Translational Results of Zo-NAnTax: A Phase II Trial of Neoadjuvant Zoledronic Acid in HER2-Positive Breast Cancer
Abstract
1. Introduction
2. Results
2.1. Clinical Results
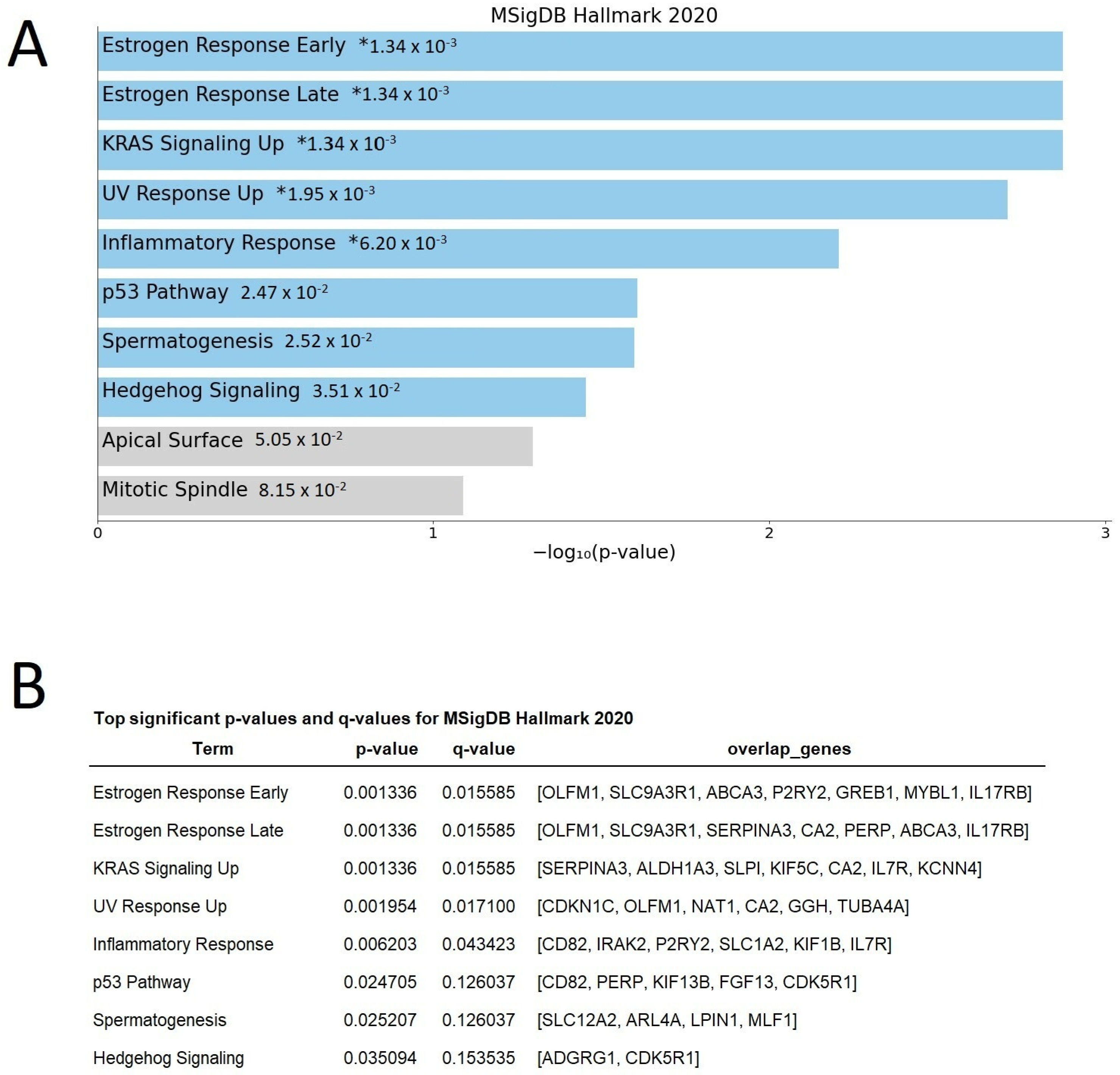
2.2. Microarray Analysis Revealed Differentially Expressed Genes at Diagnosis Related to Response
2.3. Differentially Expressed Genes Indicate Growth Factors and Metabolism Signaling as Those Pathways Related to Zo-NAnTax Response
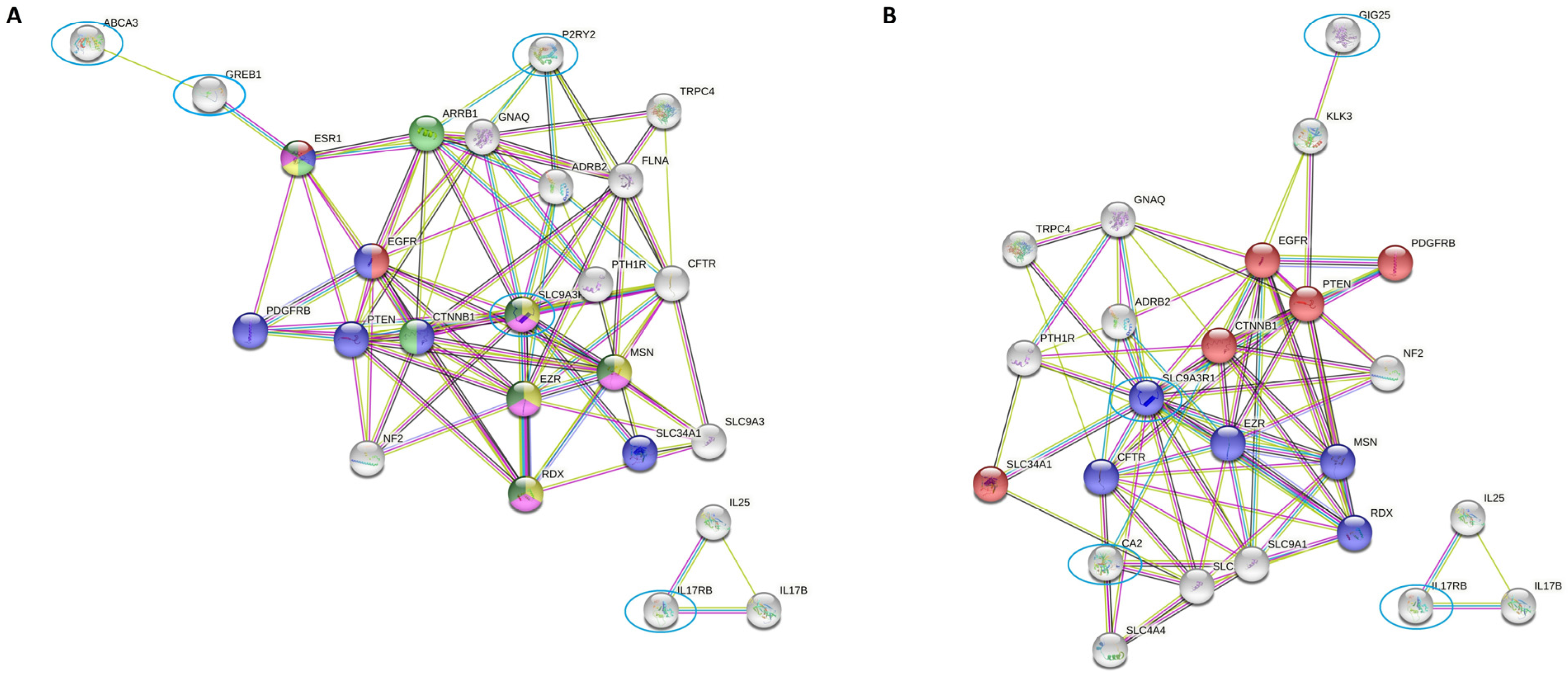
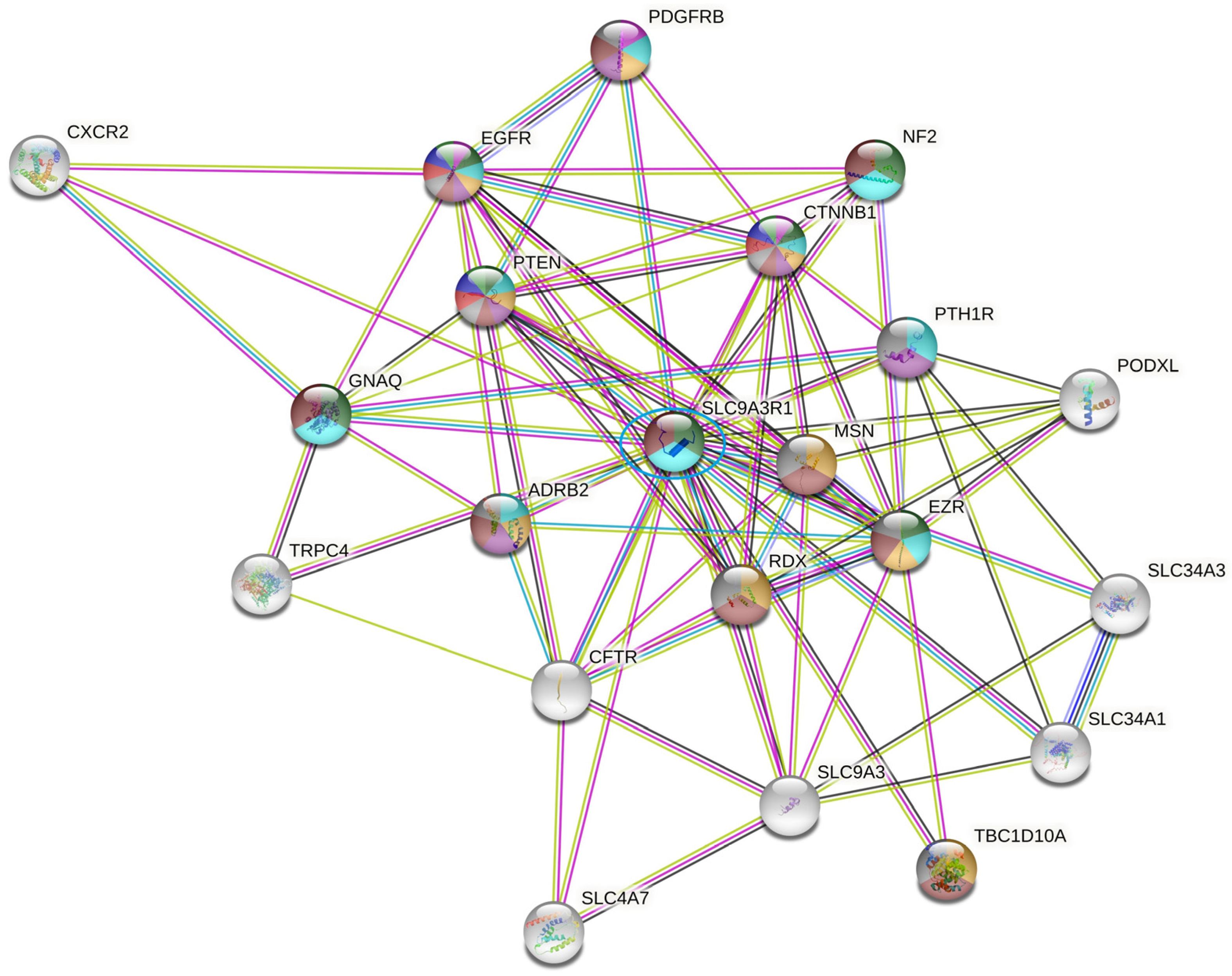
2.4. SLC9AR1 Is Highlighted as a Possible Candidate in HR-Positive/HER2-Positive Patient Outcome
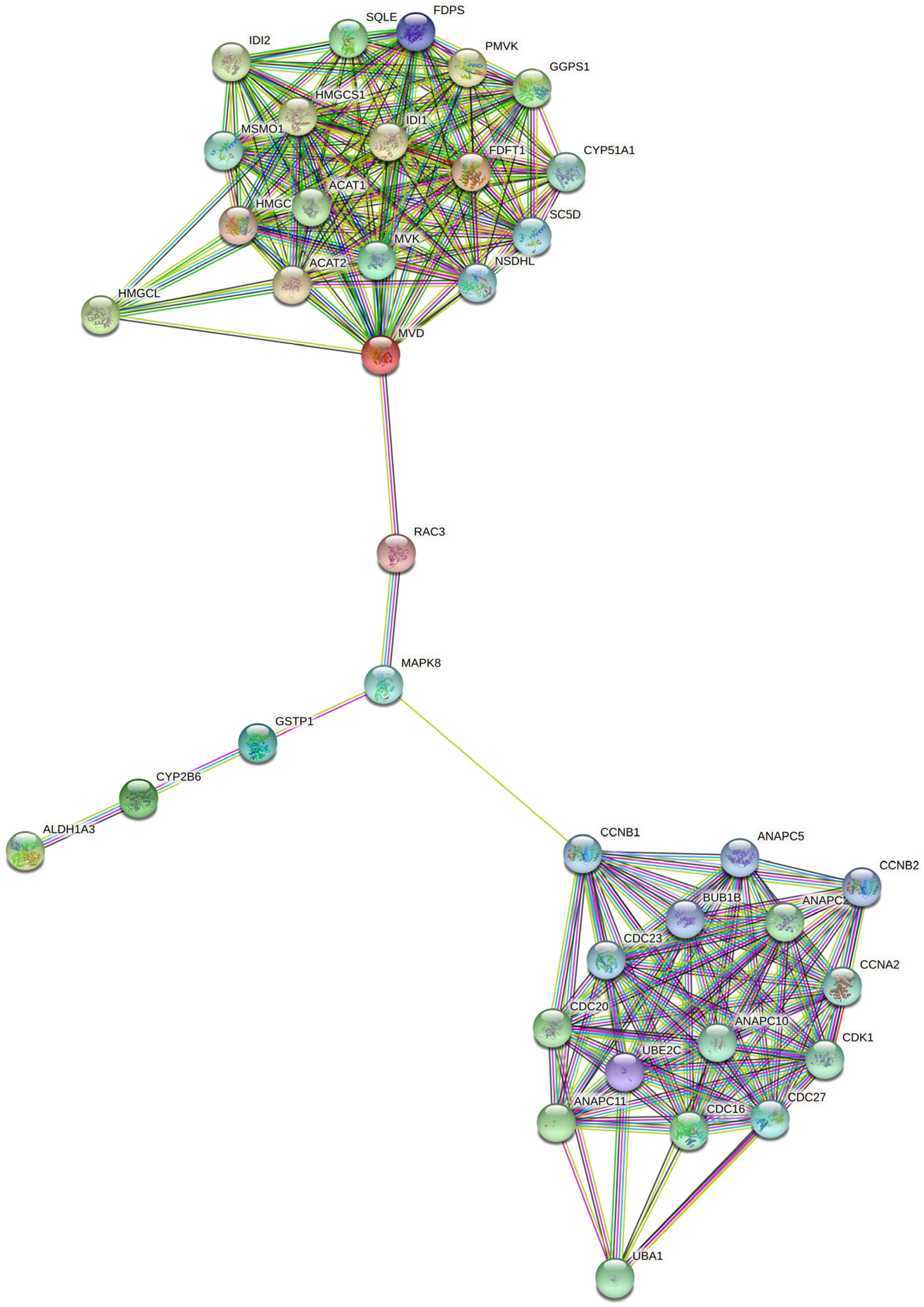
2.5. RAC3 Gene Links Growth Factor Signaling with Mevalonate Pathway
3. Discussion
4. Materials and Methods
4.1. Zo-NAnTAx Study Design
4.2. Tissue Samples
4.3. Microarray
4.4. In Silico Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Slamon, D.J.; Clark, G.M.; Wong, S.G.; Levin, W.J.; Ullrich, A.; McGuire, W.L. Human breast cancer: Correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987, 9, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Slamon, D.J.; Godolphin, W.; Jones, L.A.; Holt, J.A.; Wong, S.G.; Keith, D.E.; Levin, W.J.; Stuart, S.G.; Udove, J.; Ullrich, A.; et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science 1989, 12, 707–712. [Google Scholar] [CrossRef] [PubMed]
- Cheang, M.C.; Chia, S.K.; Voduc, D.; Gao, D.; Leung, S.; Snider, J.; Watson, M.; Davies, S.; Bernard, P.S.; Parker, J.S.; et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J. Natl. Cancer Inst. 2009, 101, 736–750. [Google Scholar] [CrossRef] [PubMed]
- Gianni, L.; Eiermann, W.; Semiglazov, V.; Lluch, A.; Tjulandin, S.; Zambetti, M.; Moliterni, A.; Vazquez, F.; Byakhov, M.J.; Lichinitser, M.; et al. Neoadjuvant and adjuvant trastuzumab in patients with HER2-positive locally advanced breast cancer (NOAH): Follow-up of a randomised controlled superiority trial with a parallel HER2-negative cohort. Lancet Oncol. 2014, 15, 640–647, Erratum in: Lancet Oncol. 2018, 19, e667. [Google Scholar] [CrossRef]
- Perez, E.A.; Romond, E.H.; Suman, V.J.; Jeong, J.H.; Sledge, G.; Geyer, C.E., Jr.; Martino, S.; Rastogi, P.; Gralow, J.; Swain, S.M.; et al. Trastuzumab plus adjuvant chemotherapy for human epidermal growth factor receptor 2-positive breast cancer: Planned joint analysis of overall survival from NSABP B-31 and NCCTG N9831. J. Clin. Oncol. 2014, 32, 3744–3752. [Google Scholar] [CrossRef]
- Hudis, C.A. Trastuzumab—mechanism of action and use in clinical practice. N. Engl. J. Med. 2007, 357, 39–51. [Google Scholar] [CrossRef]
- Rexer, B.N.; Arteaga, C.L. Intrinsic and acquired resistance to HER2-targeted therapies in HER2 gene-amplified breast cancer: Mechanisms and clinical implications. Crit. Rev. Oncog. 2012, 17, 1–16. [Google Scholar] [CrossRef]
- Wynn, C.S.; Tang, S.C. Anti-HER2 therapy in metastatic breast cancer: Many choices and future directions. Cancer Metastasis Rev. 2022, 41, 193–209. [Google Scholar] [CrossRef]
- Broglio, K.R.; Quintana, M.; Foster, M.; Olinger, M.; McGlothlin, A.; Berry, S.M.; Boileau, J.F.; Brezden-Masley, C.; Chia, S.; Dent, S.; et al. Association of Pathologic Complete Response to Neoadjuvant Therapy in HER2-Positive Breast Cancer with Long-Term Outcomes: A Meta-Analysis. JAMA Oncol. 2016, 2, 751–760. [Google Scholar] [CrossRef]
- Rala de Paula, B.H.; Crocamo, S. Aiming for the Cure in ERBB2-Positive Metastatic Breast Cancer-Should We Go “All In”? JAMA Oncol. 2022, 8, 1220–1222. [Google Scholar] [CrossRef]
- Thanopoulou, E.; Khader, L.; Caira, M.; Wardley, A.; Ettl, J.; Miglietta, F.; Neven, P.; Guarneri, V. Therapeutic Strategies for the Management of Hormone Receptor-Positive, Human Epidermal Growth Factor Receptor 2-Positive (HR+/HER2+) Breast Cancer: A Review of the Current Literature. Cancers 2020, 12, 3317. [Google Scholar] [CrossRef] [PubMed]
- Gianni, L.; Pienkowski, T.; Im, Y.H.; Tseng, L.M.; Liu, M.C.; Lluch, A.; Starostawska, E.; Haba-Rodriguez, J.; Sou, S.A.; Pedrini, J.L.; et al. Efficacy and safety of neoadjuvant pertuzumab and trastuzumab in women with locally advanced, inflammatory, or early HER2-positive breast cancer (NeoSphere): A randomised multicentre, open-label, phase 2 trial. Lancet Oncol. 2012, 13, 25–32. [Google Scholar] [CrossRef]
- de Azambuja, E.; Holmes, A.P.; Piccart-Gebhart, M.; Holmes, E.; Di Cosimo, S.; Swaby, R.F.; Untvh, M.; Jackisch, C.; Lang, I.; Smith, I.; et al. Lapatinib with trastuzumab for HER2 positive early breast cancer (NeoALTTO): Survival outcomes of a randomised, open label, multicentre phase 3 trial and their association with pathological complete response. Lancet Oncol. 2014, 15, 1137–1146. [Google Scholar] [CrossRef] [PubMed]
- Schneeweiss, A.; Chia, S.; Hickish, T.; Harvey, V.; Eniu, A.; Hegg, R.; Tausch, C.; Seo, J.H.; Tsai, Y.F.; Ratnayake, J.; et al. Pertuzumab plus trastuzumab in combination with standard neoadjuvant anthracycline-containing and anthracycline-free chemotherapy regimens in patients with HER2-positive early breast cancer: A randomized phase II cardiac safety study (TRYPHAENA). Ann. Oncol. 2013, 24, 2278–2284. [Google Scholar] [CrossRef] [PubMed]
- Carey, L.A.; Berry, D.A.; Cirrincione, C.T.; Barry, W.T.; Pitcher, B.N.; Harris, L.N.; Ollila, D.W.; Krop, I.E.; Henry, N.L.; Weckstein, D.J.; et al. Molecular heterogeneity and response to neoadjuvant human epidermal growth factor receptor 2 targeting in CALGB 40601, a randomized phase III trial of paclitaxel plus trastuzumab with or without lapatinib. J. Clin. Oncol. 2016, 34, 542–549. [Google Scholar] [CrossRef]
- Untch, M.; Fasching, P.A.; Konecny, G.E.; Hasmüller, S.; Lebeau, A.; Kreienberg, R.; Camara, O.; Müller, V.; du Bois, A.; Kühn, T.; et al. Pathologic complete response after neoadjuvant chemotherapy plus trastuzumab predicts favorable survival in human epidermal growth factor receptor 2-overexpressing breast cancer: Results from the TECHNO trial of the AGO and GBG study groups. J. Clin. Oncol. 2011, 29, 3351–3357. [Google Scholar] [CrossRef]
- Crocamo, S.; Binato, R.; de Paula, B.; Vignal, G.; Magalhães, L.; Sarmento, R.; Accioly, M.T.; Small, I.; Gioia, S.; Maroun, P.; et al. Ácido zoledrônico neoadjuvante para câncer de mama HER2-positivo: O estudo Zo-NAnTax. Ther. Adv. Med. Oncol. 2019, 11, 1758835919853971. [Google Scholar] [CrossRef]
- Juarez, D.; Fruman, D.A. Targeting the Mevalonate Pathway in Cancer. Trends Cancer 2021, 7, 525–540. [Google Scholar] [CrossRef]
- Guerra, B.; Recio, C.; Aranda-Tavío, H.; Guerra-Rodríguez, M.; García-Castellano, J.M.; Fernández-Pérez, L. The Mevalonate Pathway, a Metabolic Target in Cancer Therapy. Front. Oncol. 2021, 25, 626971. [Google Scholar] [CrossRef]
- Bathaie, S.Z.; Ashrafi, M.; Azizian, M.; Tamanoi, F. Mevalonate Pathway and Human Cancers. Curr. Mol. Pharmacol. 2017, 10, 77–85. [Google Scholar]
- Wang, L.; Fang, D.; Xu, J.; Luo, R. Various pathways of zoledronic acid against osteoclasts and bone cancer metastasis: A brief review. BMC Cancer 2020, 20, 1059. [Google Scholar] [CrossRef] [PubMed]
- Rimawi, M.F.; Schiff, R.; Osborne, C.K. Targeting HER2 for the Treatment of Breast Cancer. Annu Rev Med. 2015, 66, 111–128. [Google Scholar] [CrossRef] [PubMed]
- O’Lone, R.; Frith, M.C.; Karlsson, E.K.; Hansen, U. Genomic targets of nuclear estrogen receptors. Mol. Endocrinol. 2004, 18, 1859–1875. [Google Scholar] [CrossRef]
- Losel, R.; Wehling, M. Nongenomic actions of steroid hormones. Nat. Rev. Mol. Cell Biol. 2003, 4, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Vaz-Luis, I.; Winer, E.P.; Lin, N.U. Human epidermal growth factor receptor-2-positive breast cancer: Does estrogen receptor status define two distinct subtypes? Ann. Oncol. 2013, 24, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.C.; Morrison, G.; Gillihan, R.; Guo, J.; Ward, R.M.; Fu, X.; Botero, M.f.; Healy, N.A.; Hilsenbeck, S.G.; Phillips, G.L.; et al. Different mechanisms for resistance to trastuzumab versus lapatinib in HER2-positive breast cancers—Role of estrogen receptor and HER2 reactivation. Breast Cancer Res. 2011, 13, R121. [Google Scholar] [CrossRef]
- Guarneri, V.; Griguolo, G.; Miglietta, F.; Conte, P.F.; Dieci, M.V.; Girardi, F. Survival after neoadjuvant therapy with trastuzumab-lapatinib and chemotherapy in patients with HER2-positive early breast cancer: A meta-analysis of randomized trials. ESMO Open 2022, 7, 100433. [Google Scholar] [CrossRef]
- Schneeweiss, A.; Chia, S.; Hickish, T.; Harvey, V.; Eniu, A.; Waldron-Lynch, M.; Eng-Wong, J.; Kirk, S.; Cortés, J. Long-term efficacy analysis of the randomised, phase II TRYPHAENA cardiac safety study: Evaluating pertuzumab and trastuzumab plus standard neoadjuvant anthracycline-containing and anthracycline-free chemotherapy regimens in patients with HER2-positive early breast cancer. Eur. J. Cancer 2018, 89, 27–35. [Google Scholar]
- Oshi, M.; Takahashi, H.; Tokumaru, Y.; Yan, L.; Rashid, O.M.; Matsuyama, R.; Endo, I.; Takabe, K. G2M Cell Cycle Pathway Score as a Prognostic Biomarker of Metastasis in Estrogen Receptor (ER)-Positive Breast Cancer. Int. J. Mol. Sci. 2020, 21, 2921. [Google Scholar] [CrossRef]
- Dunbier, A.K.; Anderson, H.; Ghazoui, Z.; Folkerd, E.J.; A’Hern, R.; Crowder, R.J.; Hoog, J.; Smith, I.E.; Osin, P.; Nerurkar, A.; et al. Relationship Between Plasma Estradiol Levels and Estrogen-Responsive Gene Expression in Estrogen Receptor–Positive Breast Cancer in Postmenopausal Women. J. Clin. Oncol. 2010, 28, 1161–1167. [Google Scholar] [CrossRef]
- Mohammed, H.; D’Santos, C.; Serandour, A.A.; Ali, H.R.; Brown, G.D.; Atkins, A.; Rueda, O.M.; Holmes, K.; Theodorou, V.; Robinson, J.L.L.; et al. Endogenous Purification Reveals GREB1 as a Key Estrogen Receptor Regulatory Factor. Cell Rep. 2013, 3, 342–349. [Google Scholar] [CrossRef] [PubMed]
- Haines, C.N.; Klingensmith, H.D.; Komara, M.; Burd, C.J. GREB1 regulates PI3K/Akt signaling to control hormone-sensitive breast cancer proliferation. Carcinogenesis 2020, 41, 1660–1670. [Google Scholar] [CrossRef] [PubMed]
- Räikkönen, J.; Mönkkönen, H.; Auriola, S.; Mönkkönen, J. Mevalonate pathway intermediates downregulate zoledronic acid-induced isopentenyl pyrophosphate and ATP analog formation in human breast cancer cells. Biochem. Pharmacol. 2010, 79, 777–783. [Google Scholar] [CrossRef] [PubMed]
- Iizuka-Ohashi, M.; Watanabe, M.; Sukeno, M.; Morita, M.; Thi, N.; Hoang, H.N.T.H.; Kuchimaru, T.; Kizaka-Kondoh, S.; Sowa, Y.; Sakaguchi, K.; et al. Blockage of the mevalonate pathway overcomes the apoptotic resistance to MEK inhibitors with suppressing the activation of Akt in cancer cells. Oncotarget 2018, 9, 19597–19612. [Google Scholar] [CrossRef] [PubMed]
- Attardi, L.D.; Reczek, E.E.; Cosmas, C.; Demicco, E.G.; McCurrach, M.E.; Lowe, S.W.; Jacks, T. PERP, an apoptosis-associated target of p53, is a novel member of the PMP-22/gas3 family. Genes Dev. 2000, 14, 704–718. [Google Scholar] [CrossRef] [PubMed]
- Liou, G.Y.; Storz, P. Reactive oxygen species in cancer. Free Radic. Res. 2010, 5, 479–496. [Google Scholar] [CrossRef] [PubMed]
- Campos, L.W.; Pissinato, L.G.; Yunes, J.A. Deleterious and Oncogenic Mutations in the IL7RA. Cancers 2019, 11, 1952. [Google Scholar] [CrossRef] [PubMed]
- Cardone, R.A.; Bellizzi, A.; Busco, G.; Weinman, E.J.; Dell’Aquila, M.E.; Casavola, V.; Amalia, A.; Mangia, A.; Paradiso, A.; Reshkin, S.J. The NHERF1 PDZ2 Domain Regulates PKA–RhoA–p38-mediated NHE1 Activation and Invasion in Breast Tumor Cells. Mol. Biol Cell. 2007, 18, 1768–1780. [Google Scholar] [CrossRef]
- Paradiso, A.; Scarpi, E.; Malfettone, A.; Addati, T.; Giotta, F.; Simone, G.; Amadori, D.; Mangia, A. Nuclear NHERF1 expression as a prognostic marker in breast cancer. Cell Death Dis. 2013, 4, e904. [Google Scholar] [CrossRef]
- Liu, H.; Ma, Y.; He, H.W.; Wang, J.P.; Jiang, J.D.; Shao, R.G. SLC9A3R1 stimulates autophagy via BECN1 stabilization in breast cancer cells. Autophagy 2015, 11, 2323–2334. [Google Scholar] [CrossRef]
- Jeong, J.; Choi, J.; Kim, W.; Dann, P.; Takyar, F.; Gefter, J.; Friedman, P.A.; Wysolmerski, J. Inhibition of ezrin causes PKCα-mediated internalization of erbb2/HER2 tyrosine kinase in breast cancer cells. J. Biol. Chem. 2019, 294, 887–901. [Google Scholar] [CrossRef] [PubMed]
- Gest, C.; Joimel, U.; Huang, L.; Pritchard, L.L.; Petit, A.; Dulong, C.; Buquet, C.; Hu, C.-Q.; Miirshahi, P.; Larent, F.M.; et al. Rac3 induces a molecular pathway triggering breast cancer cell aggressiveness: Differences in MDA-MB-231 and MCF-7 breast cancer cell lines. BMC Cancer 2013, 13, 63. [Google Scholar] [CrossRef] [PubMed]
- Chalhoub, N.; Baker, S.J. PTEN and the PI3-Kinase Pathway in Cancer. Annu. Rev. Pathol. 2009, 4, 127–150. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Viciana, P.; Warne, P.H.; Vanhaesebroeck, B.; Waterfield, M.D. Activation of phosphoinositide 3-kinase by interaction with Ras and by point mutation. EMBO J. 1996, 12, 2442–2451. [Google Scholar] [CrossRef]
- Zhang, B.; Kirov, S.; Snoddy, J. WebGestalt: An integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005, 33, W741–W748. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 8, W90–W97. [Google Scholar] [CrossRef]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Alberto, S.; Tsafou, K.P.; et al. STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
| Trials | Neoadjuvant Regimen | Overall | HR-Positive pCR Rate (%) | HR-Negative pCR Rate (%) |
|---|---|---|---|---|
| pCR Rate (%) | ||||
| NEOSPHERE [12] Randomized/phase II | DocTP | 39.3 & | 26.0 Ϯ | 63.2 Ϯ |
| DocT | 21.5 & | 20.0 Ϯ | 36.8 Ϯ | |
| TP | 11.2 & | 5.9 Ϯ | 27.3 Ϯ | |
| DocP | 17.7 & | 17.4 Ϯ | 30.0 Ϯ | |
| NEOALTTO [13] | Pac * LT | 46.8 & | 41.6 Ϯ | 61.3 Ϯ |
| Randomized/ | Pac * T | 27.6 & | 22.7 Ϯ | 36.5 Ϯ |
| phase III | Pac * L | 20.0 & | 16.1 Ϯ | 33.7 Ϯ |
| TRYPHAENA [14] | FECTP→DocTP | 61.6 Ϯ | 41.1 Ϯ | 73.5 Ϯ |
| Randomized/ | FEC→DocTP | 57.3 Ϯ | 45.7 Ϯ | 62.5 Ϯ |
| phase II | DocCarbTP | 66.2 Ϯ | 47.5 Ϯ | 81.1 Ϯ |
| CALGB 40601 [15] | Pac * LT | 52.0 & | 41.0 & | 68.0 & |
| Ranzomized/ | Pac * T | 44.0 & | 39.0 & | 50.0 & |
| phase III | Pac * L | 27.0 & | 26.0 & | 30.0 & |
| TECHNOS [16] | EC→Pac ** T | 38.7 & | 35.4 & | 42.3 & |
| Single arm/ | ||||
| phase II |
| Characteristics | HR-Positive |
|---|---|
| N (%) | (n = 16) |
| Age, mean (range), y | 56 (26–74) |
| Menopausal Status, N (%) | |
| Premenopausal | 6 (37.5) |
| Postmenopausal | 10 (62.5) |
| Family History of Cancer, N (%) | |
| Breast/ovarian cancer | 4 (25) |
| Any other type of cancer | 6 (37.5) |
| No family history | 6 (37.5) |
| T size, (range), mm | 57 (30–90) |
| AJCC stage *, N (%) | |
| IIA | 4 (25) |
| IIB | 6 (37.5) |
| IIIA | 2 (12.5) |
| IIIB | 4 (25) |
| Histology, N, (%) | |
| Invasive ductal | 16 (100) |
| Histologic grade †, N (%) | |
| 2 | 10 (62.5) |
| 3 | 6 (37.5) |
| HER2, N (%) | |
| HER2 3 + (IHQ) | 12 (75) |
| HER2 2 + (FISH positive) | 4 (25) |
| TILs, N (%) | |
| Presence | 10 (62.5) |
| Absence | 6 (37.5) |
| Ki67 §, N (%) | |
| <20 | 1 (6) |
| ≥20 | 15 (84) |
| p53 §, N (%) | |
| <10 | 5 (31) |
| ≥10 | 11 (69) |
| Not performed | 0 (0) |
| Characteristics | HR-Positive |
|---|---|
| N (%) | (n = 16) |
| RCB, N (%) | |
| RCB 0 | 7 (44) |
| RCB I | 2 (12.5) |
| RCB II | 5 (31) |
| RCB III | 2 (12.5) |
| Alive at 5 years, N (%) | |
| Yes | 14 (87.5) |
| No | 2 (12.5) |
| Signaling Pathways | Databank | Overexpressed Genes | Downregulated Genes |
|---|---|---|---|
| VEGF signaling pathway | Panther | SHC2 | RAC3 |
| p53 pathway | Panther | CD82-PERP | |
| PDGF pathway | Panther | SHC2 | RERG |
| Ubiquitin proteasome pathway | Panther | UBE2C | |
| Arachidonate Epoxygenase/Epoxide Hydrolase | Wikipathway | GSTP1 | |
| Gamma-Glutamyl cycle for the biosynthesis and degradation of glutathione, including diseases | Wikipathway | GGTLC1 | |
| NRF2 pathway | Wikipathway | CBR3-FGF13-GGTLC1-GSTP1-SLC2A9 | |
| Nuclear receptors Meta-Pathway | Wikipathway | CBR3-FGF13-GGTLC1-GSTP1-SLC2A9-TNS4 | CYP2B6 |
| Metabolism of xenobiotics by cytochrome P450 | KEGG | ALDH1A3-CBR3-GSTP1 | CYP2B6-GSTT1 |
| Drug metabolism | KEGG | ALDH1A3-GSTP1 | CYP2B6-GSTT1 |
| Drug metabolism 1 | KEGG | GSTP1 | GSTT1-NAT1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Crocamo, S.; Binato, R.; dos Santos, E.C.; de Paula, B.; Abdelhay, E. Translational Results of Zo-NAnTax: A Phase II Trial of Neoadjuvant Zoledronic Acid in HER2-Positive Breast Cancer. Int. J. Mol. Sci. 2022, 23, 15515. https://doi.org/10.3390/ijms232415515
Crocamo S, Binato R, dos Santos EC, de Paula B, Abdelhay E. Translational Results of Zo-NAnTax: A Phase II Trial of Neoadjuvant Zoledronic Acid in HER2-Positive Breast Cancer. International Journal of Molecular Sciences. 2022; 23(24):15515. https://doi.org/10.3390/ijms232415515
Chicago/Turabian StyleCrocamo, Susanne, Renata Binato, Everton Cruz dos Santos, Bruno de Paula, and Eliana Abdelhay. 2022. "Translational Results of Zo-NAnTax: A Phase II Trial of Neoadjuvant Zoledronic Acid in HER2-Positive Breast Cancer" International Journal of Molecular Sciences 23, no. 24: 15515. https://doi.org/10.3390/ijms232415515
APA StyleCrocamo, S., Binato, R., dos Santos, E. C., de Paula, B., & Abdelhay, E. (2022). Translational Results of Zo-NAnTax: A Phase II Trial of Neoadjuvant Zoledronic Acid in HER2-Positive Breast Cancer. International Journal of Molecular Sciences, 23(24), 15515. https://doi.org/10.3390/ijms232415515






