The Essential Role of Prolines and Their Conformation in Allosteric Regulation of Kaiso Zinc Finger DNA-Binding Activity by the Adjacent C-Terminal Loop
Abstract
1. Introduction
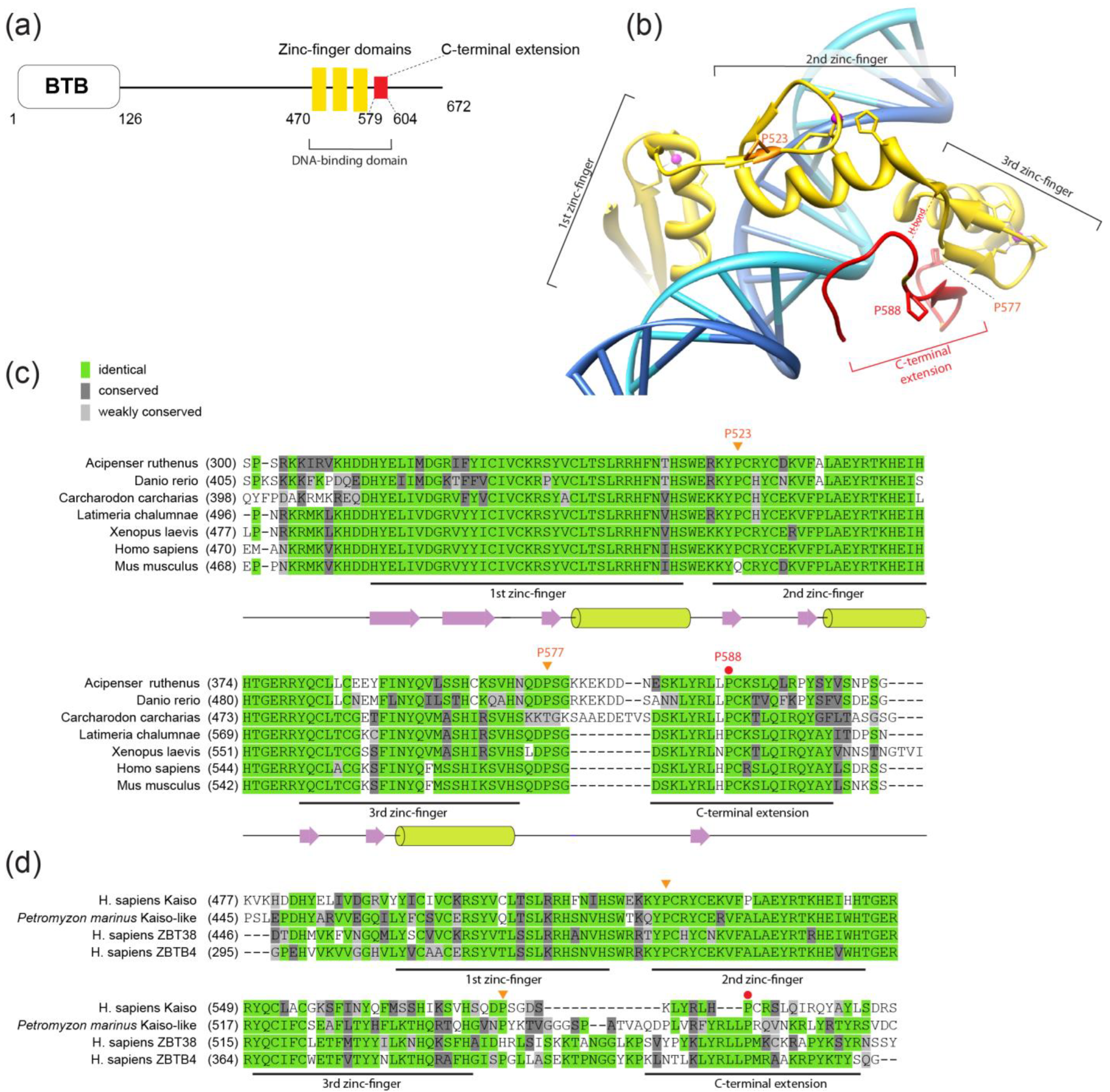
2. Results
2.1. Kaiso Has Conserved Prolines in the C-Terminal Region That are Involved in DNA Binding
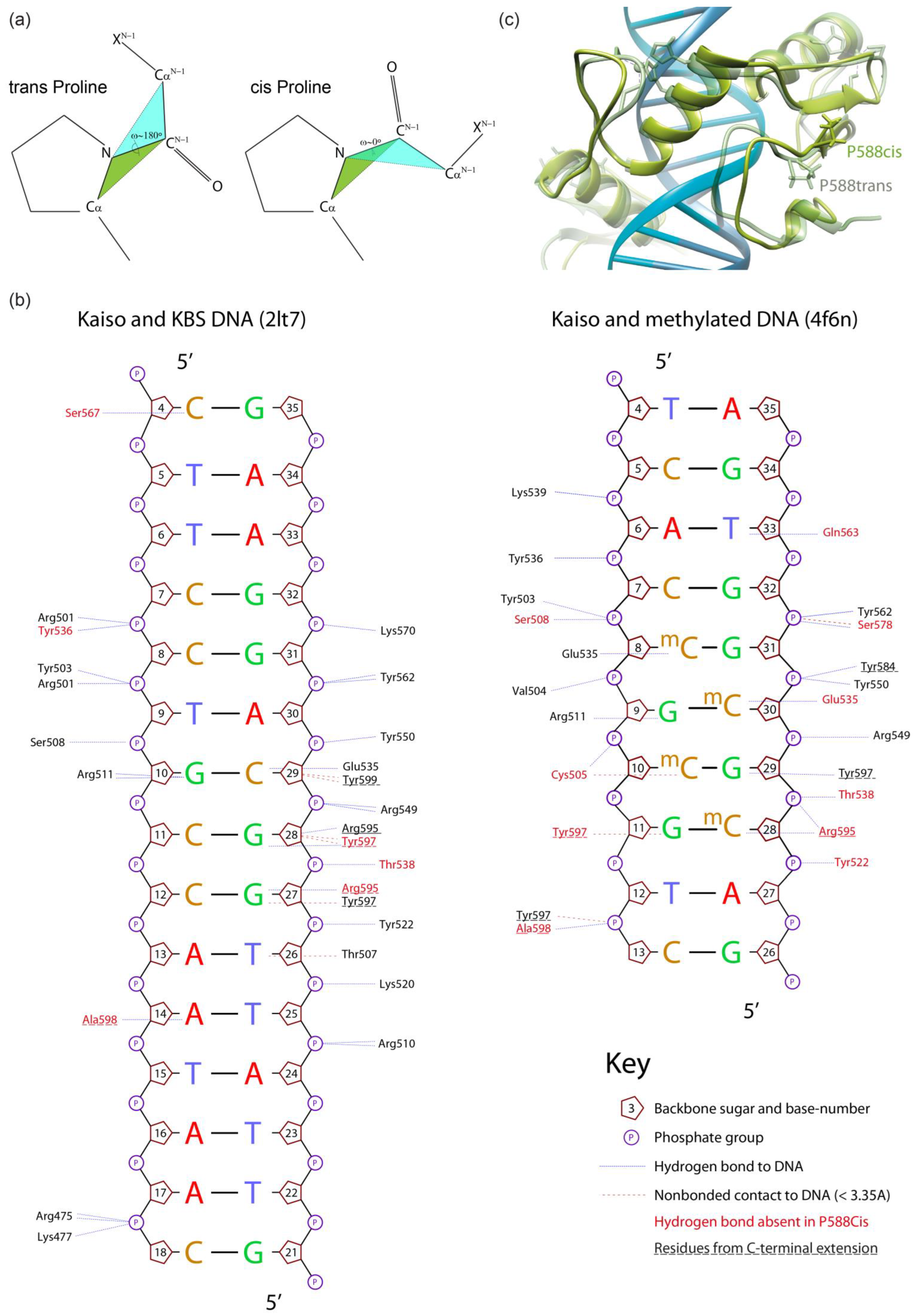
2.2. Molecular Dynamics Simulation Revealed the Importance of the Rigidity of the C-Terminal Extension Linker for the Overall Stability of the Kaiso–DNA Complex
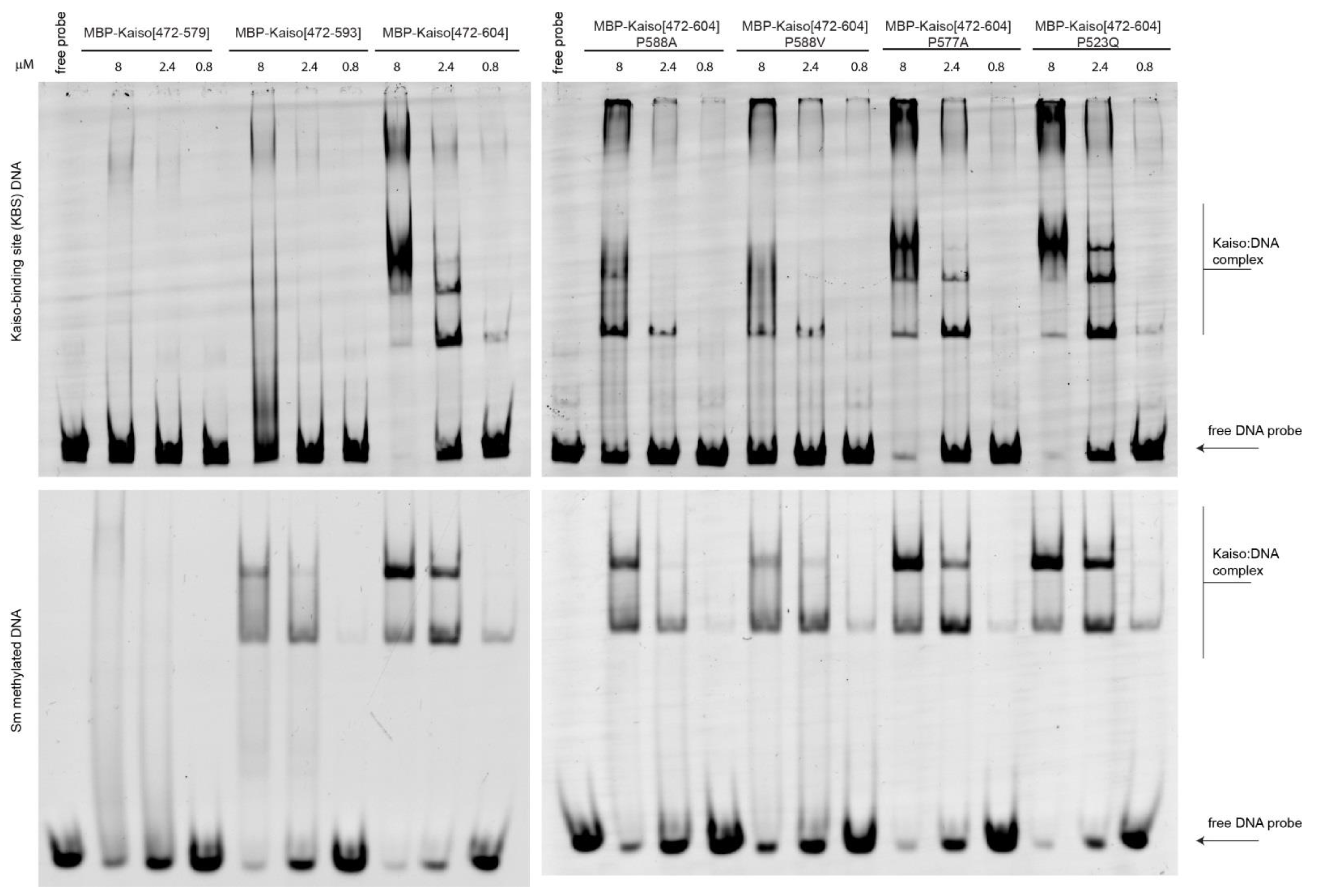
2.3. The Presence of the C-Terminal Extension and Restrained Mobility in Its Linker Are Required for Efficient Kaiso DNA Binding In Vitro
3. Discussion
4. Materials and Methods
4.1. Plasmids and Cloning
4.2. Protein Expression and Purification
4.3. Electrophoretic Mobility Shift Assay
4.4. Molecular Dynamics Simulations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lambert, S.A.; Jolma, A.; Campitelli, L.F.; Das, P.K.; Yin, Y.; Albu, M.; Chen, X.; Taipale, J.; Hughes, T.R.; Weirauch, M.T. The Human Transcription Factors. Cell 2018, 175, 598–599. [Google Scholar] [CrossRef] [PubMed]
- Razin, S.V.; Borunova, V.V.; Maksimenko, O.G.; Kantidze, O.L. Cys2His2 zinc finger protein family: Classification, functions, and major members. Biochemistry 2012, 77, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Fedotova, A.A.; Bonchuk, A.N.; Mogila, V.A.; Georgiev, P.G. C2H2 Zinc Finger Proteins: The Largest but Poorly Explored Family of Higher Eukaryotic Transcription Factors. Acta Nat. 2017, 9, 47–58. [Google Scholar] [CrossRef]
- Soto, L.F.; Li, Z.; Santoso, C.S.; Berenson, A.; Ho, I.; Shen, V.X.; Yuan, S.; Fuxman Bass, J.I. Compendium of human transcription factor effector domains. Mol. Cell 2022, 82, 514–526. [Google Scholar] [CrossRef]
- Brayer, K.J.; Segal, D.J. Keep your fingers off my DNA: Protein-protein interactions mediated by C2H2 zinc finger domains. Cell Biochem. Biophys. 2008, 50, 111–131. [Google Scholar] [CrossRef] [PubMed]
- Garton, M.; Najafabadi, H.S.; Schmitges, F.W.; Radovani, E.; Hughes, T.R.; Kim, P.M. A structural approach reveals how neighbouring C2H2 zinc fingers influence DNA binding specificity. Nucleic Acids Res. 2015, 43, 9147–9157. [Google Scholar] [CrossRef]
- Persikov, A.V.; Singh, M. De novo prediction of DNA-binding specificities for Cys2His2 zinc finger proteins. Nucleic Acids Res. 2014, 42, 97–108. [Google Scholar] [CrossRef]
- Ryan, R.F.; Darby, M.K. The role of zinc finger linkers in p43 and TFIIIA binding to 5S rRNA and DNA. Nucleic Acids Res. 1998, 26, 703–709. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.P.; Finn, G.; Lee, T.H.; Nicholson, L.K. Prolyl cis-trans isomerization as a molecular timer. Nat. Chem. Biol. 2007, 3, 619–629. [Google Scholar] [CrossRef]
- Hanes, S.D. Prolyl isomerases in gene transcription. Biochim. Biophys. Acta 2015, 1850, 2017–2034. [Google Scholar] [CrossRef]
- Hu, X.; Chen, L.F. Pinning Down the Transcription: A Role for Peptidyl-Prolyl cis-trans Isomerase Pin1 in Gene Expression. Front. Cell Dev. Biol. 2020, 8, 179. [Google Scholar] [CrossRef]
- Farrell, A.S.; Pelz, C.; Wang, X.; Daniel, C.J.; Wang, Z.; Su, Y.; Janghorban, M.; Zhang, X.; Morgan, C.; Impey, S.; et al. Pin1 regulates the dynamics of c-Myc DNA binding to facilitate target gene regulation and oncogenesis. Mol. Cell Biol. 2013, 33, 2930–2949. [Google Scholar] [CrossRef]
- Mantovani, F.; Tocco, F.; Girardini, J.; Smith, P.; Gasco, M.; Lu, X.; Crook, T.; Del Sal, G. The prolyl isomerase Pin1 orchestrates p53 acetylation and dissociation from the apoptosis inhibitor iASPP. Nat. Struct. Mol. Biol. 2007, 14, 912–920. [Google Scholar] [CrossRef]
- Buck-Koehntop, B.A.; Martinez-Yamout, M.A.; Dyson, H.J.; Wright, P.E. Kaiso uses all three zinc fingers and adjacent sequence motifs for high affinity binding to sequence-specific and methyl-CpG DNA targets. FEBS Lett. 2012, 586, 734–739. [Google Scholar] [CrossRef] [PubMed]
- Buck-Koehntop, B.A.; Stanfield, R.L.; Ekiert, D.C.; Martinez-Yamout, M.A.; Dyson, H.J.; Wilson, I.A.; Wright, P.E. Molecular basis for recognition of methylated and specific DNA sequences by the zinc finger protein Kaiso. Proc. Natl. Acad. Sci. USA 2012, 109, 15229–15234. [Google Scholar] [CrossRef]
- Prokhortchouk, A.; Hendrich, B.; Jorgensen, H.; Ruzov, A.; Wilm, M.; Georgiev, G.; Bird, A.; Prokhortchouk, E. The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev. 2001, 15, 1613–1618. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.G.; Chan, D.W.; Reynolds, A.B.; Qin, J.; Wong, J. N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Mol. Cell 2003, 12, 723–734. [Google Scholar] [CrossRef]
- Raghav, S.K.; Waszak, S.M.; Krier, I.; Gubelmann, C.; Isakova, A.; Mikkelsen, T.S.; Deplancke, B. Integrative genomics identifies the corepressor SMRT as a gatekeeper of adipogenesis through the transcription factors C/EBPbeta and KAISO. Mol. Cell 2012, 46, 335–350. [Google Scholar] [CrossRef] [PubMed]
- Kaplun, D.; Starshin, A.; Sharko, F.; Gainova, K.; Filonova, G.; Zhigalova, N.; Mazur, A.; Prokhortchouk, E.; Zhenilo, S. Kaiso Regulates DNA Methylation Homeostasis. Int. J. Mol. Sci. 2021, 22, 7587. [Google Scholar] [CrossRef]
- Bohne, F.; Langer, D.; Martine, U.; Eider, C.S.; Cencic, R.; Begemann, M.; Elbracht, M.; Bulow, L.; Eggermann, T.; Zechner, U.; et al. Kaiso mediates human ICR1 methylation maintenance and H19 transcriptional fine regulation. Clin. Epigenetics 2016, 8, 47. [Google Scholar] [CrossRef]
- Mahmood, N.; Rabbani, S.A. DNA Methylation Readers and Cancer: Mechanistic and Therapeutic Applications. Front. Oncol. 2019, 9, 489. [Google Scholar] [CrossRef] [PubMed]
- Bassey-Archibong, B.I.; Hercules, S.M.; Rayner, L.G.A.; Skeete, D.H.A.; Smith Connell, S.P.; Brain, I.; Daramola, A.; Banjo, A.A.F.; Byun, J.S.; Gardner, K.; et al. Kaiso is highly expressed in TNBC tissues of women of African ancestry compared to Caucasian women. Cancer Causes Control 2017, 28, 1295–1304. [Google Scholar] [CrossRef] [PubMed]
- Bassey-Archibong, B.I.; Rayner, L.G.; Hercules, S.M.; Aarts, C.W.; Dvorkin-Gheva, A.; Bramson, J.L.; Hassell, J.A.; Daniel, J.M. Kaiso depletion attenuates the growth and survival of triple negative breast cancer cells. Cell Death Dis. 2017, 8, e2689. [Google Scholar] [CrossRef] [PubMed]
- Kwiecien, J.M.; Bassey-Archibong, B.I.; Dabrowski, W.; Rayner, L.G.; Lucas, A.R.; Daniel, J.M. Loss of Kaiso expression in breast cancer cells prevents intra-vascular invasion in the lung and secondary metastasis. PLoS ONE 2017, 12, e0183883. [Google Scholar] [CrossRef] [PubMed]
- Singhal, S.K.; Byun, J.S.; Park, S.; Yan, T.; Yancey, R.; Caban, A.; Hernandez, S.G.; Hewitt, S.M.; Boisvert, H.; Hennek, S.; et al. Kaiso (ZBTB33) subcellular partitioning functionally links LC3A/B, the tumor microenvironment, and breast cancer survival. Commun. Biol. 2021, 4, 150. [Google Scholar] [CrossRef]
- Pierre, C.C.; Longo, J.; Mavor, M.; Milosavljevic, S.B.; Chaudhary, R.; Gilbreath, E.; Yates, C.; Daniel, J.M. Kaiso overexpression promotes intestinal inflammation and potentiates intestinal tumorigenesis in Apc(Min/+) mice. Biochim. Biophys. Acta 2015, 1852, 1846–1855. [Google Scholar] [CrossRef]
- Daniel, J.M.; Spring, C.M.; Crawford, H.C.; Reynolds, A.B.; Baig, A. The p120(ctn)-binding partner Kaiso is a bi-modal DNA-binding protein that recognizes both a sequence-specific consensus and methylated CpG dinucleotides. Nucleic Acids Res. 2002, 30, 2911–2919. [Google Scholar] [CrossRef] [PubMed]
- Hudson, N.O.; Whitby, F.G.; Buck-Koehntop, B.A. Structural insights into methylated DNA recognition by the C-terminal zinc fingers of the DNA reader protein ZBTB38. J. Biol. Chem. 2018, 293, 19835–19843. [Google Scholar] [CrossRef]
- Carnahan, R.H.; Rokas, A.; Gaucher, E.A.; Reynolds, A.B. The molecular evolution of the p120-catenin subfamily and its functional associations. PLoS ONE 2010, 5, e15747. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2002, 1, 2–3. [Google Scholar] [CrossRef] [PubMed]
- Kim, P.S.; Baldwin, R.L. Specific intermediates in the folding reactions of small proteins and the mechanism of protein folding. Annu. Rev. Biochem. 1982, 51, 459–489. [Google Scholar] [CrossRef]
- Luscombe, N.M.; Laskowski, R.A.; Thornton, J.M. NUCPLOT: A program to generate schematic diagrams of protein-nucleic acid interactions. Nucleic Acids Res. 1997, 25, 4940–4945. [Google Scholar] [CrossRef] [PubMed]
- Nikolova, E.N.; Stanfield, R.L.; Dyson, H.J.; Wright, P.E. A Conformational Switch in the Zinc Finger Protein Kaiso Mediates Differential Readout of Specific and Methylated DNA Sequences. Biochemistry 2020, 59, 1909–1926. [Google Scholar] [CrossRef] [PubMed]
- Nikolova, E.N.; Stanfield, R.L.; Dyson, H.J.; Wright, P.E. CH...O Hydrogen Bonds Mediate Highly Specific Recognition of Methylated CpG Sites by the Zinc Finger Protein Kaiso. Biochemistry 2018, 57, 2109–2120. [Google Scholar] [CrossRef] [PubMed]
- Galindo-Murillo, R.; Robertson, J.C.; Zgarbova, M.; Sponer, J.; Otyepka, M.; Jurecka, P.; Cheatham, T.E., 3rd. Assessing the Current State of Amber Force Field Modifications for DNA. J. Chem. Theory Comput. 2016, 12, 4114–4127. [Google Scholar] [CrossRef]
- Dovat, S.; Ronni, T.; Russell, D.; Ferrini, R.; Cobb, B.S.; Smale, S.T. A common mechanism for mitotic inactivation of C2H2 zinc finger DNA-binding domains. Genes Dev. 2002, 16, 2985–2990. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.; Murtola, T.; Schulz, R.; Pall, S.; Smith, J.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Ponder, J.W.; Case, D.A. Force fields for protein simulations. Adv. Protein Chem. 2003, 66, 27–85. [Google Scholar] [CrossRef]
- Ivani, I.; Dans, P.D.; Noy, A.; Perez, A.; Faustino, I.; Hospital, A.; Walther, J.; Andrio, P.; Goni, R.; Balaceanu, A.; et al. Parmbsc1: A refined force field for DNA simulations. Nat. Methods 2016, 13, 55–58. [Google Scholar] [CrossRef]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmuller, H.; MacKerell, A.D., Jr. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38, 27–38. [Google Scholar] [CrossRef] [PubMed]
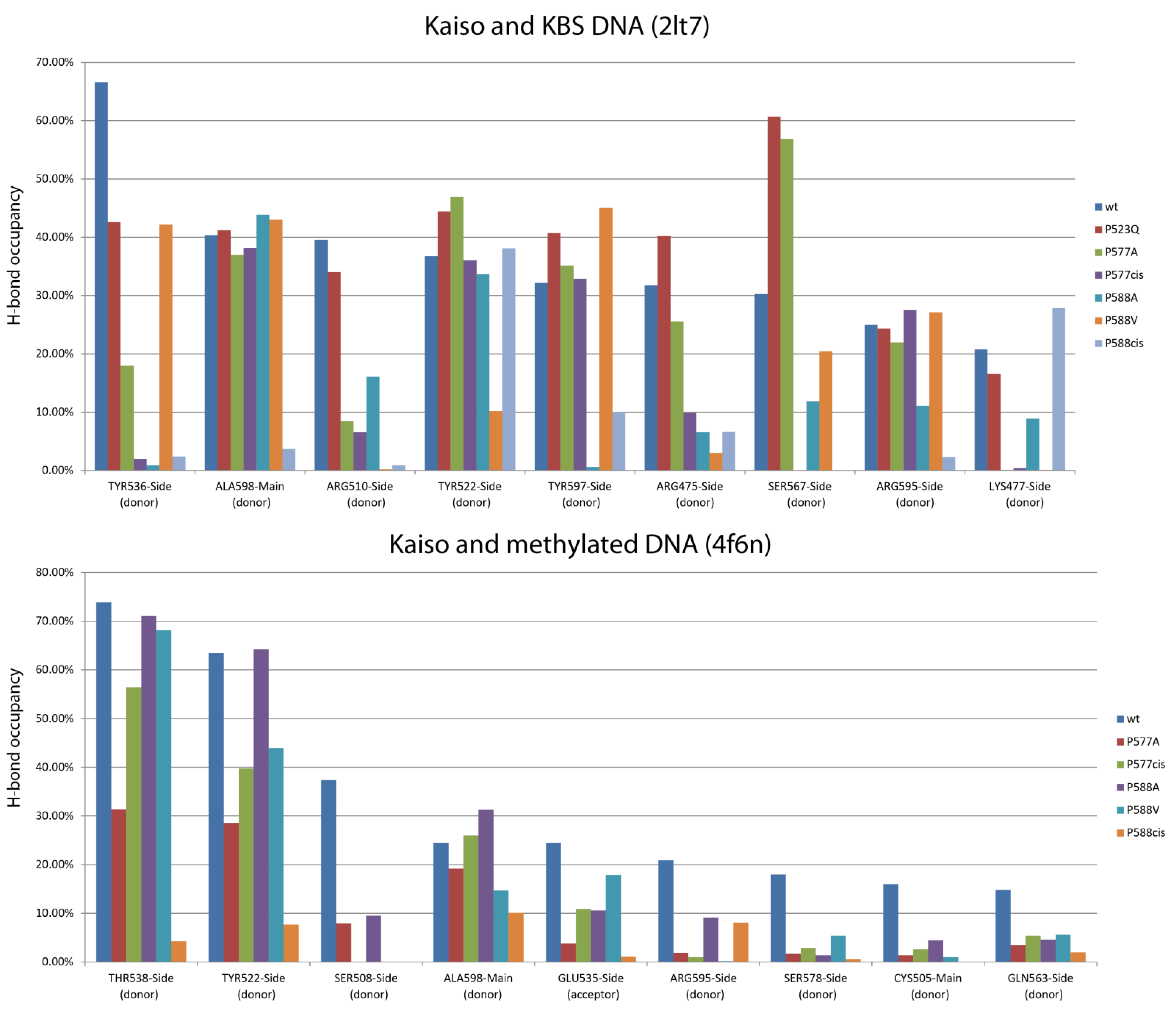

| Donor | Acceptor | wt | P523Q | P577cis | P588A | P588V | P588cis |
|---|---|---|---|---|---|---|---|
| KBS DNA | |||||||
| SER591-Main | GLU547-Side | 33.87% | 39.76% | 35.46% | 32.77% | 33.71% | 1.60% |
| SER591-Side | GLU547-Side | 77.52% | 60.34% | 63.94% | 48.15% | 70.32% | 26.65% |
| ARG590-Main | GLU547-Side | 29.67% | 50.65% | 37.36% | 43.16% | 39.25% | 0.10% |
| methylated DNA | |||||||
| SER591-Main | GLU547-Side | 46.35% | ND | 27.07% | 48.55% | 45.95% | 0.10% |
| SER591-Side | GLU547-Side | 80.72% | ND | 70.13% | 83.22% | 79.12% | 10.29% |
| ARG590-Main | GLU547-Side | 36.06% | ND | 32.67% | 31.07% | 35.86% | 2.30% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Belova, E.; Maksimenko, O.; Georgiev, P.; Bonchuk, A. The Essential Role of Prolines and Their Conformation in Allosteric Regulation of Kaiso Zinc Finger DNA-Binding Activity by the Adjacent C-Terminal Loop. Int. J. Mol. Sci. 2022, 23, 15494. https://doi.org/10.3390/ijms232415494
Belova E, Maksimenko O, Georgiev P, Bonchuk A. The Essential Role of Prolines and Their Conformation in Allosteric Regulation of Kaiso Zinc Finger DNA-Binding Activity by the Adjacent C-Terminal Loop. International Journal of Molecular Sciences. 2022; 23(24):15494. https://doi.org/10.3390/ijms232415494
Chicago/Turabian StyleBelova, Elena, Oksana Maksimenko, Pavel Georgiev, and Artem Bonchuk. 2022. "The Essential Role of Prolines and Their Conformation in Allosteric Regulation of Kaiso Zinc Finger DNA-Binding Activity by the Adjacent C-Terminal Loop" International Journal of Molecular Sciences 23, no. 24: 15494. https://doi.org/10.3390/ijms232415494
APA StyleBelova, E., Maksimenko, O., Georgiev, P., & Bonchuk, A. (2022). The Essential Role of Prolines and Their Conformation in Allosteric Regulation of Kaiso Zinc Finger DNA-Binding Activity by the Adjacent C-Terminal Loop. International Journal of Molecular Sciences, 23(24), 15494. https://doi.org/10.3390/ijms232415494







