PubMed]
- Baena-Diez, J.M.; Penafiel, J.; Subirana, I.; Ramos, R.; Elosua, R.; Marin-Ibanez, A.; Guembe, M.J.; Rigo, F.; Tormo-Diaz, M.J.; Moreno-Iribas, C.; et al. Risk of Cause-Specific Death in Individuals With Diabetes: A Competing Risks Analysis. Diabetes Care 2016, 39, 1987–1995. [Google Scholar] [CrossRef]
- Livingstone, S.J.; Looker, H.C.; Hothersall, E.J.; Wild, S.H.; Lindsay, R.S.; Chalmers, J.; Cleland, S.; Leese, G.P.; McKnight, J.; Morris, A.D.; et al. Risk of cardiovascular disease and total mortality in adults with type 1 diabetes: Scottish registry linkage study. PLoS Med. 2012, 9, e1001321. [Google Scholar] [CrossRef]
- Ahmed, F.W.; Rider, R.; Glanville, M.; Narayanan, K.; Razvi, S.; Weaver, J.U. Metformin improves circulating endothelial cells and endothelial progenitor cells in type 1 diabetes: MERIT study. Cardiovasc. Diabetol. 2016, 15, 116. [Google Scholar] [CrossRef] [PubMed]
- Sibal, L.; Aldibbiat, A.; Agarwal, S.C.; Mitchell, G.; Oates, C.; Razvi, S.; Weaver, J.U.; Shaw, J.A.; Home, P.D. Circulating endothelial progenitor cells, endothelial function, carotid intima-media thickness and circulating markers of endothelial dysfunction in people with type 1 diabetes without macrovascular disease or microalbuminuria. Diabetologia 2009, 52, 1464–1473. [Google Scholar] [CrossRef]
- West, D.J.; Campbell, M.D.; Gonzalez, J.T.; Walker, M.; Stevenson, E.J.; Ahmed, F.W.; Wijaya, S.; Shaw, J.A.; Weaver, J.U. The inflammation, vascular repair and injury responses to exercise in fit males with and without Type 1 diabetes: An observational study. Cardiovasc. Diabetol. 2015, 14, 71. [Google Scholar] [CrossRef]
- Werner, N.; Kosiol, S.; Schiegl, T.; Ahlers, P.; Walenta, K.; Link, A.; Bohm, M.; Nickenig, G. Circulating endothelial progenitor cells and cardiovascular outcomes. N. Engl. J. Med. 2005, 353, 999–1007. [Google Scholar] [CrossRef]
- Savoia, C.; Sada, L.; Zezza, L.; Pucci, L.; Lauri, F.M.; Befani, A.; Alonzo, A.; Volpe, M. Vascular inflammation and endothelial dysfunction in experimental hypertension. Int. J. Hypertens. 2011, 2011, 281240. [Google Scholar] [CrossRef]
- Haller, M.J.; Samyn, M.; Nichols, W.W.; Brusko, T.; Wasserfall, C.; Schwartz, R.F.; Atkinson, M.; Shuster, J.J.; Pierce, G.L.; Silverstein, J.H. Radial artery tonometry demonstrates arterial stiffness in children with type 1 diabetes. Diabetes Care 2004, 27, 2911–2917. [Google Scholar] [CrossRef]
- Urbina, E.M.; Wadwa, R.P.; Davis, C.; Snively, B.M.; Dolan, L.M.; Daniels, S.R.; Hamman, R.F.; Dabelea, D. Prevalence of increased arterial stiffness in children with type 1 diabetes mellitus differs by measurement site and sex: The SEARCH for Diabetes in Youth Study. J. Pediatr. 2010, 156, 731–737.e1. [Google Scholar] [CrossRef] [PubMed]
- Jarvisalo, M.J.; Putto-Laurila, A.; Jartti, L.; Lehtimaki, T.; Solakivi, T.; Ronnemaa, T.; Raitakari, O.T. Carotid artery intima-media thickness in children with type 1 diabetes. Diabetes 2002, 51, 493–498. [Google Scholar] [CrossRef] [PubMed]
- Krantz, J.S.; Mack, W.J.; Hodis, H.N.; Liu, C.R.; Liu, C.H.; Kaufman, F.R. Early onset of subclinical atherosclerosis in young persons with type 1 diabetes. J. Pediatr. 2004, 145, 452–457. [Google Scholar] [CrossRef] [PubMed]
- Rabago Rodriguez, R.; Gomez-Diaz, R.A.; Tanus Haj, J.; Avelar Garnica, F.J.; Ramirez Soriano, E.; Nishimura Meguro, E.; Aguilar-Salinas, C.A.; Wacher, N.H. Carotid intima-media thickness in pediatric type 1 diabetic patients. Diabetes Care 2007, 30, 2599–2602. [Google Scholar] [CrossRef]
- Urbina, E.M.; Dabelea, D.; D’Agostino, R.B., Jr.; Shah, A.S.; Dolan, L.M.; Hamman, R.F.; Daniels, S.R.; Marcovina, S.; Wadwa, R.P. Effect of type 1 diabetes on carotid structure and function in adolescents and young adults: The SEARCH CVD study. Diabetes Care 2013, 36, 2597–2599. [Google Scholar] [CrossRef]
- Gourgari, E.; Dabelea, D.; Rother, K. Modifiable Risk Factors for Cardiovascular Disease in Children with Type 1 Diabetes: Can Early Intervention Prevent Future Cardiovascular Events? Curr. Diab. Rep. 2017, 17, 134. [Google Scholar] [CrossRef]
- Lerman, A.; Zeiher, A.M. Endothelial function: Cardiac events. Circulation 2005, 111, 363–368. [Google Scholar] [CrossRef]
- Taddei, S.; Virdis, A.; Ghiadoni, L.; Sudano, I.; Salvetti, A. Effects of antihypertensive drugs on endothelial dysfunction: Clinical implications. Drugs 2002, 62, 265–284. [Google Scholar] [CrossRef]
- Versari, D.; Daghini, E.; Virdis, A.; Ghiadoni, L.; Taddei, S. Endothelial dysfunction as a target for prevention of cardiovascular disease. Diabetes Care 2009, 32, S314–S321. [Google Scholar] [CrossRef]
- Widlansky, M.E.; Gokce, N.; Keaney, J.F., Jr.; Vita, J.A. The clinical implications of endothelial dysfunction. J. Am. Coll. Cardiol. 2003, 42, 1149–1160. [Google Scholar] [CrossRef]
- Rabar, S.; Harker, M.; O’Flynn, N.; Wierzbicki, A.S.; Guideline Development, G. Lipid modification and cardiovascular risk assessment for the primary and secondary prevention of cardiovascular disease: Summary of updated NICE guidance. BMJ 2014, 349, g4356. [Google Scholar] [CrossRef] [PubMed]
- Ekezue, B.F.; Laditka, S.B.; Laditka, J.N.; Studnicki, J.; Blanchette, C.M. Diabetes complications and adverse health outcomes after coronary revascularization. Diabetes Res. Clin. Pract. 2014, 103, 530–537. [Google Scholar] [CrossRef] [PubMed]
- Piccolo, R.; Galasso, G.; Iversen, A.Z.; Eitel, I.; Dominguez-Rodriguez, A.; Gu, Y.L.; de Smet, B.J.; Mahmoud, K.D.; Abreu-Gonzalez, P.; Trimarco, B.; et al. Effects of baseline coronary occlusion and diabetes mellitus in patients with ST-segment elevation myocardial infarction undergoing primary percutaneous coronary intervention. Am. J. Cardiol. 2014, 114, 1145–1150. [Google Scholar] [CrossRef] [PubMed]
- Creemers, E.E.; Tijsen, A.J.; Pinto, Y.M. Circulating microRNAs: Novel biomarkers and extracellular communicators in cardiovascular disease? Circ. Res. 2012, 110, 483–495. [Google Scholar] [CrossRef]
- Reddy, M.A.; Jin, W.; Villeneuve, L.; Wang, M.; Lanting, L.; Todorov, I.; Kato, M.; Natarajan, R. Pro-inflammatory role of microrna-200 in vascular smooth muscle cells from diabetic mice. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 721–729. [Google Scholar] [CrossRef]
- Singh, G.B.; Raut, S.K.; Khanna, S.; Kumar, A.; Sharma, S.; Prasad, R.; Khullar, M. MicroRNA-200c modulates DUSP-1 expression in diabetes-induced cardiac hypertrophy. Mol. Cell. Biochem. 2017, 424, 1–11. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, J.; Qu, D.; Wang, L.; Luo, J.Y.; Lau, C.W.; Liu, P.; Gao, Z.; Tipoe, G.L.; Lee, H.K.; et al. Inhibition of miR-200c Restores Endothelial Function in Diabetic Mice Through Suppression of COX-2. Diabetes 2016, 65, 1196–1207. [Google Scholar] [CrossRef]
- Ray, S.L.; Coulson, D.J.; Yeoh, M.L.Y.; Tamara, A.; Latief, J.S.; Bakhashab, S.; Weaver, J.U. The Role of miR-342 in Vascular Health. Study in Subclinical Cardiovascular Disease in Mononuclear Cells, Plasma, Inflammatory Cytokines and PANX2. Int. J. Mol. Sci. 2020, 21, 7217. [Google Scholar] [CrossRef]
- Borilova Linhartova, P.; Kavrikova, D.; Tomandlova, M.; Poskerova, H.; Rehka, V.; Dusek, L.; Izakovicova Holla, L. Differences in Interleukin-8 Plasma Levels between Diabetic Patients and Healthy Individuals Independently on Their Periodontal Status. Int. J. Mol. Sci. 2018, 19, 3214. [Google Scholar] [CrossRef]
- Ji, T.; Xu, C.; Sun, L.; Yu, M.; Peng, K.; Qiu, Y.; Xiao, W.; Yang, H. Aryl Hydrocarbon Receptor Activation Down-Regulates IL-7 and Reduces Inflammation in a Mouse Model of DSS-Induced Colitis. Dig. Dis. Sci. 2015, 60, 1958–1966. [Google Scholar] [CrossRef]
- Gaal, E.I.; Tammela, T.; Anisimov, A.; Marbacher, S.; Honkanen, P.; Zarkada, G.; Leppanen, V.M.; Tatlisumak, T.; Hernesniemi, J.; Niemela, M.; et al. Comparison of vascular growth factors in the murine brain reveals placenta growth factor as prime candidate for CNS revascularization. Blood 2013, 122, 658–665. [Google Scholar] [CrossRef]
- Liu, S.; Li, H. Down-regulation of microRNA-28 in peripheral blood mononuclear cell plays a role in pathogenesis of type 1 diabetes. Int. J. Clin. Exp. Med. 2017, 10, 10021–10030. [Google Scholar]
- Yang, Z.; Zeng, B.; Tang, X.; Wang, H.; Wang, C.; Yan, Z.; Huang, P.; Pan, Y.; Xu, B. MicroRNA-146a and miR-99a are potential biomarkers for disease activity and clinical efficacy assessment in psoriasis patients treated with traditional Chinese medicine. J. Ethnopharmacol. 2016, 194, 727–732. [Google Scholar] [CrossRef] [PubMed]
- Merkerova, M.; Vasikova, A.; Belickova, M.; Bruchova, H. MicroRNA expression profiles in umbilical cord blood cell lineages. Stem Cells Dev. 2010, 19, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Magenta, A.; D’Agostino, M.; Sileno, S.; Di Vito, L.; Uras, C.; Abeni, D.; Martino, F.; Barilla, F.; Madonna, S.; Albanesi, C.; et al. The Oxidative Stress-Induced miR-200c Is Upregulated in Psoriasis and Correlates with Disease Severity and Determinants of Cardiovascular Risk. Oxid. Med. Cell. Longev. 2019, 2019, 8061901. [Google Scholar] [CrossRef]
- Krieglstein, C.F.; Granger, D.N. Adhesion molecules and their role in vascular disease. Am. J. Hypertens. 2001, 14, 44S–54S. [Google Scholar] [CrossRef]
- von Andrian, U.H.; Mackay, C.R. T-cell function and migration. Two sides of the same coin. N. Engl. J. Med. 2000, 343, 1020–1034. [Google Scholar] [CrossRef]
- Liu, Y.; Jiang, Y.; Li, W.; Han, C.; Zhou, L.; Hu, H. MicroRNA-200c-3p inhibits proliferation and migration of renal artery endothelial cells by directly targeting ZEB2. Exp. Cell Res. 2020, 387, 111778. [Google Scholar] [CrossRef]
- Wang, G.; Kwan, B.C.; Lai, F.M.; Choi, P.C.; Chow, K.M.; Li, P.K.; Szeto, C.C. Intrarenal expression of microRNAs in patients with IgA nephropathy. Lab. Investig. 2010, 90, 98–103. [Google Scholar] [CrossRef]
- Baseler, W.A.; Thapa, D.; Jagannathan, R.; Dabkowski, E.R.; Croston, T.L.; Hollander, J.M. miR-141 as a regulator of the mitochondrial phosphate carrier (Slc25a3) in the type 1 diabetic heart. Am. J. Physiol. Cell Physiol. 2012, 303, C1244–C1251. [Google Scholar] [CrossRef]
- Saito, S.; Thuc, L.C.; Teshima, Y.; Nakada, C.; Nishio, S.; Kondo, H.; Fukui, A.; Abe, I.; Ebata, Y.; Saikawa, T.; et al. Glucose Fluctuations Aggravate Cardiac Susceptibility to Ischemia/Reperfusion Injury by Modulating MicroRNAs Expression. Circ. J. 2016, 80, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Magenta, A.; Cencioni, C.; Fasanaro, P.; Zaccagnini, G.; Greco, S.; Sarra-Ferraris, G.; Antonini, A.; Martelli, F.; Capogrossi, M.C. miR-200c is upregulated by oxidative stress and induces endothelial cell apoptosis and senescence via ZEB1 inhibition. Cell Death Differ. 2011, 18, 1628–1639. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Guan, Q.; Jin, X. miR-200c serves an important role in H5V endothelial cells in high glucose by targeting Notch1. Mol. Med. Rep. 2017, 16, 2149–2155. [Google Scholar] [CrossRef] [PubMed]
- Abdolahi, S.; Hosseini, M.; Rezaei, R.; Mohebbi, S.R.; Rostami-Nejad, M.; Mojarad, E.N.; Mirjalali, H.; Yadegar, A.; Asadzadeh Aghdaei, H.; Zali, M.R.; et al. Evaluation of miR-200c-3p and miR-421-5p levels during immune responses in the admitted and recovered COVID-19 subjects. Infect. Genet. Evol. 2022, 98, 105207. [Google Scholar] [CrossRef]
- Song, Y.; Tran, M.; Wang, L.; Shin, D.J.; Wu, J. MiR-200c-3p targets SESN1 and represses the IL-6/AKT loop to prevent cholangiocyte activation and cholestatic liver fibrosis. Lab. Investig. 2022, 102, 485–493. [Google Scholar] [CrossRef]
- Van der Goten, J.; Vanhove, W.; Lemaire, K.; Van Lommel, L.; Machiels, K.; Wollants, W.J.; De Preter, V.; De Hertogh, G.; Ferrante, M.; Van Assche, G.; et al. Integrated miRNA and mRNA expression profiling in inflamed colon of patients with ulcerative colitis. PLoS ONE 2014, 9, e116117. [Google Scholar]
- Urbich, C.; Dimmeler, S. Endothelial progenitor cells functional characterization. Trends Cardiovasc. Med. 2004, 14, 318–322. [Google Scholar] [CrossRef]
- Wendlandt, E.B.; Graff, J.W.; Gioannini, T.L.; McCaffrey, A.P.; Wilson, M.E. The role of microRNAs miR-200b and miR-200c in TLR4 signaling and NF-kappaB activation. Innate Immun. 2012, 18, 846–855. [Google Scholar] [CrossRef]
- Li, F.; Han, F.; Li, H.; Zhang, J.; Qiao, X.; Shi, J.; Yang, L.; Dong, J.; Luo, M.; Wei, J.; et al. Human placental mesenchymal stem cells of fetal origins-alleviated inflammation and fibrosis by attenuating MyD88 signaling in bleomycin-induced pulmonary fibrosis mice. Mol. Immunol. 2017, 90, 11–21. [Google Scholar] [CrossRef]
- Xia, P.; Gu, R.; Zhang, W.; Shao, L.; Li, F.; Wu, C.; Sun, Y. MicroRNA-200c promotes osteogenic differentiation of human bone mesenchymal stem cells through activating the AKT/beta-Catenin signaling pathway via downregulating Myd88. J. Cell. Physiol. 2019, 234, 22675–22686. [Google Scholar] [CrossRef]
- Hill, J.M.; Zalos, G.; Halcox, J.P.; Schenke, W.H.; Waclawiw, M.A.; Quyyumi, A.A.; Finkel, T. Circulating endothelial progenitor cells, vascular function, and cardiovascular risk. N. Engl. J. Med. 2003, 348, 593–600. [Google Scholar] [CrossRef] [PubMed]
- Hortenhuber, T.; Rami-Mehar, B.; Satler, M.; Nagl, K.; Hobaus, C.; Hollerl, F.; Koppensteiner, R.; Schernthaner, G.; Schober, E.; Schernthaner, G.H. Endothelial progenitor cells are related to glycemic control in children with type 1 diabetes over time. Diabetes Care 2013, 36, 1647–1653. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Loomans, C.J.; de Koning, E.J.; Staal, F.J.; Rookmaaker, M.B.; Verseyden, C.; de Boer, H.C.; Verhaar, M.C.; Braam, B.; Rabelink, T.J.; van Zonneveld, A.J. Endothelial progenitor cell dysfunction: A novel concept in the pathogenesis of vascular complications of type 1 diabetes. Diabetes 2004, 53, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Colagiuri, S.; Lee, C.M.; Wong, T.Y.; Balkau, B.; Shaw, J.E.; Borch-Johnsen, K.; Group, D.-C.W. Glycemic thresholds for diabetes-specific retinopathy: Implications for diagnostic criteria for diabetes. Diabetes Care 2011, 34, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Blondal, T.; Jensby Nielsen, S.; Baker, A.; Andreasen, D.; Mouritzen, P.; Wrang Teilum, M.; Dahlsveen, I.K. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods 2013, 59, S1–S6. [Google Scholar] [CrossRef] [PubMed]
- Coulson, D.J.; Bakhashab, S.; Latief, J.S.; Weaver, J.U. MiR-126, IL-7, CXCR1/2 receptors, inflammation and circulating endothelial progenitor cells: The study on targets for treatment pathways in a model of subclinical cardiovascular disease (type 1 diabetes mellitus). J. Transl. Med. 2021, 19, 140. [Google Scholar] [CrossRef]
- Karagkouni, D.; Paraskevopoulou, M.D.; Chatzopoulos, S.; Vlachos, I.S.; Tastsoglou, S.; Kanellos, I.; Papadimitriou, D.; Kavakiotis, I.; Maniou, S.; Skoufos, G.; et al. DIANA-TarBase v8: A decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res 2018, 46, D239–D245. [Google Scholar] [CrossRef]
- Vasa, M.; Fichtlscherer, S.; Aicher, A.; Adler, K.; Urbich, C.; Martin, H.; Zeiher, A.M.; Dimmeler, S. Number and migratory activity of circulating endothelial progenitor cells inversely correlate with risk factors for coronary artery disease. Circ. Res. 2001, 89, e1–e7. [Google Scholar] [CrossRef]

| Target Gene | Representative Transcript | Gene Name | Transcript Position | Predicted Consequential Pairing of Target Region in Transcript (Top) and miRNA (Bottom) | Site Type |
|---|---|---|---|---|---|
| EIF2S1 | ENSG00000134001 | Eukaryotic Translation initiation factor 2 Subunit α | 58–85 3′UTR | (Transcript) 5′AUCCUAGACUUGAAAGUUUUCCAGUAUUG3′ (miRNA) 3′AGGUAGUAAUGGGCCGUCAUAAU5′ | 7mer |
| IKBKB | ENSG00000104365 | Inhibitor of Nuclear Factor Kappa B Kinase Subunit β | 2719–2739 3′UTR | (Transcript) 5′CCUGUCUCUCACAGCAUCUACAGUAUUA3′ (miRNA) 3′AGGUAGUAAUGGGCCGUCAUAAU5′ | 9mer |
| JUN | ENSG00000177606 | Jun Proto-Oncogene, AP-1 Transcription Factor Subunit | 944–970 3′UTR | (Transcript) 5′GACCUAACAUUCGAUCUCAUUCAGUAUUA3′ (miRNA) 3′AGGUAGUAAUGGGCCGUCAUAAU5′ | 8mer |
| KDR | ENSG00000128052 | Kinase insert domain receptor (VEGFR-2) | 427–445 3′UTR | (Transcript) 5′CACUCUCACCCCGCAACCCAUCAGUAUUU3′ (miRNA) 3′AGGUAGUAAUGGGCCGUCAUAAU5′ | 7mer |
| MAP3K5 | ENSG00000197442 | Mitogen-Activated Protein Kinase Kinase Kinase 5 | 595–617 3′UTR | (Transcript) 5′AAAGGCGCUGCACUUUAAAUCCAGUAUUU3′ (miRNA) 3′AGUAAUGGGCCGUCAUAAU5′ | 7mer |
| PIK3CA | ENSG00000121879 | Protein Phosphatase 2 Catalytic Subunit Alpha | 702–708 3′UTR | (Transcript) 5’UUUUUUUCUUCUGGACAGUAUUU3′ (miRNA) 3’AGGUAGUAAUGGGCCGUCAUAAU5′ | 7mer-8m |
| PLCG1 | ENSG00000124181 | Phospholipase C Gamma 1 | 618–640 3′UTR | (Transcript) 5′AGGGGAUCAUGUUAAAAAUAGCAGUAUUA3′ (miRNA) 3′AGGUAGUAAUGGGCCGUCAUAAU5′ | 9mer |
| PPP2CA | ENSG00000113575 | Protein Phosphatase 2 Catalytic Subunit Alpha | 19–25 3′UTR | (Transcript) 5’AAUUUUAAACUUGUACAGUAUUG3′ (miRNA) 3’AGGUAGUAAUGGGCCGUCAUAAU5′ | 7mer-m8 |
| RAP1B | ENSG00000127314 | RAP1B, Member Of RAS Oncogene Family | 235–256 3′UTR | (Transcript) 5′GGAAAACAGAGGCUACAUCCAGUAUUA3′ (miRNA) 3′AGGUAGUAAUGGGCCGUCAUAAU5′ | 8mer |
| RPS6KB1 | ENSG00000108443 | Ribosomal protein S6 kinase B1 | 291–298 3′UTR | (Transcript) 5’AUGACUCGAAACUGACAGUAUUA3′ (miRNA) 3’AGGUAGUAAUGGGCCGUCAUAAU5′ | 8mer |
| VEGFA | ENSG00000112715 | Vascular endothelial growth factor A | 1284–1303 3′UTR | (Transcript) 5′UUAAAGAGUAGGGUUUUUUUUCAGUAUUC3′ (miRNA) 3′GGUAGUAAUGGGCCGUCAUAAU5′ | 7mer |
| YWHAG | ENSG00000170027 | Tyrosine 3-Monooxygenase/Tryptophan 5-Monooxygenase | 564–571 3′UTR | (Transcript) 5’UCUCUGAUUAGUUGACAGUAUUA3′ (miRNA) 3’AGGUAGUAAUGGGCCGUCAUAAU5′ | 8mer |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bakhashab, S.; Yuen Yeoh, M.L.; Coulson, D.J.; Steel, S.C.; Ray, S.L.; Weaver, J.U. Deciphering the Role of miR-200c-3p in Type 1 Diabetes (Subclinical Cardiovascular Disease) and Its Correlation with Inflammation and Vascular Health. Int. J. Mol. Sci. 2022, 23, 15659. https://doi.org/10.3390/ijms232415659
Bakhashab S, Yuen Yeoh ML, Coulson DJ, Steel SC, Ray SL, Weaver JU. Deciphering the Role of miR-200c-3p in Type 1 Diabetes (Subclinical Cardiovascular Disease) and Its Correlation with Inflammation and Vascular Health. International Journal of Molecular Sciences. 2022; 23(24):15659. https://doi.org/10.3390/ijms232415659
Chicago/Turabian StyleBakhashab, Sherin, Megan Li Yuen Yeoh, David J. Coulson, Samuel Christian Steel, Sabina L. Ray, and Jolanta U. Weaver. 2022. "Deciphering the Role of miR-200c-3p in Type 1 Diabetes (Subclinical Cardiovascular Disease) and Its Correlation with Inflammation and Vascular Health" International Journal of Molecular Sciences 23, no. 24: 15659. https://doi.org/10.3390/ijms232415659
APA StyleBakhashab, S., Yuen Yeoh, M. L., Coulson, D. J., Steel, S. C., Ray, S. L., & Weaver, J. U. (2022). Deciphering the Role of miR-200c-3p in Type 1 Diabetes (Subclinical Cardiovascular Disease) and Its Correlation with Inflammation and Vascular Health. International Journal of Molecular Sciences, 23(24), 15659. https://doi.org/10.3390/ijms232415659





