Conservation Study of Imprinted Genes in Maize Triparental Heterozygotic Kernels
Abstract
:1. Introduction
2. Results
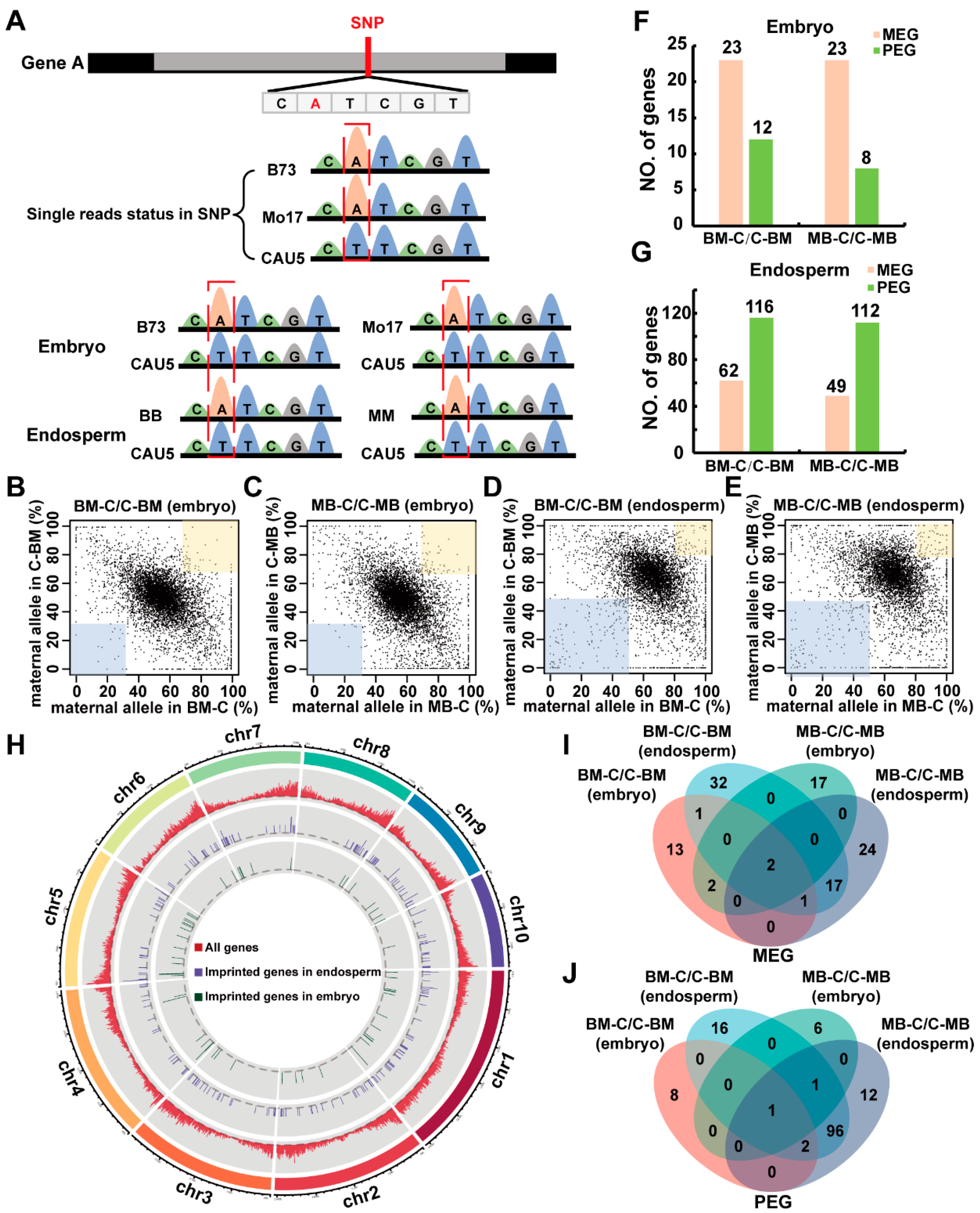
2.1. Analysis of Gene Expression and Identification of Imprinted Genes in Reciprocal Crosses
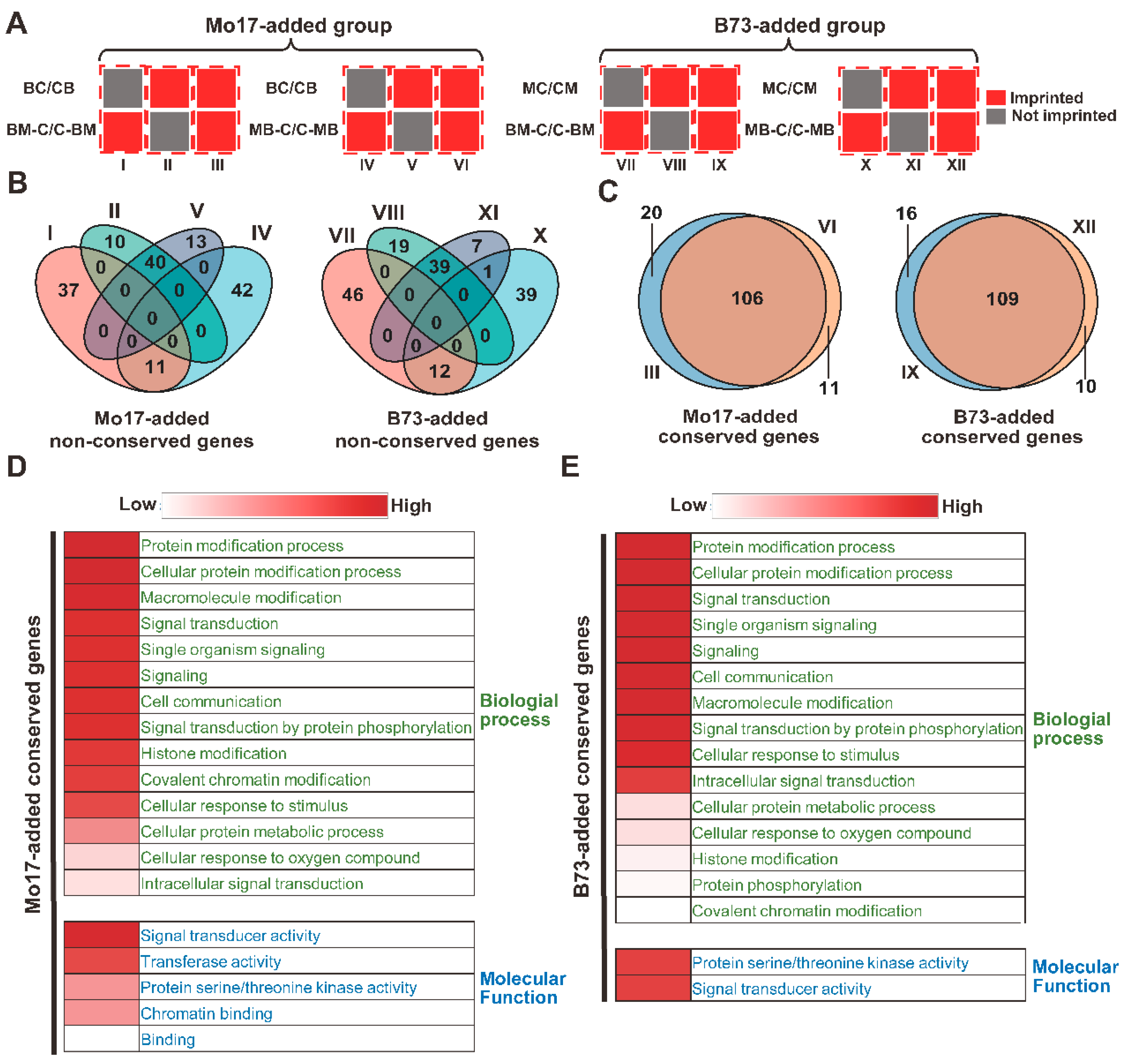
2.2. Conservation and GO Analysis of the Non-Conserved Genes
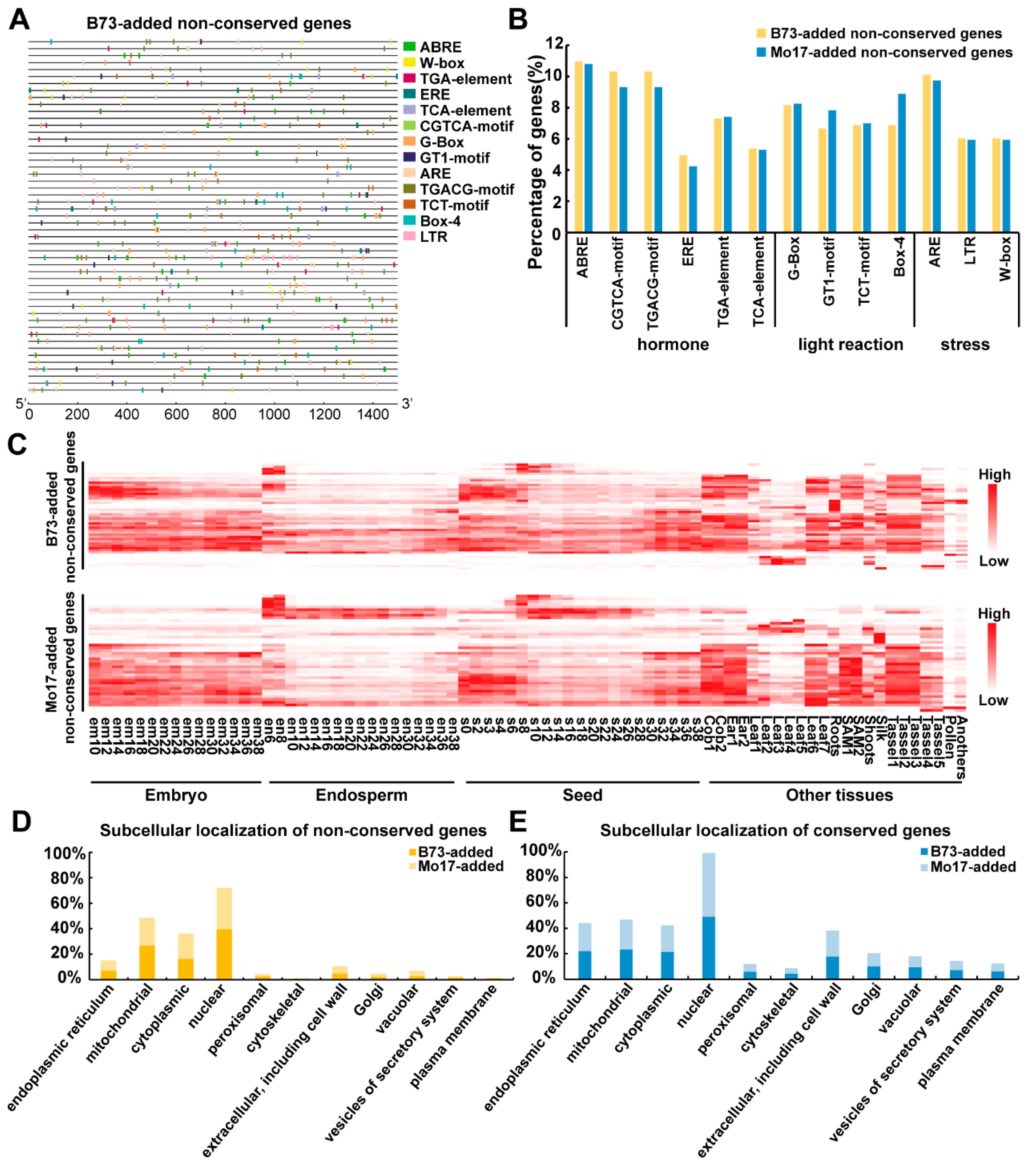
2.3. Cis-Element Distribution, Expression Pattern and Subcellular Location of Proteins from the Conserved and Non-Conserved Imprinted Genes
2.4. Extremely Non-Conserved Imprinted Genes Detected in Different Species
2.5. Phenotype Analysis of the Non-Conserved PEG Zm00001d020769 Transgenic Line
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Library Construction for RNA-Seq
4.3. Read Mapping, Gene Expression Analysis and SNP Calling
4.4. Measuring Allelic Expression and Identification of Imprinted Genes
4.5. GO Term Enrichment and Functional Category Analysis
4.6. Validation of Imprinted Genes
4.7. Genetic Transformation of Maize
4.8. Primers
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Kelsey, G.; Feil, R. New insights into establishment and maintenance of DNA methylation imprints in mammals. Philos. Trans. R. Soc. B 2013, 368, 20110336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kermicle, J.L. Dependence of the R-mottled aleurone phenotype in maize on mode of sexual transmission. Genetics 1970, 66, 69–85. [Google Scholar] [CrossRef] [PubMed]
- Gehring, M. Genomic imprinting: Insights from plants. Annu. Rev. Genet. 2013, 47, 187–208. [Google Scholar] [CrossRef] [PubMed]
- Selinger, D.A.; Chandler, V.L. B-Bolivia, an allele of the maize b1 gene with variable expression, contains a high copy retrotransposon-related sequence immediately upstream. Plant Physiol. 2001, 125, 1363–1379. [Google Scholar] [CrossRef] [Green Version]
- Dumas, C.; Mogensen, H.L. Gametes and fertilization: Maize as a model system for experimental embryogenesis in flowering plants. Plant Cell 1993, 5, 1337–1348. [Google Scholar] [CrossRef] [Green Version]
- Méchin, V.; Thévenot, C.; Le Guilloux, M.; Prioul, J.L.; Damerval, C. Developmental analysis of maize endosperm proteome suggests a pivotal role for pyruvate orthophosphate dikinase. Plant Physiol. 2007, 143, 1203–1219. [Google Scholar] [CrossRef] [Green Version]
- Feng, J.; Xu, X.; Liu, X.; Zhang, C.; Liang, X.; Wu, W. Embryogenesis, germination, structure and cotyledon dimorphism of Zea mays embryo. Acta Bot. Sin. 2003, 45, 712–723. [Google Scholar]
- Chen, J.; Zeng, B.; Zhang, M.; Xie, S.; Wang, G.; Hauck, A.; Lai, J. Dynamic transcriptome landscape of maize embryo and endosperm development. Plant Physiol. 2014, 166, 252–264. [Google Scholar] [CrossRef] [Green Version]
- Zhan, J.; Thakare, D.; Ma, C.; Lloyd, A.; Nixon, N.M.; Arakaki, A.M.; Burnett, W.J.; Logan, K.O.; Wang, D.; Wang, X.; et al. RNA sequencing of laser-capture microdissected compartments of the maize kernel identifies regulatory modules associated with endosperm cell differentiation. Plant Cell 2015, 27, 513–531. [Google Scholar] [CrossRef]
- Hsieh, T.F.; Shin, J.; Uzawa, R.; Silva, P.; Cohen, S.; Bauer, M.J.; Hashimoto, M.; Kirkbride, R.C.; Harada, J.J.; Zilberman, D.; et al. Regulation of imprinted gene expression in Arabidopsis endosperm. Proc. Natl. Acad. Sci. USA 2011, 108, 1755–1762. [Google Scholar] [CrossRef] [Green Version]
- Waters, A.J.; Makarevitch, I.; Eichten, S.R.; Swanson-Wagner, R.A.; Yeh, C.T.; Xu, W.; Schnable, P.S.; Vaughn, M.W.; Gehring, M.; Springer, N.M. Parent-of-origin effects on gene expression and DNA methylation in the maize endosperm. Plant Cell 2011, 23, 4221–4233. [Google Scholar] [CrossRef] [PubMed]
- Xin, M.; Yang, R.; Li, G.; Chen, H.; Laurie, J.; Ma, C.; Wang, D.; Yao, Y.; Larkins, B.A.; Sun, Q.; et al. Dynamic Expression of imprinted genes associates with maternally controlled nutrient allocation during maize endosperm development. Plant Cell 2013, 25, 3212–3227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pignatta, D.; Erdmann, R.M.; Scheer, E.; Picard, C.L.; Bell, G.W.; Gehring, M. Natural epigenetic polymorphisms lead to intraspecific variation in Arabidopsis gene imprinting. Elife 2014, 3, e03198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, W.; Dai, M.; Li, F.; Liu, A. Genomic imprinting, methylation and parent-of-origin effects in reciprocal hybrid endosperm of castor bean. Nucleic Acids Res. 2014, 42, 6987–6998. [Google Scholar] [CrossRef] [Green Version]
- Hatorangan, M.R.; Laenen, B.; Steige, K.A.; Slotte, T.; Köhler, C. Rapid evolution of genomic imprinting in two species of the brassicaceae. Plant Cell 2016, 28, 1815–1827. [Google Scholar] [CrossRef] [Green Version]
- Klosinska, M.; Picard, C.L.; Gehring, M. Conserved imprinting associated with unique epigenetic signatures in the Arabidopsis genus. Nat. Plants 2016, 2, 16145. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Li, N.; He, W.; Zhang, H.; Yang, W.; Liu, B. Genome-wide screen of genes imprinted in sorghum endosperm, and the roles of allelic differential cytosine methylation. Plant J. 2016, 85, 424–436. [Google Scholar] [CrossRef] [Green Version]
- Meng, D.; Zhao, J.; Zhao, C.; Luo, H.; Xie, M.; Liu, R.; Lai, J.; Zhang, X.; Jin, W. Sequential Gene Activation and Gene Imprinting during Early Embryo Development in Maize. Plant J. 2018, 93, 445–459. [Google Scholar] [CrossRef] [Green Version]
- Bernardi, J.; Lanubile, A.; Li, Q.B.; Kumar, D.; Kladnik, A.; Cook, S.D.; Ross, J.J.; Marocco, A.; Chourey, P.S. Impaired auxin biosynthesis in the defective endosperm18 mutant is due to mutational loss of expression in the ZmYuc1 gene encoding endosperm-specific yucca1 protein in maize. Plant Physiol. 2012, 160, 1318–1328. [Google Scholar] [CrossRef] [Green Version]
- Gerald, J.N.F.; Hui, P.S.; Berger, F. Polycomb group-dependent imprinting of the actin regulator ATFH5 regulates morphogenesis in Arabidopsis thaliana. Development 2009, 136, 3399–3404. [Google Scholar] [CrossRef] [Green Version]
- Patten, M.M.; Ross, L.; Curley, J.P.; Queller, D.C.; Bonduriansky, R.; Wolf, J.B. The evolution of genomic imprinting: Theories, predictions and empirical tests. Heredity 2014, 113, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Haig, D.; Westoby, M. Parent-specific gene expression and the triploid endosperm. Am. Nat. 1989, 134, 147–155. [Google Scholar] [CrossRef] [Green Version]
- Haig, D. The kinship theory of genomic imprinting. Annu. Rev. Ecol. Syst. 2000, 31, 9–32. [Google Scholar] [CrossRef] [Green Version]
- Haig, D. Genomic imprinting and kinship: How good is the evidence? Annu. Rev. Genet. 2004, 38, 553–585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Day, T.; Bonduriansky, R. Intralocus Sexual Conflict Can Drive the Evolution of Genomic Imprinting. Genetics 2004, 167, 1537–1546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolf, J.B.; Hager, R. A Maternal-offspring coadaptation theory for the evolution of genomic imprinting. PLoS Biol. 2006, 4, 2238–2243. [Google Scholar] [CrossRef] [Green Version]
- Wolf, J.B.; Hager, R. Selective abortion and the evolution of genomic imprinting. J. Evol. Biol. 2009, 22, 2519–2523. [Google Scholar] [CrossRef]
- Wolf, J.B. Evolution of genomic imprinting as a coordinator of coadapted gene expression. Proc. Natl. Acad. Sci. USA 2013, 110, 5085–5090. [Google Scholar] [CrossRef] [Green Version]
- Lizé, A.; Cortesero, A.M.; Atlan, A.; Poinsot, D. Kin recognition in aleochara bilineata could support the kinship theory of genomic imprinting. Genetics 2007, 175, 1735–1740. [Google Scholar] [CrossRef] [Green Version]
- Pegoraro, M.; Marshall, H.; Lonsdale, Z.N.; Mallon, E.B. Do social insects support haig’s kin theory for the evolution of genomic imprinting? Epigenetics 2017, 12, 725–742. [Google Scholar] [CrossRef]
- Luo, M.; Taylor, J.M.; Spriggs, A.; Zhang, H.; Wu, X.; Russell, S.; Singh, M.; Koltunow, A. A genome-wide survey of imprinted genes in rice seeds reveals imprinting primarily occurs in the endosperm. PLoS Genet. 2011, 7, e1002125. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Li, T.; Zhu, S.; Liu, Z.; Shi, Z.; Zheng, X.; Chen, R.; Huang, J.; Shen, Y.; Luo, S.; et al. Characterization of imprinted genes in rice reveals conservation of regulation and imprinting with other plant species. Plant Physiol. 2018, 177, 1754–1771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gehring, M.; Missirian, V.; Henikoff, S. Genomic analysis of parent-of-origin allelic expression in Arabidopsis thaliana seeds. PLoS ONE 2011, 6, e23687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.; Zhao, H.; Xie, S.; Chen, J.; Xu, Y.; Wang, K.; Zhao, H.; Guan, H.; Hu, X.; Jiao, Y.; et al. Extensive, clustered parental imprinting of protein-coding and noncoding RNAs in developing maize endosperm. Proc. Natl. Acad. Sci. USA 2011, 108, 20042–20047. [Google Scholar] [CrossRef] [Green Version]
- Waters, A.J.; Bilinski, P.; Eichten, S.R.; Vaughn, M.W.; Ross-Ibarra, J.; Gehring, M.; Springer, N.M. Comprehensive analysis of imprinted genes in maize reveals allelic variation for imprinting and limited conservation with other species. Proc. Natl. Acad. Sci. USA 2013, 110, 19639–19644. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Xing, F.; He, Q.; Qamar, M.T.U.; Chen, L.L.; Xing, Y. Conserved imprinted genes between intra-subspecies and inter-subspecies are involved in energy metabolism and seed development in rice. Int. J. Mol. Sci. 2020, 21, 9618. [Google Scholar] [CrossRef]
- MacDonald, W.A. Epigenetic mechanisms of genomic imprinting: Common themes in the regulation of imprinted regions in mammals, plants, and insects. Genet. Res. Int. 2012, 2012, 585024. [Google Scholar] [CrossRef] [Green Version]
- Vaughn, M.W.; Tanurdžić, M.; Lippman, Z.; Jiang, H.; Carrasquillo, R.; Rabinowicz, P.D.; Dedhia, N.; McCombie, W.R.; Agier, N.; Bulski, A.; et al. Epigenetic natural variation in Arabidopsis thaliana. PLoS Biol. 2007, 5, e174. [Google Scholar] [CrossRef]
- Groszmann, M.; Greaves, I.K.; Albertyn, Z.I.; Scofield, G.N.; Peacock, W.J.; Dennis, E.S. Changes in 24-Nt SiRNA levels in Arabidopsis hybrids suggest an epigenetic contribution to hybrid vigor. Proc. Natl. Acad. Sci. USA 2011, 108, 2617–2622. [Google Scholar] [CrossRef] [Green Version]
- He, G.; Chen, B.; Wang, X.; Li, X.; Li, J.; He, H.; Yang, M.; Lu, L.; Qi, Y.; Wang, X.; et al. Conservation and Divergence of transcriptomic and epigenomic variation in maize hybrids. Genome Biol. 2013, 14, R57. [Google Scholar] [CrossRef] [Green Version]
- Raissig, M.T.; Bemer, M.; Baroux, C.; Grossniklaus, U. Genomic imprinting in the Arabidopsis embryo is partly regulated by PRC2. PLoS Genet. 2013, 9, e1003862. [Google Scholar] [CrossRef] [PubMed]
- Meyer, S.; Pospisil, H.; Scholten, S. Heterosis associated gene expression in maize embryos 6 days after fertilization exhibits additive, dominant and overdominant pattern. Plant Mol. Biol. 2007, 63, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Xie, S.; Dong, X.; Zhao, X.; Zeng, B.; Chen, J.; Li, H.; Yang, W.; Zhao, H.; Wang, G.; et al. Genome-wide high resolution parental-specific DNA and histone methylation maps uncover patterns of imprinting regulation in maize. Genome Res. 2014, 24, 167–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, S.; Scholten, S. Equivalent parental contribution to early plant zygotic development. Curr. Biol. 2007, 17, 1686–1691. [Google Scholar] [CrossRef] [Green Version]
- Chodavarapu, R.K.; Feng, S.; Ding, B.; Simon, S.A.; Lopez, D.; Jia, Y.; Wang, G.L.; Meyers, B.C.; Jacobsen, S.E.; Pellegrini, M. Transcriptome and methylome interactions in rice hybrids. Proc. Natl. Acad. Sci. USA 2012, 109, 12040–12045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greaves, I.K.; Groszmann, M.; Ying, H.; Taylor, J.M.; Peacock, W.J.; Dennis, E.S. Trans chromosomal methylation in Arabidopsis hybrids. Proc. Natl. Acad. Sci. USA 2012, 109, 3570–3575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, H.; He, H.; Li, J.; Chen, W.; Wang, X.; Guo, L.; Peng, Z.; He, G.; Zhong, S.; Qi, Y.; et al. Genome-wide analysis of DNA methylation and gene expression changes in two Arabidopsis ecotypes and their reciprocal hybrids. Plant Cell 2012, 24, 875–892. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotech. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential Gene and Transcript Expression Analysis of RNA-Seq Experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Durbin, R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. AgriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishida, Y.; Hiei, Y.; Komari, T. Agrobacterium-mediated transformation of maize. Nat. Protoc. 2007, 2, 1614–1621. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, X.; Luo, H.; Yao, J.; Guo, Q.; Yu, S.; Zhang, X.; Li, F.; Ruan, Y.; Jin, W.; Meng, D. Conservation Study of Imprinted Genes in Maize Triparental Heterozygotic Kernels. Int. J. Mol. Sci. 2022, 23, 15424. https://doi.org/10.3390/ijms232315424
Dong X, Luo H, Yao J, Guo Q, Yu S, Zhang X, Li F, Ruan Y, Jin W, Meng D. Conservation Study of Imprinted Genes in Maize Triparental Heterozygotic Kernels. International Journal of Molecular Sciences. 2022; 23(23):15424. https://doi.org/10.3390/ijms232315424
Chicago/Turabian StyleDong, Xiaomei, Haishan Luo, Jiabin Yao, Qingfeng Guo, Shuai Yu, Xiaoyu Zhang, Fenghai Li, Yanye Ruan, Weiwei Jin, and Dexuan Meng. 2022. "Conservation Study of Imprinted Genes in Maize Triparental Heterozygotic Kernels" International Journal of Molecular Sciences 23, no. 23: 15424. https://doi.org/10.3390/ijms232315424
APA StyleDong, X., Luo, H., Yao, J., Guo, Q., Yu, S., Zhang, X., Li, F., Ruan, Y., Jin, W., & Meng, D. (2022). Conservation Study of Imprinted Genes in Maize Triparental Heterozygotic Kernels. International Journal of Molecular Sciences, 23(23), 15424. https://doi.org/10.3390/ijms232315424





