Distribution of KIR Genes and Their HLA Ligands in Different Viral Infectious Diseases: Frequency Study in Sicilian Population
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Studied Populations
4.2. Molecular Analysis
4.3. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Middleton, D.; Gonzelez, F. The extensive polymorphism of KIR genes. Immunology 2010, 129, 8–19. [Google Scholar] [CrossRef]
- Shilling, H.G.; Young, N.; Guethlein, L.A.; Cheng, N.W.; Gardiner, C.M.; Tyan, D.; Parham, P. Genetic control of human NK cell repertoire. J. Immunol. 2002, 169, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.P.; Carrington, M. KIR locus polymorphisms: Genotyping and disease association analysis. Methods Mol. Biol. 2008, 415, 49–64. [Google Scholar] [CrossRef]
- IPD-KIR Database. Available online: https://www.ebi.ac.uk/ipd/kir/about/statistics/ (accessed on 22 July 2022).
- Middleton, D.; Gonzalezm, A.; Gilmore, P.M. Studies on the expression of the deleted KIR2DS4*003 gene product and distribution of KIR2DS4 deleted and nondeleted versions in different populations. Hum. Immunol. 2007, 68, 128–134. [Google Scholar] [CrossRef]
- Bontadini, A.; Testi, M.; Cuccia, M.C.; Martinetti, M.; Carcassi, C.; Chiesa, A.; Cosentini, E.; Dametto, E.; Frison, S.; Iannone, A.M.; et al. Distribution of killer cell immunoglobulin-like receptors genes in the Italian Caucasian population. J. Transl. Med. 2006, 4, 44. [Google Scholar] [CrossRef][Green Version]
- Lanier, L.L. Up on the tightrope: Natural killer cell activation and inhibition. Nat. Immunol. 2008, 9, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Littera, R.; Piredda, G.; Argiolas, D.; Lai, S.; Congeddu, E.; Ragatzu, P.; Melis, M.; Carta, E.; Michittu, M.B.; Valentini, D.; et al. KIR and their HLA Class I ligands: Two more pieces towards completing the puzzle of chronic rejection and graft loss in kidney transplantation. PLoS ONE 2017, 12, e0180831. [Google Scholar] [CrossRef]
- Wu, X.; Yao, Y.; Bao, X.; Zhou, H.; Tang, X.; Han, Y.; Ma, X.; Liu, Y.; Chen, J.; Zhou, H.; et al. KIR2DS4 and Its Variant KIR1D Are Associated with Acute Graft-versus-Host Disease, Cytomegalovirus, and Overall Survival after Sibling-Related HLA-Matched Transplantation in Patients with Donors with KIR Gene Haplotype A. Biol. Blood Marrow Transplant. 2016, 22, 220–225. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.A.; Paximadis, M.; Gray, G.E.; Kuhn, L.; Tiemessen, C.T. KIR2DS4 allelic variants: Differential effects on in utero and intrapartum HIV-1 mother-to-child transmission. Clin. Immunol. 2013, 149, 498–508. [Google Scholar] [CrossRef]
- Rizzo, C.; Accardi, G.; Caruso, C. Genetic variation in human leukocyte antigen and susceptibility to acute myeloid leukemia. Acta Haematol. 2015, 133, 162–163. [Google Scholar] [CrossRef]
- Rajalingam, R. Human diversity of killer cell immunoglobulin-like receptors and disease. Korean J. Hematol. 2011, 46, 216–228. [Google Scholar] [CrossRef] [PubMed]
- Jamil, K.M.; Khakoo, S.I. KIR/HLA interactions and pathogen immunity. J. Biomed. Biotechnol. 2011, 2011, 298348. [Google Scholar] [CrossRef]
- Cerwenka, A.; Lanier, L.L. Ligands for natural killer cell receptors: Redundancy or specificity. Immunol. Rev. 2001, 181, 158–169. [Google Scholar] [CrossRef] [PubMed]
- Aiello, A.; Candore, G.; Accardi, G.; Caruso, C.; Colomba, C.; Duro, G.; Gambino, C.M.; Ligotti, M.E.; Di Bona, D. Translation of Basic Research into Clinics: Killer Immunoglobulin-like Receptors Genes in Autoimmune and Infectious Diseases. Curr. Pharm. Des. 2018, 24, 3113–3122. [Google Scholar] [CrossRef] [PubMed]
- Aiello, A.; Accardi, G.; Candore, G.; Caruso, C.; Colomba, C.; Di Bona, D.; Duro, G.; Gambino, C.M.; Ligotti, M.E.; Pandey, J.P. Role of Immunogenetics in the Outcome of HCMV Infection: Implications for Ageing. Int. J. Mol. Sci. 2019, 20, 685. [Google Scholar] [CrossRef]
- Tuttolomondo, A.; Colomba, C.; Di Bona, D.; Casuccio, A.; Di Raimondo, D.; Clemente, G.; Arnao, V.; Pecoraro, R.; Ragonese, P.; Aiello, A.; et al. HLA and killer cell immunoglobulin-like receptor (KIRs) genotyping in patients with acute viral encephalitis. Oncotarget 2018, 9, 17523–17532. [Google Scholar] [CrossRef] [PubMed]
- Di Bona, D.; Aiello, A.; Colomba, C.; Bilancia, M.; Accardi, G.; Rubino, R.; Giannitrapani, L.; Tuttolomondo, A.; Cascio, A.; Caiaffa, M.F.; et al. KIR2DL3 and the KIR ligand groups HLA-A-Bw4 and HLA-C2 predict the outcome of hepatitis B virus infection. J. Viral Hepat. 2017, 24, 768–775. [Google Scholar] [CrossRef] [PubMed]
- Beksac, M.; Akin, H.Y.; Gencer-Oncul, E.B.; Yousefzadeh, M.; Cengiz Seval, G.; Gulten, E.; Akdemir Kalkan, I.; Cinar, G.; Memikoglu, O.; Karaagaoglu, E.; et al. A model integrating Killer Immunoglobulin-like Receptor (KIR) haplotypes for risk prediction of COVID-19 clinical disease severity. Immunogenetics 2021, 73, 449–458. [Google Scholar] [CrossRef]
- Pojero, F.; Candore, G.; Caruso, C.; Di Bona, D.; Groneberg, D.A.; Ligotti, M.E.; Accardi, G.; Aiello, A. The Role of Immunogenetics in COVID-19. Int. J. Mol. Sci. 2021, 22, 2636. [Google Scholar] [CrossRef] [PubMed]
- Guinan, K.J.; Cunningham, R.T.; Meenagh, A.; Dring, M.M.; Middleton, D.; Gardiner, C.M. Receptor systems controlling natural killer cell function are genetically stratified in Europe. Genes Immun. 2010, 11, 67–78. [Google Scholar] [CrossRef]
- Fasano, M.E.; Rendine, S.; Pasi, A.; Bontadini, A.; Cosentini, E.; Carcassi, C.; Capittini, C.; Cornacchini, G.; Espadas de Arias, A.; Garbarino, L.; et al. The distribution of KIR-HLA functional blocks is different from north to south of Italy. Tissue Antigens 2014, 83, 168–173. [Google Scholar] [CrossRef]
- Capittini, C.; Messina, F.; Puglisi, F.; Azzaro, M.; Toscano, S.; De Silvestri, A.; Tinelli, C.; Sortino, G. An historical approach to the genetic distribution of KIR and HLA ligands in Eastern Sicilians compared to modern descendants of their invaders. Hum. Immunol. 2018, 79, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, S.; Martin, M.P.; Carrington, M. The Yin and Yang of HLA and KIR in human disease. Semin. Immunol. 2008, 20, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.P.; Gao, X.; Lee, J.H.; Nelson, G.W.; Detels, R.; Goedert, J.J.; Buchbinder, S.; Hoots, K.; Vlahov, D.; Trowsdale, J.; et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat. Genet. 2002, 31, 429–434. [Google Scholar] [CrossRef]
- Qi, Y.; Martin, M.P.; Gao, X.; Jacobson, L.; Goedert, J.J.; Buchbinder, S.; Kirk, G.D.; O’Brien, S.J.; Trowsdale, J.; Carrington, M. KIR/HLA pleiotropism: Protection against both HIV and opportunistic infections. PLoS Pathog. 2006, 2, e79. [Google Scholar] [CrossRef]
- Hens, J.; Jennes, W.; Kestens, L. The role of NK cells in HIV-1 protection: Autologous, allogeneic or both? AIDS Res Ther. 2016, 13, 15. [Google Scholar] [CrossRef][Green Version]
- Martin, M.P.; Qi, Y.; Gao, X.; Yamada, E.; Martin, J.N.; Pereyra, F.; Colombo, S.; Brown, E.E.; Shupert, W.L.; Phair, J.; et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat. Genet. 2007, 39, 733–740. [Google Scholar] [CrossRef]
- Pende, D.; Falco, M.; Vitale, M.; Cantoni, C.; Vitale, C.; Munari, E.; Bertaina, A.; Moretta, F.; Del Zotto, G.; Pietra, G.; et al. Killer Ig-Like Receptors (KIRs): Their Role in NK Cell Modulation and Developments Leading to Their Clinical Exploitation. Front. Immunol. 2019, 10, 1179. [Google Scholar] [CrossRef] [PubMed]
- Mori, M.; Wichukchinda, N.; Miyahara, R.; Rojanawiwat, A.; Pathipvanich, P.; Tsuchiya, N.; Miura, T.; Yasunami, M.; Ariyoshi, K.; Sawanpanyalert, P. The effect of KIR2D-HLA-C receptor-ligand interactions on clinical outcome in a HIV-1 CRF01_AE-infected Thai population. AIDS 2015, 29, 1607–1615. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Jiao, Y.; Wang, L.; Liu, X.; Sun, W.; Cui, B.; Chen, Z.; Zhao, Y. Inhibitory KIR and specific HLA-C gene combinations confer susceptibility to or protection against chronic hepatitis B. Clin. Immunol. 2010, 137, 139–146. [Google Scholar] [CrossRef]
- Littera, R.; Chessa, L.; Deidda, S.; Angioni, G.; Campagna, M.; Lai, S.; Melis, M.; Cipri, S.; Firinu, D.; Santus, S.; et al. Natural killer-cell immunoglobulin-like receptors trigger differences in immune response to SARS-CoV-2 infection. PLoS ONE 2021, 16, e0255608. [Google Scholar] [CrossRef] [PubMed]
- Saresella, M.; Trabattoni, D.; Marventano, I.; Piancone, F.; La Rosa, F.; Caronni, A.; Lax, A.; Bianchi, L.; Banfi, P.; Navarro, J.; et al. NK Cell Subpopulations and Receptor Expression in Recovering SARS-CoV-2 Infection. Mol. Neurobiol. 2021, 58, 6111–6120. [Google Scholar] [CrossRef] [PubMed]
- Charan, J.; Biswas, T. How to calculate sample size for different study designs in medical research? Indian J. Psychol. Med. 2013, 35, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Sarno, S.; Boattini, A.; Carta, M.; Ferri, G.; Alù, M.; Yao, D.Y.; Ciani, G.; Pettener, D.; Luiselli, D. An ancient Mediterranean melting pot: Investigating the uniparental genetic structure and population history of sicily and southern Italy. PLoS ONE 2014, 9, e96074. [Google Scholar] [CrossRef] [PubMed]
- Piazza, A.; Olivetti, E.; Griffo, R.M.; Rendine, S.; Amoroso, A.; Barbanti, M.; Caruso, C.; Conighi, C.; Conte, R.; Favoino, B.; et al. The distribution of HLA antigens in Italy. Gene Geogr. 1989, 3, 141–164. [Google Scholar] [PubMed]
- Listì, F.; Caruso, C.; Colonna-Romano, G.; Lio, D.; Nuzzo, D.; Candore, G. HLA and KIR frequencies in Sicilian Centenarians. Rejuvenation Res. 2010, 13, 314–318. [Google Scholar] [CrossRef] [PubMed]
- Allele Frequency Database. Available online: http://www.allelefrequencies.net (accessed on 18 July 2022).
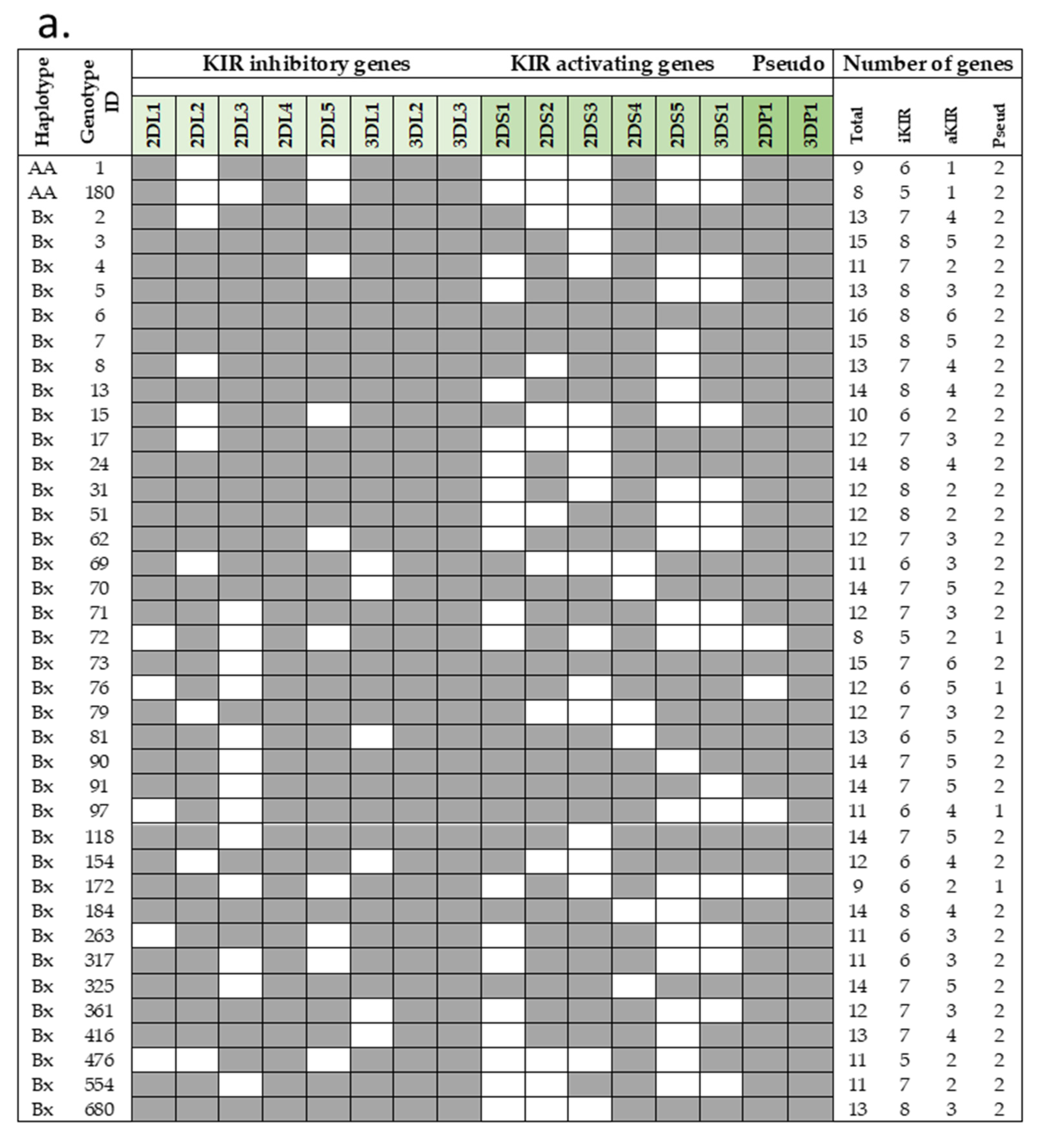
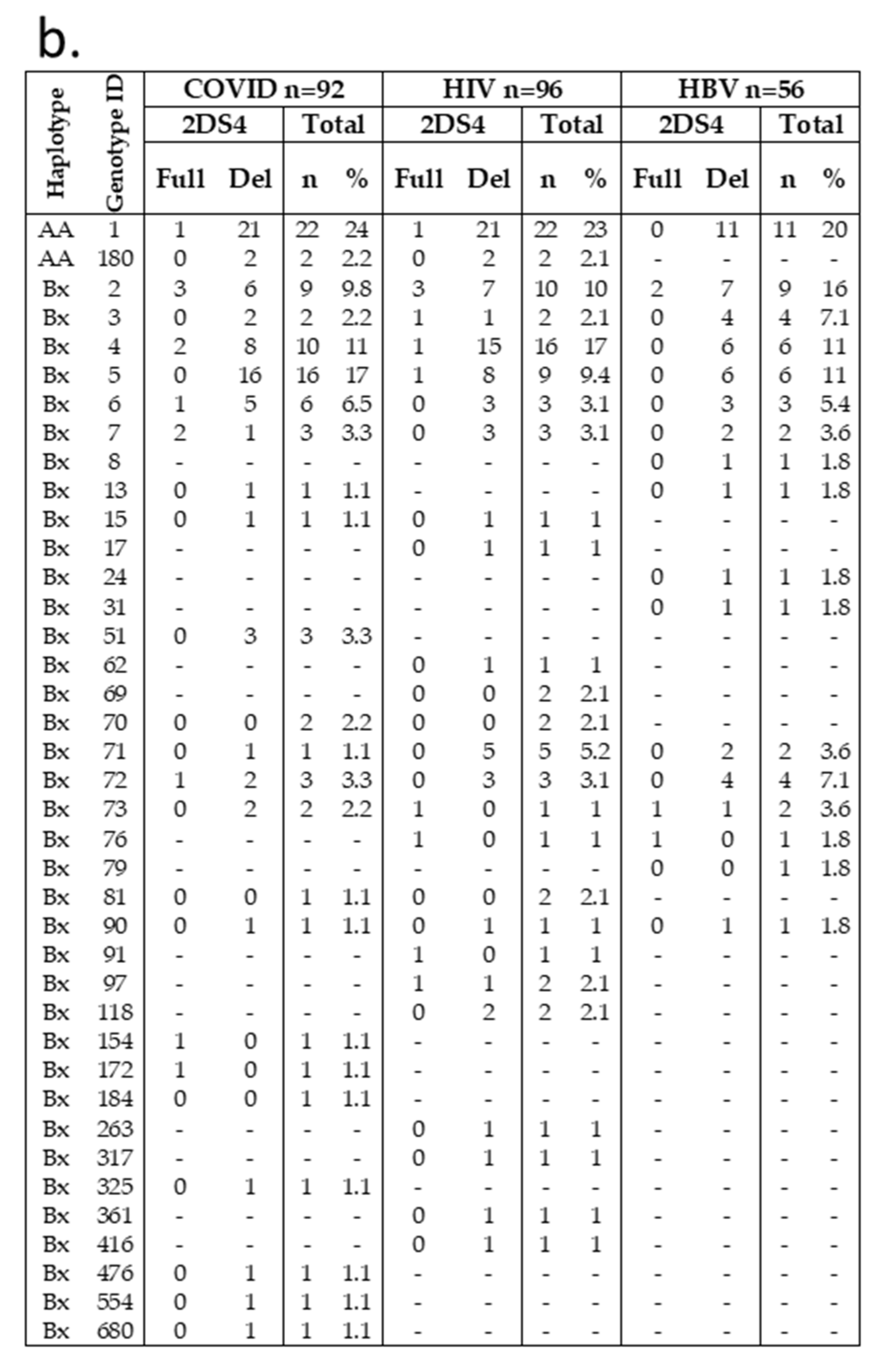
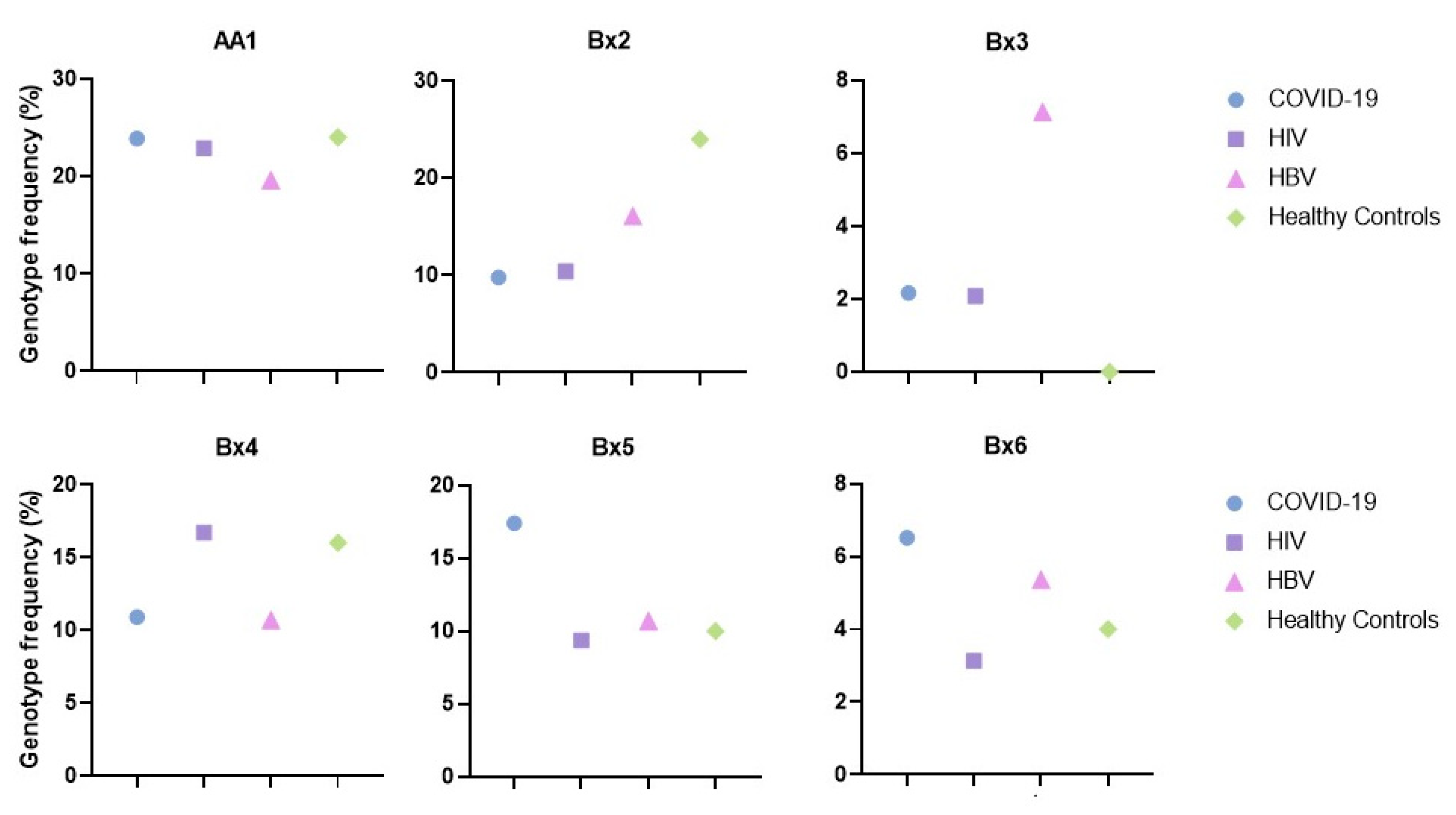
| COVID-19 Ntot = 92 N (%) | HIV Ntot = 96 N (%) | HBV Ntot = 56 N (%) | |
|---|---|---|---|
| Haplotypes | |||
| AA | 24 (26) | 24 (25) | 11 (20) |
| AB + BB | 68 (74) | 72 (75) | 45 (80) |
| KIR activating genes | |||
| 2DS1 | 30 (33) | 33 (34) | 25 (45) |
| 2DS2 | 50 (54) | 58 (60) | 33 (59) |
| 2DS3 | 38 (41) | 34 (35) | 18 (32) |
| 2DS4 Full | 13 (14) | 10 (10) | 4 (7) |
| 2DS4 Del | 74 (80) | 79 (82) | 51 (91) |
| 2DS5 | 25 (21) | 27 (28) | 21 (38) |
| 3DS1 | 32 (35) | 31 (32) | 26 (46) |
| KIR inhibitory genes | |||
| 2DL1 | 88 (96) | 89 (93) | 53 (95) |
| 2DL2 | 55 (60) | 58 (60) | 33 (59) |
| 2DL3 | 79 (86) | 73 (76) | 47 (84) |
| 2DL5A | 30 (33) | 31 (32) | 26 (46) |
| 2DL5B | 32 (35) | 30 (31) | 18 (32) |
| 3DL1 | 88 (96) | 88 (92) | 56 (100) |
| HLA groups | |||
| HLA-C1 | 77 (84) | 75 (75) | 36 (64) |
| HLA-C2 | 64 (70) | 64 (67) | 48 (86) |
| HLA Bw4 | 71 (77) | 77 (80) | 50 (89) |
| 20 (22) | 23 (24) | 14 (25) |
| 50 (54) | 52 (54) | 31 (55) |
| 22 (24) | 41 (32) | 32 (57) |
| HLA-C1/HLA-C1 | 28 (30) | 32 (33) | 7 (13) |
| HLA-C2/HLA-C2 | 15 (16) | 24(25) | 19 (34) |
| HLA-C1/HLA-C2 | 49 (53) | 40 (42) | 29 (52) |
| KIR–HLA combinations | |||
| 3DL1/HLA Bw4 | 67 (73) | 70 (73) | 50 (89) |
| 46 (50) | 44 (46) | 31 (55) |
| 18 (20) | 21 (22) | 14 (25) |
| 21 (23) | 29 (30) | 32 (57) |
| 3DS1/HLA Bw4 | 24 (26) | 23 (24) | 18 (32) |
| 20 (22) | 16 (80) | 12 (21) |
| 8 (9) | 10 (10) | 13 (23) |
| 2DL1/HLA-C2 | 60 (65) | 57 (39) | 46 (82) |
| 2DL2/HLA-C1 | 43 (47) | 39 (41) | 20 (36) |
| 2DL3/HLA-C1 | 64 (70) | 49 (51) | 30 (54) |
| 2DS1/HLA-C2 | 60 (21) | 15 (16) | 23 (41) |
| 2DS2/HLA-C1 | 38 (41) | 39 (41) | 20 (36) |
| HC Ntot = 50 N (%) | COVID-19 vs. HC Crude-OR | p-Value | HIV vs. HC Crude-OR | p-Value | HBV vs. HC Crude-OR | p-Value | |
|---|---|---|---|---|---|---|---|
| KIR activating genes | |||||||
| 2DS1 | 22 (44) | 1.62 | ns | 0.77 | ns | 1.03 | ns |
| 2DS2 | 27 (54) | 1.01 | ns | 1.30 | ns | 1.22 | ns |
| 2DS3 | 22 (44) | 0.89 | ns | 0.70 | ns | 0.60 | ns |
| 2DS4 | 46 (92) | 1.51 | ns | 1.10 | ns | 4.78 | ns |
| 2DS5 | 14 (28) | 0.96 | ns | 1.0 | ns | 1.54 | ns |
| 3DS1 | 20 (40) | 0.80 | ns | 0.71 | ns | 1.30 | ns |
| KIR inhibitory genes | |||||||
| 2DL1 | 50 (100) | 0.00 | ns | 0.00 | ns | 0.00 | ns |
| 2DL2 | 30 (60) | 0.99 | ns | 1.02 | ns | 0.96 | ns |
| 2DL3 | 44 (88) | 0.83 | ns | 0.43 | ns | 0.71 | ns |
| 2DL5A | 26 (52) | 0.44 | 0.031 | 0.44 | 0.031 | 0.80 | ns |
| 2DL5B | 21 (42) | 0.74 | ns | 0.63 | ns | 0.65 | ns |
| 3DL1 | 45 (90) | 2.44 | ns | 1.22 | ns | ∞ | 0.021 |
| HLA groups | |||||||
| HLA-C1 | 27 (54) | 4.37 | 0.0003 | 2.55 | 0.015 | 1.53 | ns |
| HLA-C2 | 23 (46) | 2.68 | 0.007 | 2.34 | 0.021 | 7.04 | <0.0001 |
| HLA-B Bw4 | 17 (34) | 4.43 | <0.0001 | 3.88 | 0.0002 | 10.14 | <0.0001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ligotti, M.E.; Aiello, A.; Accardi, G.; Calabrò, A.; Ciaccio, M.; Colomba, C.; Di Bona, D.; Lo Sasso, B.; Pojero, F.; Tuttolomondo, A.; et al. Distribution of KIR Genes and Their HLA Ligands in Different Viral Infectious Diseases: Frequency Study in Sicilian Population. Int. J. Mol. Sci. 2022, 23, 15466. https://doi.org/10.3390/ijms232415466
Ligotti ME, Aiello A, Accardi G, Calabrò A, Ciaccio M, Colomba C, Di Bona D, Lo Sasso B, Pojero F, Tuttolomondo A, et al. Distribution of KIR Genes and Their HLA Ligands in Different Viral Infectious Diseases: Frequency Study in Sicilian Population. International Journal of Molecular Sciences. 2022; 23(24):15466. https://doi.org/10.3390/ijms232415466
Chicago/Turabian StyleLigotti, Mattia Emanuela, Anna Aiello, Giulia Accardi, Anna Calabrò, Marcello Ciaccio, Claudia Colomba, Danilo Di Bona, Bruna Lo Sasso, Fanny Pojero, Antonino Tuttolomondo, and et al. 2022. "Distribution of KIR Genes and Their HLA Ligands in Different Viral Infectious Diseases: Frequency Study in Sicilian Population" International Journal of Molecular Sciences 23, no. 24: 15466. https://doi.org/10.3390/ijms232415466
APA StyleLigotti, M. E., Aiello, A., Accardi, G., Calabrò, A., Ciaccio, M., Colomba, C., Di Bona, D., Lo Sasso, B., Pojero, F., Tuttolomondo, A., Caruso, C., Candore, G., & Duro, G. (2022). Distribution of KIR Genes and Their HLA Ligands in Different Viral Infectious Diseases: Frequency Study in Sicilian Population. International Journal of Molecular Sciences, 23(24), 15466. https://doi.org/10.3390/ijms232415466












