Meta-Analysis of Extracellular Matrix Dynamics after Myocardial Infarction Using RNA-Sequencing Transcriptomic Database
Abstract
1. Introduction
2. Results
2.1. Characteristics of the Selected Studies
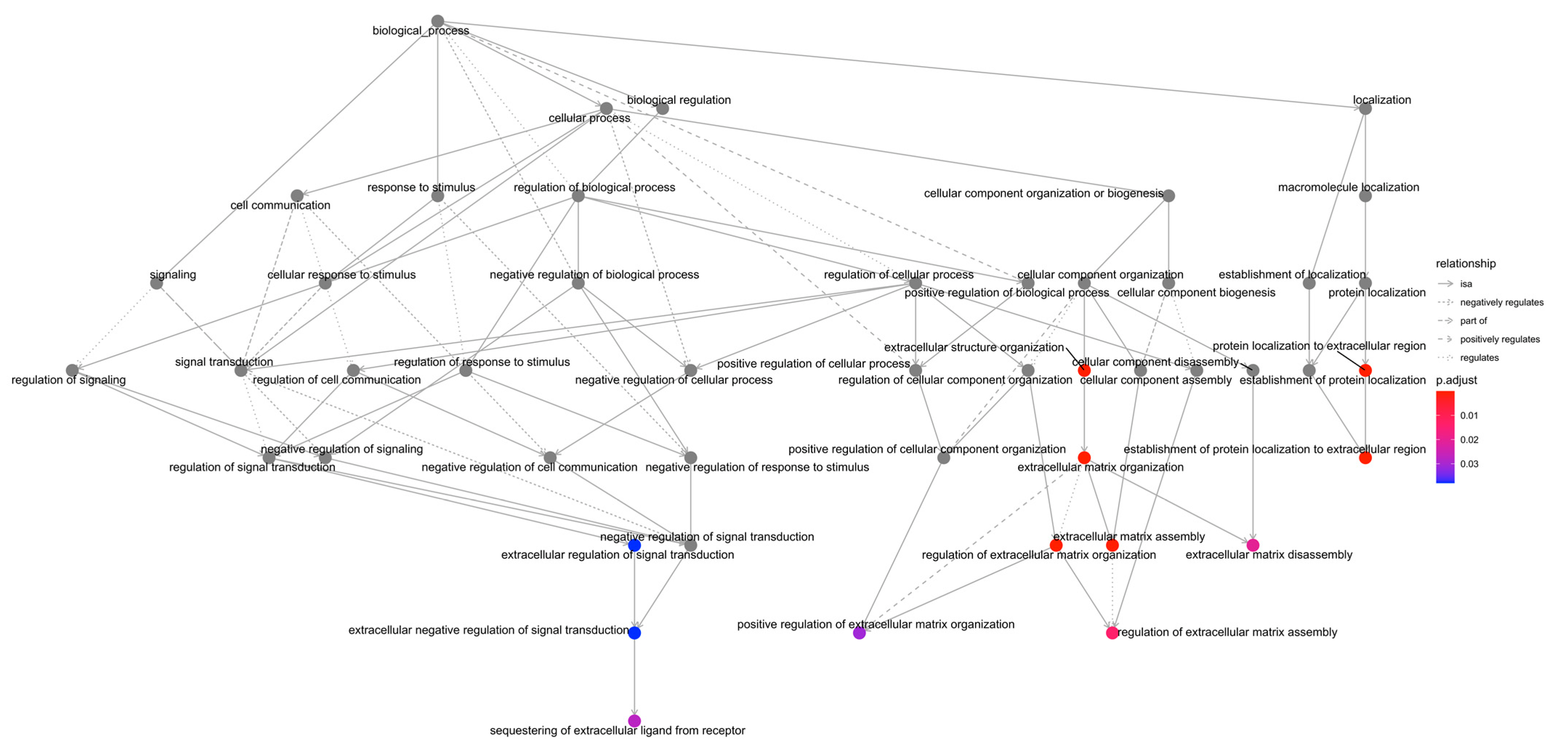
2.2. GO Enrichment Analysis
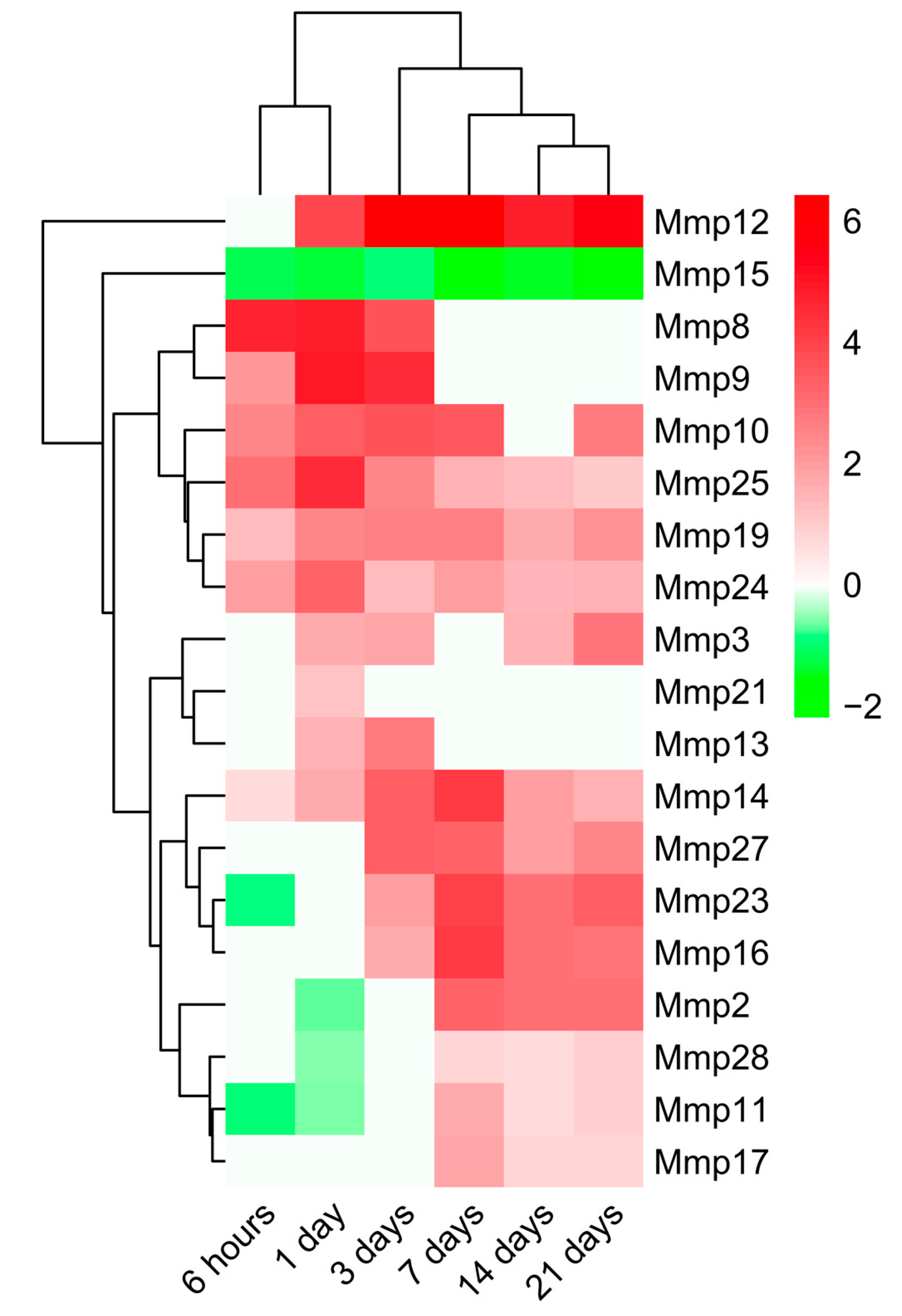
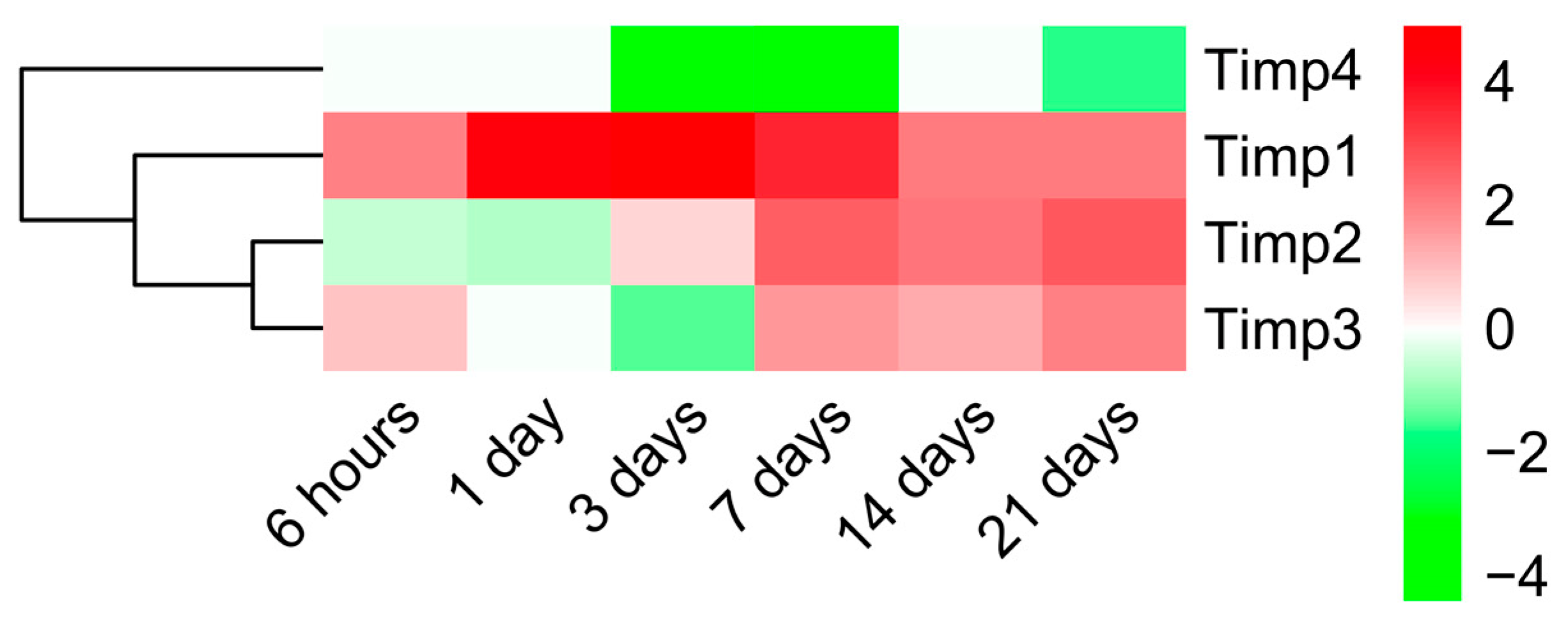
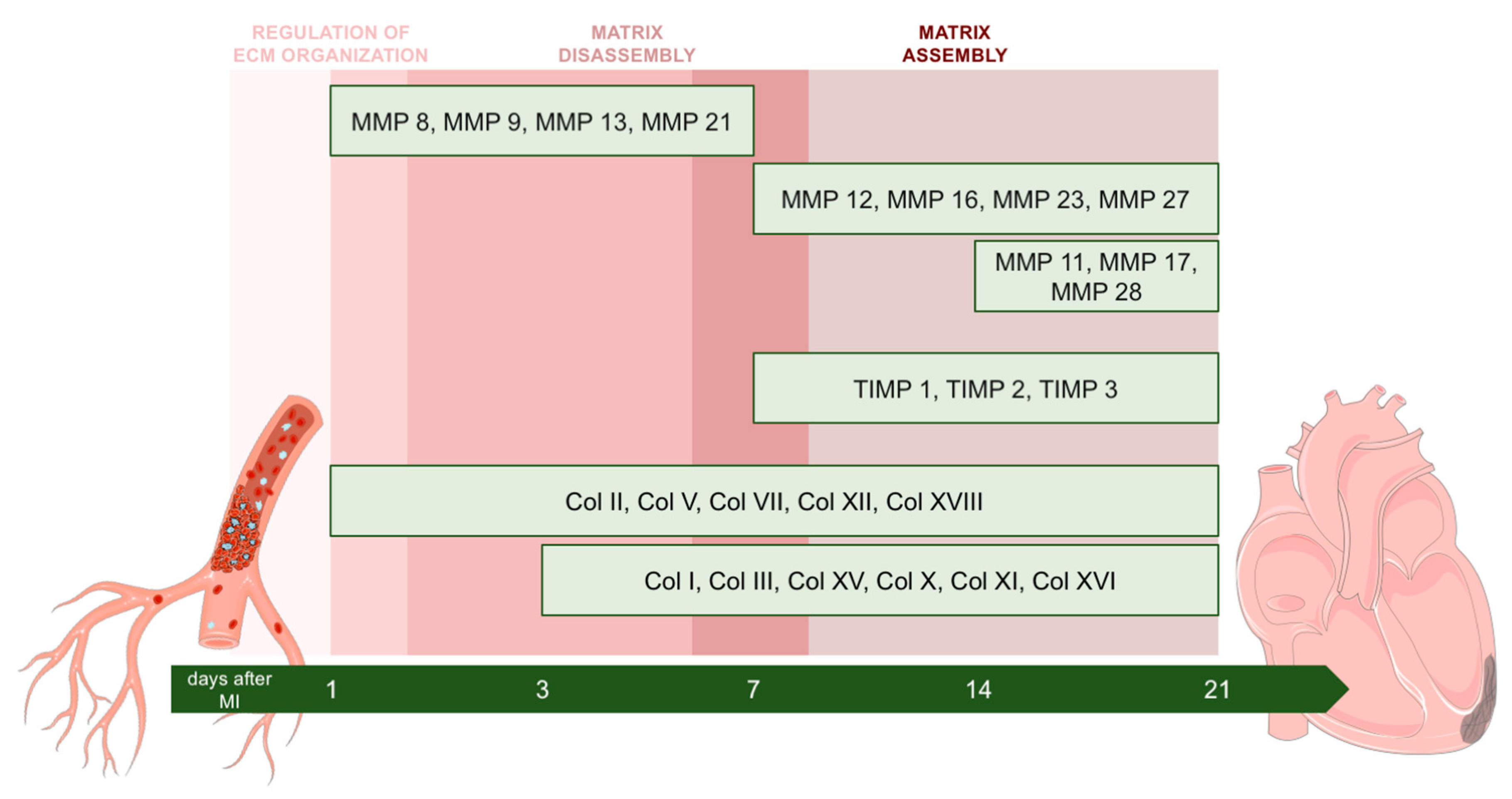
2.3. Dynamic Changes in Genes Encoding MMPs and TIMPs in the Infarcted Area Post-MI
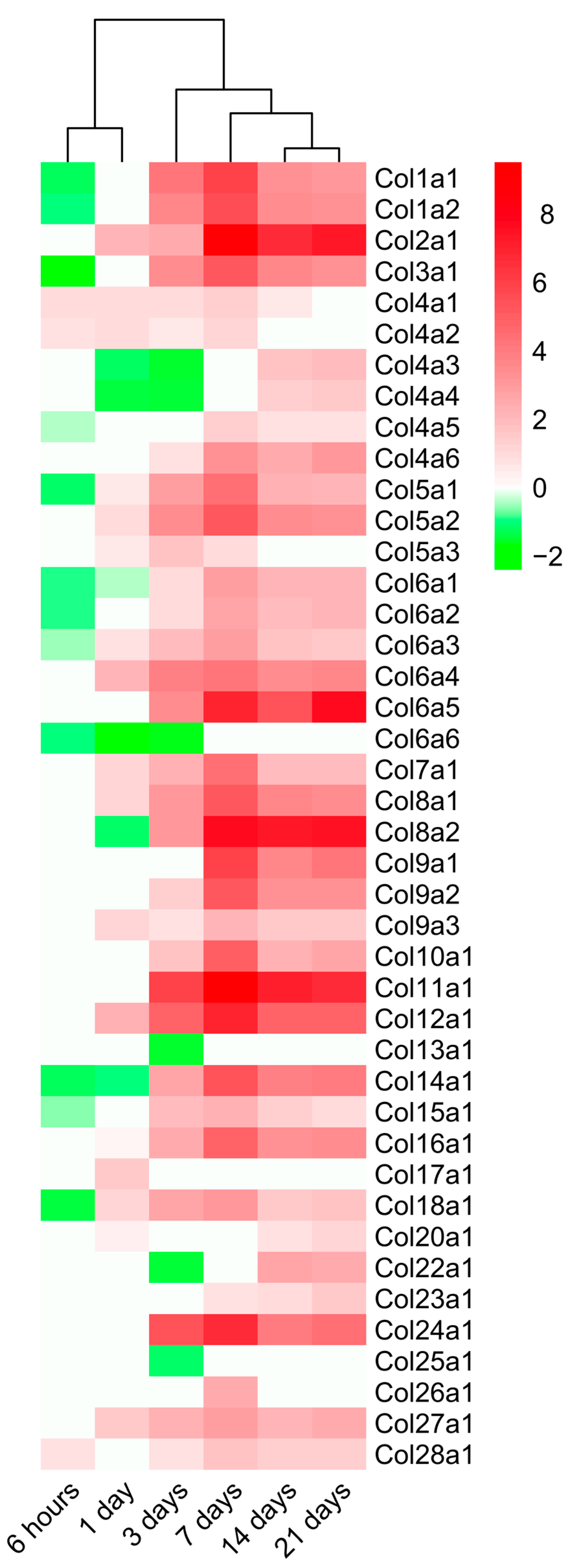
2.4. Dynamic Alterations in Transcriptomic Analysis of Genes Encoding Collagen Subunits
3. Discussion
3.1. Role of Cardiac Interstitium Following MI
3.2. RNA-Sequencing Technology to Further Comprehend Mechanisms Underlying ECM Changes
3.3. Alterations in MMP and TIMP Expression after MI
3.4. Dynamic Changes in the Expression of Genes Implicated in Collagen Synthesis
3.5. Limitations of the Study
4. Materials and Methods
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Frangogiannis, N.G. The extracellular matrix in ischemic and nonischemic heart failure. Circ. Res. 2019, 125, 117–146. [Google Scholar] [CrossRef] [PubMed]
- Theocharis, A.D.; Manou, D.; Karamanos, N.K. The extracellular matrix as a multitasking player in disease. FEBS J. 2019, 286, 2830–2869. [Google Scholar] [CrossRef]
- Rios-Navarro, C.; Ortega, M.; Marcos-Garces, V.; Gavara, J.; de Dios, E.; Perez-Sole, N.; Chorro, F.J.; Bodi, V.; Ruiz-Sauri, A. Interstitial changes after reperfused myocardial infarction in swine: Morphometric and genetic analysis. BMC Vet. Res. 2020, 16, 262–266. [Google Scholar] [CrossRef] [PubMed]
- Ibanez, B.; Heusch, G.; Ovize, M.; Van de Werf, F. Evolving therapies for myocardial ischemia/reperfusion injury. J. Am. Coll. Cardiol. 2015, 65, 1454–1471. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, S.H.; Mouton, A.J.; DeLeon-Pennell, K.Y.; Genovese, F.; Karsdal, M.; Lindsey, M.L. Understanding cardiac extracellular matrix remodeling to develop biomarkers of myocardial infarction outcomes. Matrix Biol. 2019, 75–76, 43–57. [Google Scholar] [CrossRef]
- Hervas, A.; Ruiz-Sauri, A.; Gavara, J.; Monmeneu, J.V.; de Dios, E.; Rios-Navarro, C.; Perez-Sole, N.; Perez, I.; Monleon, D.; Morales, J.M.; et al. A Multidisciplinary Assessment of Remote Myocardial Fibrosis After Reperfused Myocardial Infarction in Swine and Patients. J. Cardiovasc. Transl. Res. 2016, 9, 321–333. [Google Scholar] [CrossRef]
- Frangogiannis, N.G. The extracellular matrix in myocardial injury, repair, and remodeling. J. Clin. Investig. 2017, 127, 1600–1612. [Google Scholar] [CrossRef] [PubMed]
- Yokota, T.; McCourt, J.; Ma, F.; Ren, S.; Li, S.; Kim, T.H.; Kurmangaliyev, Y.Z.; Nasiri, R.; Ahadian, S.; Nguyen, T.; et al. Type V Collagen in Scar Tissue Regulates the Size of Scar after Heart Injury. Cell 2020, 182, 545–562. [Google Scholar] [CrossRef]
- Liu, W.; Shen, J.; Li, Y.; Wu, J.; Luo, X.; Yu, Y.; Zhang, Y.; Gu, L.; Zhang, X.; Jiang, C.; et al. Pyroptosis inhibition improves the symptom of acute myocardial infarction. Cell Death Dis. 2021, 12, 852–853. [Google Scholar] [CrossRef] [PubMed]
- Park, T.J.; Park, J.H.; Lee, G.S.; Lee, J.Y.; Shin, J.H.; Kim, M.W.; Kim, Y.S.; Kim, J.Y.; Oh, K.J.; Han, B.S.; et al. Quantitative proteomic analyses reveal that GPX4 downregulation during myocardial infarction contributes to ferroptosis in cardiomyocytes. Cell Death Dis. 2019, 10, 835–838. [Google Scholar] [CrossRef]
- Arif, M.; Klevstig, M.; Benfeitas, R.; Doran, S.; Turkez, H.; Uhlen, M.; Clausen, M.; Wikstrom, J.; Etal, D.; Zhang, C.; et al. Integrative transcriptomic analysis of tissue-specific metabolic crosstalk after myocardial infarction. Elife 2021, 10, e66921. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.L.; Walton, C.B.; Pinell, B.; Khadka, V.S.; Dunn, B.; Lee, K.; Anagaran, M.C.T.; Avelar, A.; Shohet, R.V. Ischemic heart injury leads to HIF1-dependent differential splicing of CaMK2gamma. Sci. Rep. 2021, 11, 13116. [Google Scholar] [CrossRef]
- Olsen, M.B.; Hildrestrand, G.A.; Scheffler, K.; Vinge, L.E.; Alfsnes, K.; Palibrk, V.; Wang, J.; Neurauter, C.G.; Luna, L.; Johansen, J.; et al. NEIL3-Dependent Regulation of Cardiac Fibroblast Proliferation Prevents Myocardial Rupture. Cell Rep. 2017, 18, 82–92. [Google Scholar] [CrossRef]
- Niccoli, G.; Montone, R.A.; Ibanez, B.; Thiele, H.; Crea, F.; Heusch, G.; Bulluck, H.; Hausenloy, D.J.; Berry, C.; Stiermaier, T.; et al. Optimized Treatment of ST-Elevation Myocardial Infarction. Circ. Res. 2019, 125, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Crespo-Avilan, G.E.; Mukhametshina, R.T.; Kwek, X.Y.; Cabrera-Fuentes, H.A.; Hausenloy, D.J. Inflammation following acute myocardial infarction: Multiple players, dynamic roles, and novel therapeutic opportunities. Pharmacol. Ther. 2018, 186, 73–87. [Google Scholar]
- Prabhu, S.D.; Frangogiannis, N.G. The Biological Basis for Cardiac Repair After Myocardial Infarction: From Inflammation to Fibrosis. Circ. Res. 2016, 119, 91–112. [Google Scholar] [CrossRef] [PubMed]
- Wight, T.N.; Vernon, R.B. Targeting the extracellular matrix promotes healing following myocardial infarction. Circ. Res. 2019, 1255, 802–804. [Google Scholar] [CrossRef]
- Yan, X.; Anzai, A.; Katsumata, Y.; Matsuhashi, T.; Ito, K.; Endo, J.; Yamamoto, T.; Takeshima, A.; Shinmura, K.; Shen, W.; et al. Temporal dynamics of cardiac immune cell accumulation following acute myocardial infarction. J. Mol. Cell. Cardiol. 2013, 62, 24–35. [Google Scholar] [CrossRef]
- Arpino, V.; Brock, M.; Gill, S.E. The role of TIMPs in regulation of extracellular matrix proteolysis. Matrix Biol. 2015, 44–46, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Cabral-Pacheco, G.A.; Garza-Veloz, I.; Castruita-De la Rosa, C.; Ramirez-Acuña, J.M.; Perez-Romero, B.A.; Guerrero-Rodriguez, J.F.; Martinez-Avila, N.; Martinez-Fierro, M.L. The roles of matrix metalloproteinases and their inhibitors in human diseases. Int. J. Mol. Sci. 2020, 21, 9739. [Google Scholar] [CrossRef] [PubMed]
- Kaminski, A.R.; Moore, E.T.; Daseke, M.J.; Valerio, F.M.; Flynn, E.R.; Lindsey, M.L. The compendium of matrix metalloproteinase expression in the left ventricle of mice following myocardial infarction. Am. J. Physiol. Heart Circ. Physiol. 2020, 318, H706–H714. [Google Scholar] [CrossRef] [PubMed]
- DeLeon-Pennell, K.Y.; Meschiari, C.A.; Jung, M.; Lindsey, M.L. Matrix Metalloproteinases in Myocardial Infarction and Heart Failure. Prog. Mol. Biol. Transl. Sci. 2017, 147, 75–100. [Google Scholar] [PubMed]
- Chen, J.; Tung, C.H.; Allport, J.R.; Chen, S.; Weissleder, R.; Huang, P.L. Near-infrared fluorescent imaging of matrix metalloproteinase activity after myocardial infarction. Circulation 2005, 111, 1800–1805. [Google Scholar] [CrossRef] [PubMed]
- Iyer, R.P.; Jung, M.; Lindsey, M.L. MMP-9 signaling in the left ventricle following myocardial infarction. Am. J. Physiol. Heart Circ. Physiol. 2016, 311, 190. [Google Scholar] [CrossRef]
- Lindsey, M.L. Assigning matrix metalloproteinase roles in ischaemic cardiac remodelling. Nat. Rev. Cardiol. 2018, 15, 471–479. [Google Scholar] [CrossRef]
- Ma, Y.; Halade, G.V.; Zhang, J.; Ramirez, T.A.; Levin, D.; Voorhees, A.; Jin, Y.F.; Han, H.C.; Manicone, A.M.; Lindsey, M.L. Matrix metalloproteinase-28 deletion exacerbates cardiac dysfunction and rupture after myocardial infarction in mice by inhibiting M2 macrophage activation. Circ. Res. 2013, 112, 675–688. [Google Scholar] [CrossRef]
- Talman, V.; Ruskoaho, H. Cardiac fibrosis in myocardial infarction—From repair and remodeling to regeneration. Cell Tissue Res. 2016, 365, 563–581. [Google Scholar] [CrossRef]
- Marcos-Garcés, V.; Rios-Navarro, C.; Gómez-Torres, F.; Gavara, J.; de Dios, E.; Diaz, A.; Miñana, G.; Chorro, F.J.; Bodi, V.; Ruiz-Sauri, A. Fourier analysis of collagen bundle orientation in myocardial infarction scars. Histochem. Cell Biol. 2022, 158, 471–483. [Google Scholar] [CrossRef]
- Hervas, A.; Ruiz-Sauri, A.; de Dios, E.; Forteza, M.J.; Minana, G.; Nunez, J.; Gomez, C.; Bonanad, C.; Perez-Sole, N.; Gavara, J.; et al. Inhomogeneity of collagen organization within the fibrotic scar after myocardial infarction: Results in a swine model and in human samples. J. Anat. 2016, 228, 47–58. [Google Scholar] [CrossRef]
- Barallobre-Barreiro, J.; Didangelos, A.; Schoendube, F.A.; Drozdov, I.; Yin, X.; Fernandez Caggiano, M.; Willeit, P.; Puntmann, V.O.; Aldama-Lopez, G.; Shah, A.M.; et al. Proteomics analysis of cardiac extracellular matrix remodeling in a porcine model of ischemia/reperfusion injury. Circulation 2012, 125, 789–802. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- RStudio Team. RStudio: Integrated Development for R; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550–558. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; Available online: https://ggplot2tidyverse.org (accessed on 11 November 2021).
- Conway, J.R.; Lex, A.; Gehlenborg, N. UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics 2017, 33, 2938–2940. [Google Scholar] [CrossRef]
- Kolde, R. Pheatmap: Pretty Heatmaps; R Foundation for Statistical Computing: Vienna, Austria, 2015. [Google Scholar]
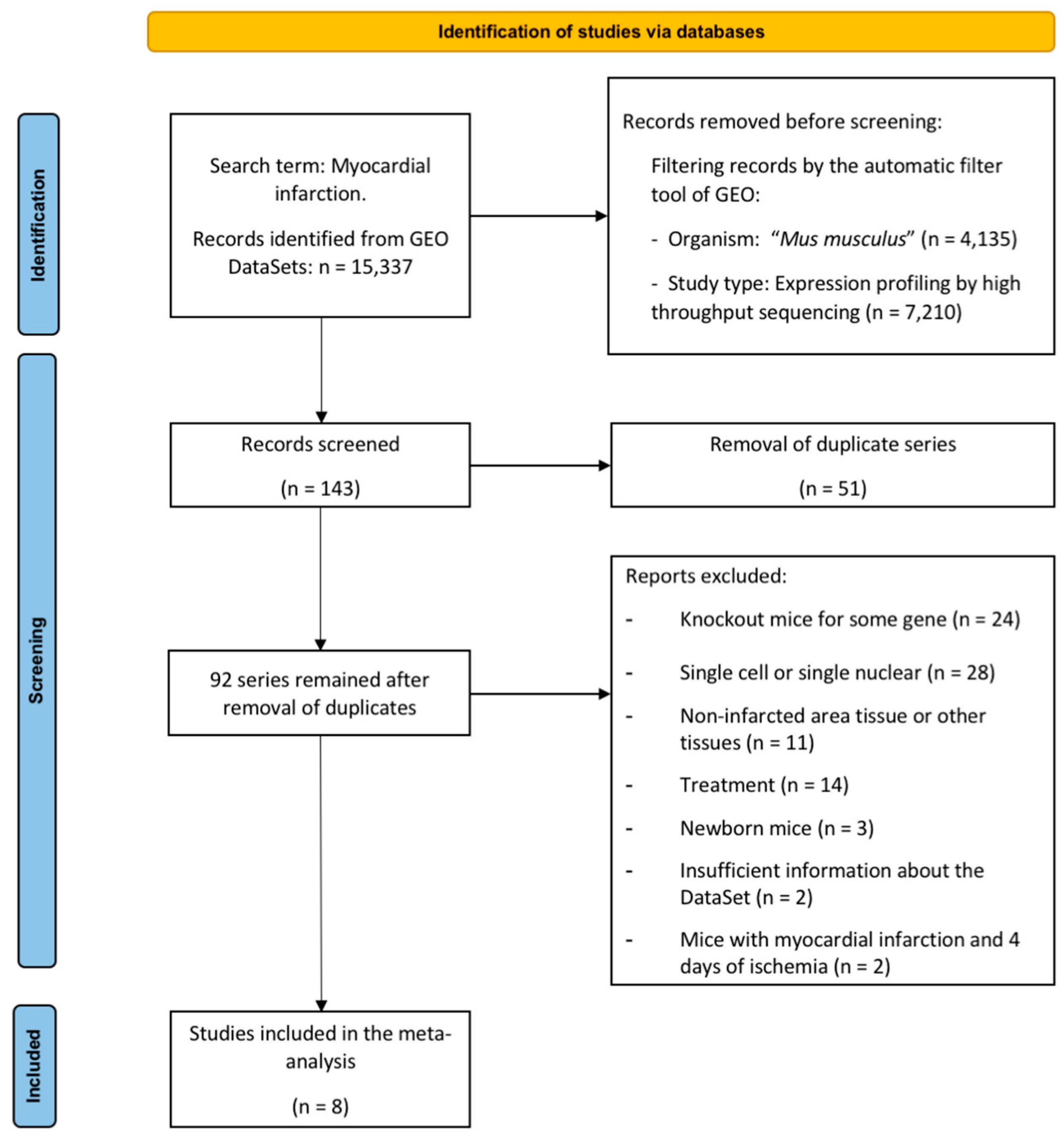

| GEODatasets | Reference | Number of Samples | ||||||
|---|---|---|---|---|---|---|---|---|
| Sham | 6 h | 1 Day | 3 Days | 7 Days | 14 Days | 21 Days | ||
| GSE153494 | [9] | 3 | 3 | 3 | 3 | |||
| GSE153493 | N/A | 3 | 3 | 3 | ||||
| GSE151834 | [8] | 4 | 4 | 4 | 8 | |||
| GSE114695 | [10] | 9 | 3 | 3 | 3 | |||
| GSE153485 | [11] | 10 | 5 | 5 | ||||
| GSE154072 | N/A | 3 | ||||||
| GSE104187 | [12] | 4 | 2 | 2 | ||||
| GSE83350 | [13] | 1 | 1 | |||||
| Total | 30 | 8 | 16 | 10 | 7 | 10 | 11 | |
| Gene | Name | 6 h | 1 Day | 3 Days | 7 Days | 14 Days | 21 Days | |
|---|---|---|---|---|---|---|---|---|
| MATRIX DISASSEMBLY | Fgfr4 | Tyrosine Kinase Related To Fibroblast Growth Factor Receptor | X | |||||
| Il6 | Interleukin-6 | X | X | X | ||||
| Pdpn | Glycoprotein 36 | X | X | X | ||||
| Tgfb2 | Transforming Growth Factor Beta 2 | X | X | |||||
| Lcp1 | Lymphocyte Cytosolic Protein 1 | X | X | |||||
| Fscn1 | Fascin Actin-Bundling Protein 1 | X | ||||||
| Ctss | Cathepsin S | X | X | |||||
| Sh3pxd2b | Tyrosine Kinase Substrate With Four SH3 Domains | X | X | |||||
| Mmp13 | Matrix Metallopeptidase 13 | X | ||||||
| Fap | Fibroblast Activation Protein Alpha | X | ||||||
| Kif9 | Kinesin Family Member 9 | X | ||||||
| MATRIX ASSEMBLY | Sox9 | SRY-Box Transcription Factor 9 | X | X | X | |||
| Fkbp10 | FKBP Prolyl Isomerase 10 | X | ||||||
| Fbln5 | Fibulin 5 | X | X | X | ||||
| Has2 | Hyaluronan Synthase 2 | X | ||||||
| Lox | Lysyl Oxidase | X | X | X | ||||
| Efemp2 | EGF Containing Fibulin Extracellular Matrix Protein 2 | X | ||||||
| Ltbp3 | Latent Transforming Growth Factor Beta Binding Protein 3 | X | X | X | ||||
| Emilin1 | Elastin Microfibril Interfacer 1 | X | ||||||
| Col1a2 | Collagen Type I Alpha 2 Chain | X | X | X | ||||
| Phldb2 | Pleckstrin homology Like Domain Family B Member 2 | X | ||||||
| Antxr1 | ANTXR Cell Adhesion Molecule 1 | X | X | X | ||||
| Mfap4 | Microfibril Associated Protein 4 | X | X | X | ||||
| Gpm6b | Glycoprotein M6B | X |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ortega, M.; Ríos-Navarro, C.; Gavara, J.; de Dios, E.; Perez-Solé, N.; Marcos-Garcés, V.; Ferrández-Izquierdo, A.; Bodí, V.; Ruiz-Saurí, A. Meta-Analysis of Extracellular Matrix Dynamics after Myocardial Infarction Using RNA-Sequencing Transcriptomic Database. Int. J. Mol. Sci. 2022, 23, 15615. https://doi.org/10.3390/ijms232415615
Ortega M, Ríos-Navarro C, Gavara J, de Dios E, Perez-Solé N, Marcos-Garcés V, Ferrández-Izquierdo A, Bodí V, Ruiz-Saurí A. Meta-Analysis of Extracellular Matrix Dynamics after Myocardial Infarction Using RNA-Sequencing Transcriptomic Database. International Journal of Molecular Sciences. 2022; 23(24):15615. https://doi.org/10.3390/ijms232415615
Chicago/Turabian StyleOrtega, María, César Ríos-Navarro, Jose Gavara, Elena de Dios, Nerea Perez-Solé, Victor Marcos-Garcés, Antonio Ferrández-Izquierdo, Vicente Bodí, and Amparo Ruiz-Saurí. 2022. "Meta-Analysis of Extracellular Matrix Dynamics after Myocardial Infarction Using RNA-Sequencing Transcriptomic Database" International Journal of Molecular Sciences 23, no. 24: 15615. https://doi.org/10.3390/ijms232415615
APA StyleOrtega, M., Ríos-Navarro, C., Gavara, J., de Dios, E., Perez-Solé, N., Marcos-Garcés, V., Ferrández-Izquierdo, A., Bodí, V., & Ruiz-Saurí, A. (2022). Meta-Analysis of Extracellular Matrix Dynamics after Myocardial Infarction Using RNA-Sequencing Transcriptomic Database. International Journal of Molecular Sciences, 23(24), 15615. https://doi.org/10.3390/ijms232415615








