A Single Variant in Pri-miRNA-155 Associated with Susceptibility to Hereditary Breast Cancer Promotes Aggressiveness in Breast Cancer Cells
Abstract
1. Introduction
2. Results
2.1. Variant Identification in miRNA Genes of BC Patients
2.2. Association Study of Variants rs376491654, rs755634302, and rs190708267
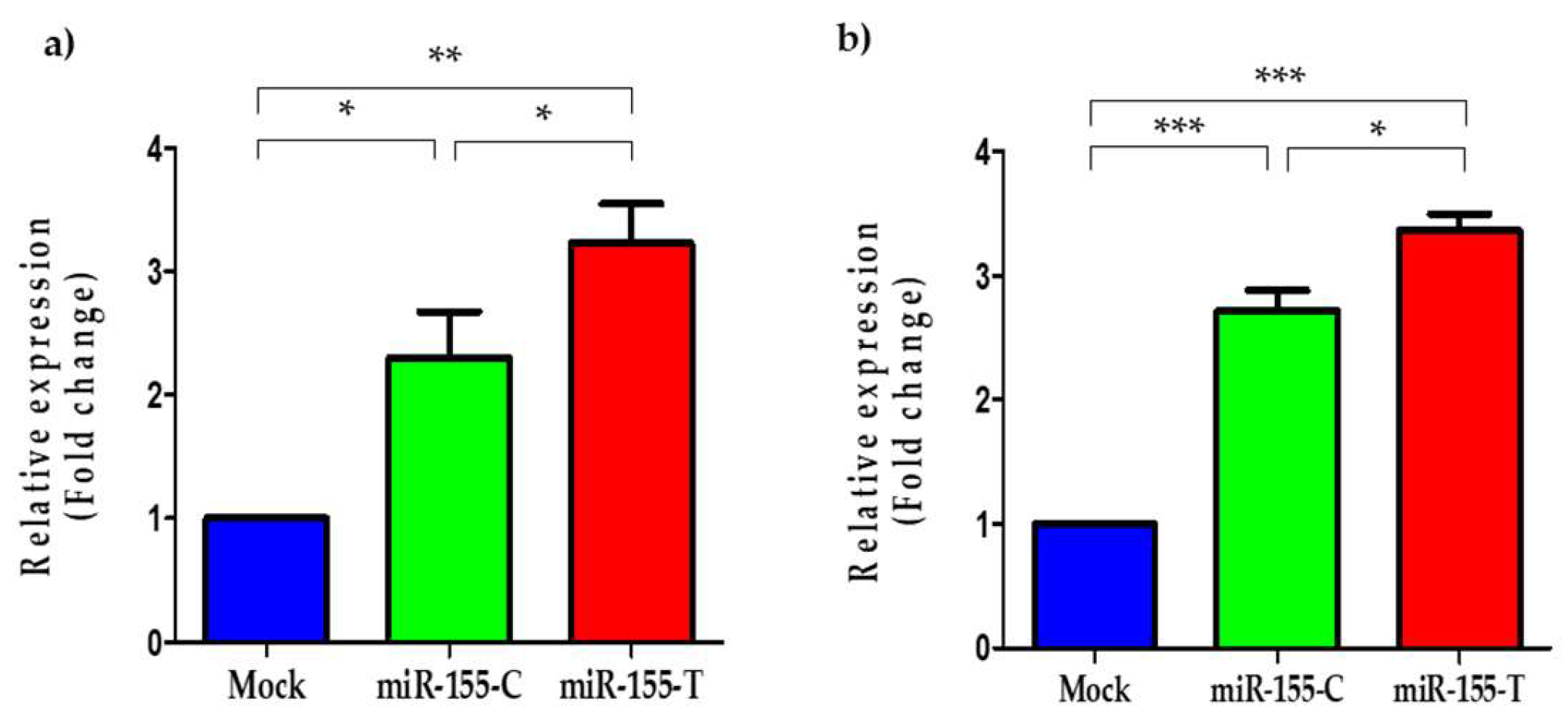
2.3. The miR-155-T Allele Increases Levels of the Mature miR-155-5p in BC Cells
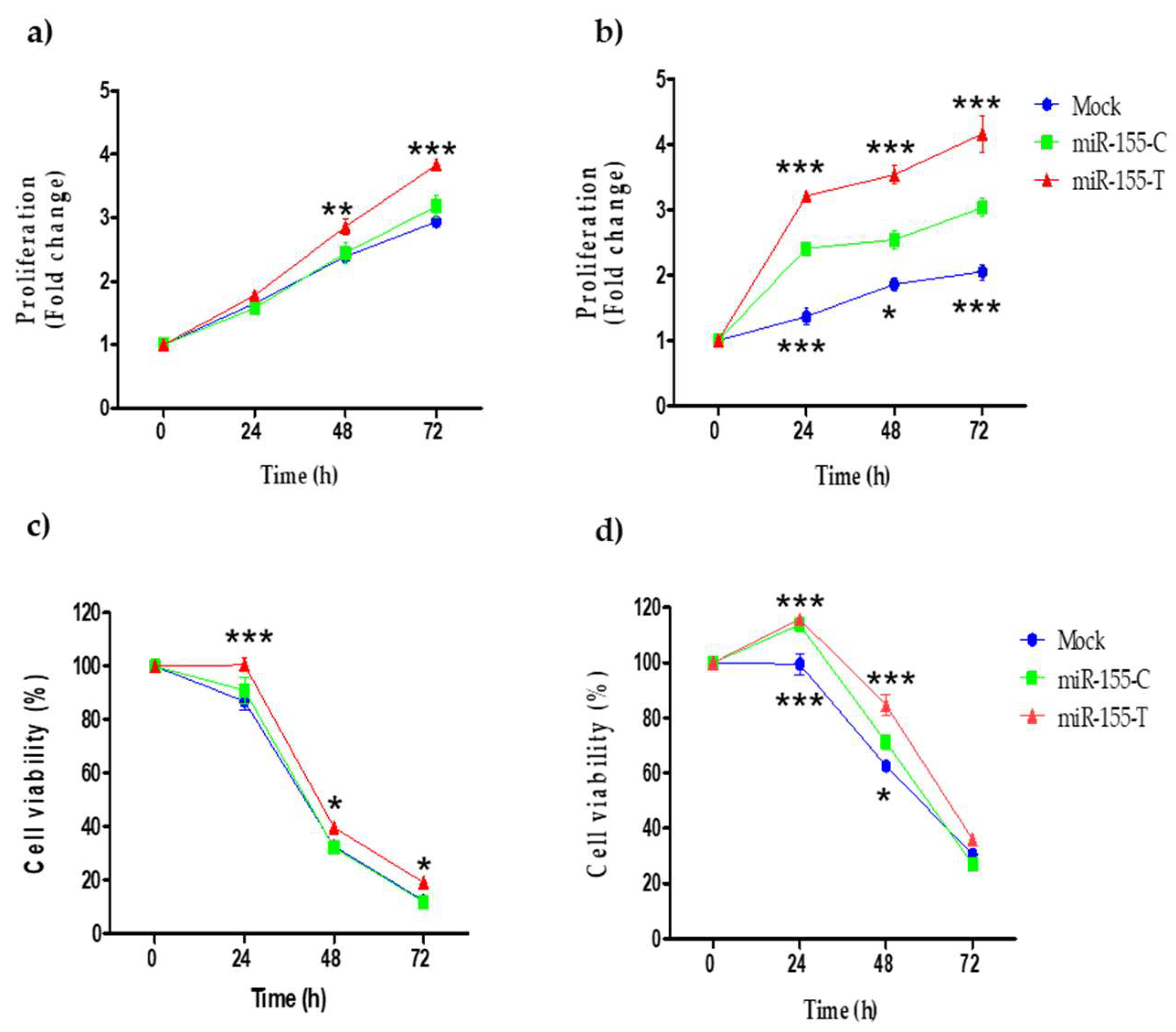
2.4. Proliferation and Resistance to Cisplatin Are Promoted by the miR-155-T Allele
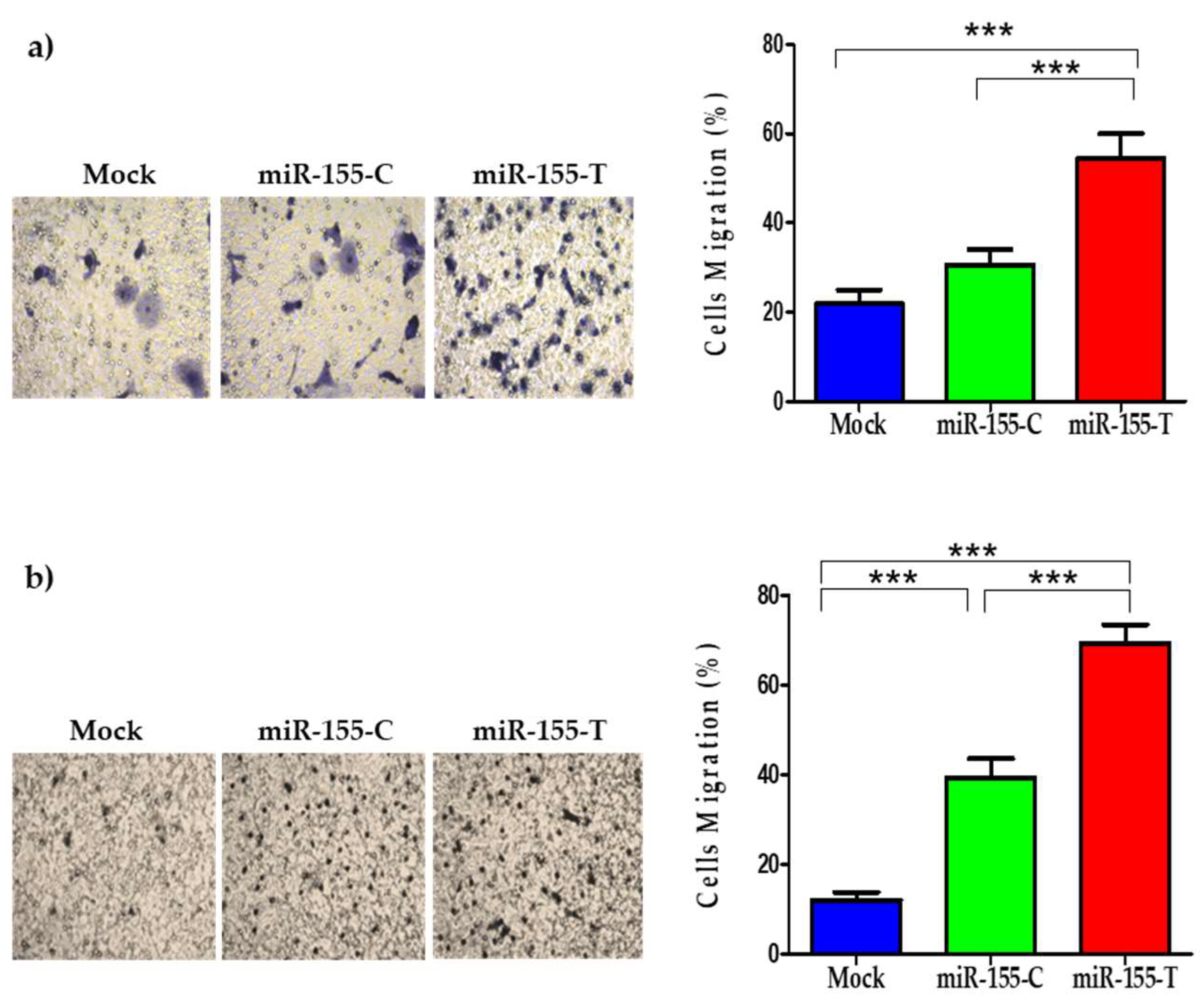
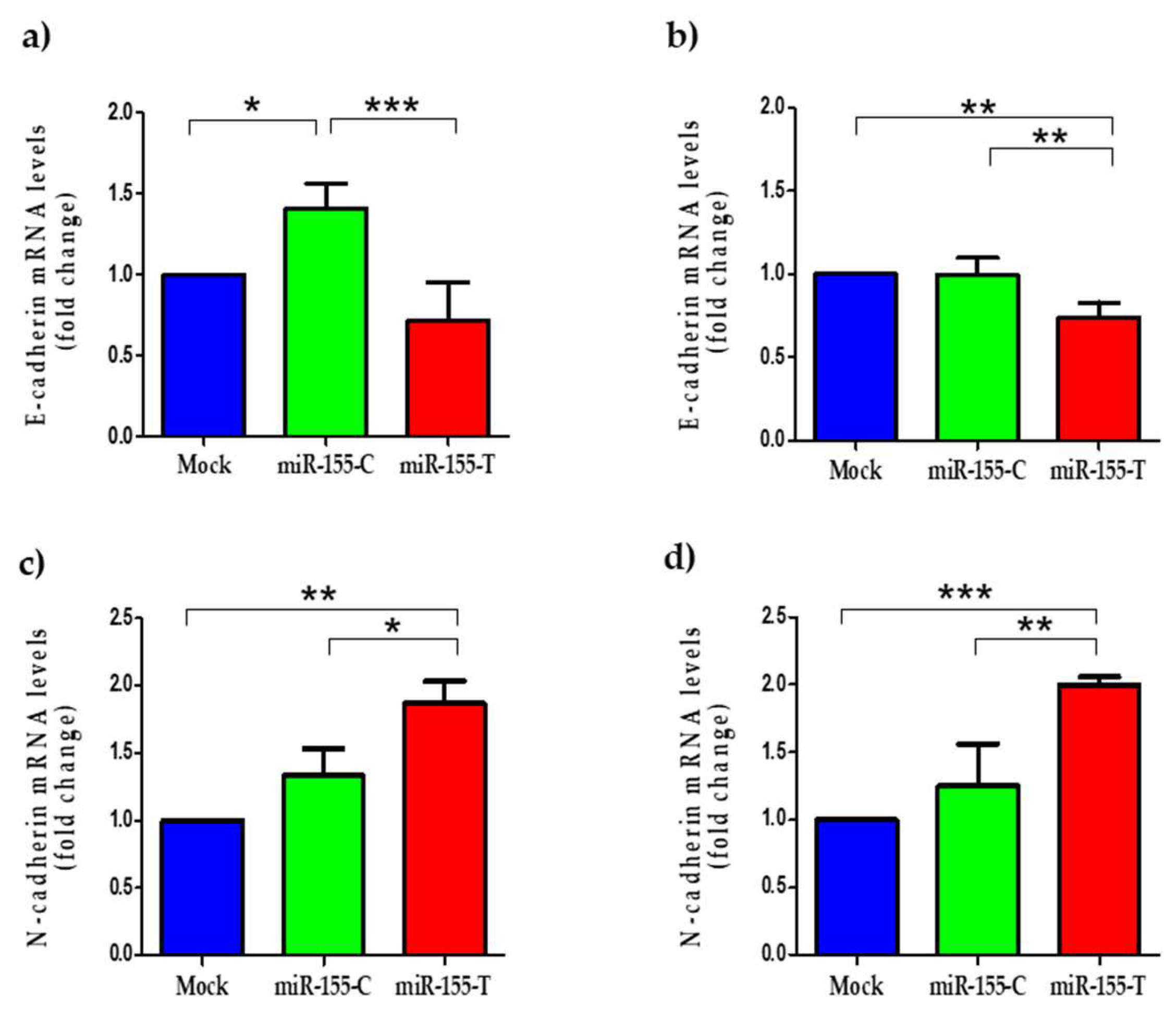
2.5. The miR-155-T Allele Favors Migration and Expression of EMT-Marker Genes
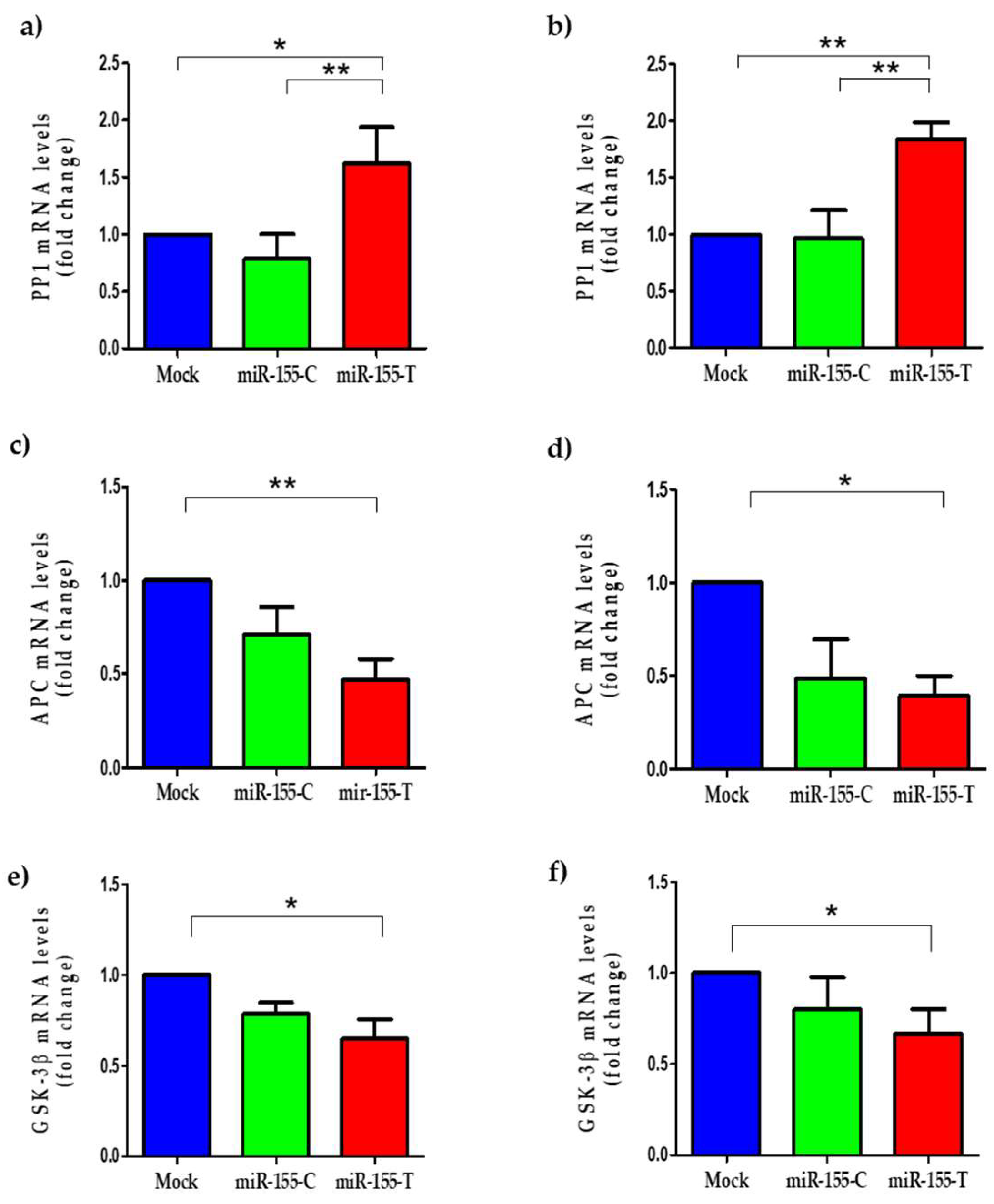
2.6. The miR-155-T Allele Modulates Some Canonical Wnt Pathway Regulators
3. Discussion
4. Materials and Methods
4.1. BC Population Study
4.2. Selection and Sequencing of Deregulated microRNA Genes in BRCA Negative Patients
4.3. In Silico Analysis of Secondary Structures of the Pri-microRNAs
4.4. Genotyping of the Selected Variants
4.5. Cloning of the Polymorphic miR-155 Sequences
4.6. Cell Cultures and Transfection of Polymorphic microRNAs
4.7. RNA Extraction and RT-qPCR
4.8. Transwell Migration Assay
4.9. Cell Proliferation and Cisplatin Resistance
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef]
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Couch, F.J.; Nathanson, K.L.; Offit, K. Two Decades After BRCA: Setting Paradigms in Personalized Cancer Care and Prevention. Science 2014, 343, 1466–1470. [Google Scholar] [CrossRef]
- Pharoah, P.D.P.; Dunning, A.M.; Ponder, B.A.J.; Easton, D.F. Association studies for finding cancer-susceptibility genetic variants. Nat. Rev. Cancer 2004, 4, 850–860. [Google Scholar] [CrossRef]
- Iorio, M.V.; Ferracin, M.; Liu, C.-G.; Veronese, A.; Spizzo, R.; Sabbioni, S.; Magri, E.; Pedriali, M.; Fabbri, M.; Campiglio, M.; et al. MicroRNA Gene Expression Deregulation in Human Breast Cancer. Cancer Res. 2005, 65, 7065–7070. [Google Scholar] [CrossRef]
- Tahiri, A.; Aure, M.R.; Kristensen, V.N. MicroRNA Networks in Breast Cancer Cells. Methods Mol. Biol. 2018, 1711, 55–81. [Google Scholar]
- Tekiner, T.A.; Basaga, H. Role of microRNA deregulation in breast cancer cell chemoresistance and stemness. Curr. Med. Chem. 2013, 20, 3358–3369. [Google Scholar] [CrossRef] [PubMed]
- Sabit, H.; Cevik, E.; Tombuloglu, H.; Abdel-Ghany, S.; Tombuloglu, G.; Esteller, M. Triple negative breast cancer in the era of miRNA. Crit. Rev. Oncol./Hematol. 2021, 157, 103196. [Google Scholar] [CrossRef] [PubMed]
- Balkrishna, A.; Mittal, R.; Arya, V. Potential Role of miRNA in Metastatic Cascade of Triple-Negative Breast Cancer. Curr. Cancer Drug Targets 2021, 21, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Aigner, A. MicroRNAs (miRNAs) in cancer invasion and metastasis: Therapeutic approaches based on metastasis-related miRNAs. J. Mol. Med. 2011, 89, 445–457. [Google Scholar] [CrossRef]
- van Schooneveld, E.; Wildiers, H.; Vergote, I.; Vermeulen, P.B.; Dirix, L.Y.; Van Laere, S.J. Dysregulation of microRNAs in breast cancer and their potential role as prognostic and predictive biomarkers in patient management. Breast Cancer Res. 2015, 17, 21. [Google Scholar] [CrossRef]
- Bertoli, G.; Cava, C.; Castiglioni, I. MicroRNAs: New Biomarkers for Diagnosis, Prognosis, Therapy Prediction and Therapeutic Tools for Breast Cancer. Theranostics 2015, 5, 1122–1243. [Google Scholar] [CrossRef]
- Mashima, R. Physiological roles of miR-155. Immunology 2015, 145, 323–333. [Google Scholar] [CrossRef] [PubMed]
- Hsin, J.-P.; Lu, Y.; Loeb, G.B.; Leslie, C.S.; Rudensky, A.Y. The effect of cellular context on miR-155-mediated gene regulation in four major immune cell types. Nat. Immunol. 2018, 19, 1137–1145. [Google Scholar] [CrossRef] [PubMed]
- Metzler, M.; Wilda, M.; Busch, K.; Viehmann, S.; Borkhardt, A. High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer 2004, 39, 167–169. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.M.; Zhao, J.; Deng, H.Y. MiR-155 promotes proliferation of human breast cancer MCF-7 cells through targeting tumor protein 53-induced nuclear protein 1. J. Biomed. Sci. 2013, 20, 79. [Google Scholar] [CrossRef] [PubMed]
- Zuo, J.; Yu, Y.; Zhu, M.; Jing, W.; Yu, M.; Chai, H.; Liang, C.; Tu, J. Inhibition of miR-155, a therapeutic target for breast cancer, prevented in cancer stem cell formation. Cancer Biomarkers 2018, 21, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Van Roosbroeck, K.; Fanini, F.; Setoyama, T.; Ivan, C.; Rodriguez-Aguayo, C.; Fuentes-Mattei, E.; Xiao, L.; Vannini, I.; Redis, R.S.; D’Abundo, L.; et al. Combining Anti-Mir-155 with Chemotherapy for the Treatment of Lung Cancers. Clin. Cancer Res. 2017, 23, 2891–2904. [Google Scholar] [CrossRef] [PubMed]
- Lei, C.; Wang, Y.; Huang, Y.; Yu, H.; Huang, Y.; Wu, L.; Huang, L. Up-regulated miR155 reverses the epithelial-mesenchymal transition induced by EGF and increases chemo-sensitivity to cisplatin in human Caski cervical cancer cells. PLoS ONE 2012, 7, e52310. [Google Scholar] [CrossRef]
- Han, M.; Zheng, Y. Comprehensive analysis of single nucleotide polymorphisms in human microRNAs. PLoS ONE 2013, 8, e78028. [Google Scholar] [CrossRef]
- Sun, G.; Yan, J.; Noltner, K.; Feng, J.; Li, H.; Sarkis, D.A.; Sommer, S.S.; Rossi, J.J. SNPs in human miRNA genes affect biogenesis and function. RNA 2009, 15, 1640–1651. [Google Scholar] [CrossRef]
- Ryan, B.M.; Robles, A.I.; Harris, C.C. Genetic variation in microRNA networks: The implications for cancer research. Nat. Rev. Cancer 2010, 10, 389–402. [Google Scholar] [CrossRef] [PubMed]
- Bahreini, F.; Rayzan, E.; Rezaei, N. microRNA-related single-nucleotide polymorphisms and breast cancer. J. Cell. Physiol. 2021, 236, 1593–1605. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; He, L.; Coppola, M.; Guo, J.; Esposito, N.N.; Coppola, D.; Cheng, J.Q. MicroRNA-155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J. Biol. Chem. 2010, 285, 17869–17879. [Google Scholar] [CrossRef]
- Bayraktar, R.; Van Roosbroeck, K. miR-155 in cancer drug resistance and as target for miRNA-based therapeutics. Cancer Metastasis Rev. 2018, 37, 33–44. [Google Scholar] [CrossRef]
- Zang, Y.-S.; Zhong, Y.-F.; Fang, Z.; Li, B.; An, J. MiR-155 inhibits the sensitivity of lung cancer cells to cisplatin via negative regulation of Apaf-1 expression. Cancer Gene Ther. 2012, 19, 773–778. [Google Scholar] [CrossRef]
- Gasparini, P.; Lovat, F.; Fassan, M.; Casadei, L.; Cascione, L.; Jacob, N.K.; Carasi, S.; Palmieri, D.; Costinean, S.; Shapiro, C.L.; et al. Protective role of miR-155 in breast cancer through RAD51 targeting impairs homologous recombination after irradiation. Proc. Natl. Acad. Sci. USA 2014, 111, 4536–4541. [Google Scholar] [CrossRef]
- Shen, R.; Wang, Y.; Wang, C.-X.; Yin, M.; Liu, H.-L.; Chen, J.-P.; Han, J.-Q.; Wang, W.-B. MiRNA-155 mediates TAM resistance by modulating SOCS6-STAT3 signalling pathway in breast cancer. Am. J. Transl. Res. 2015, 7, 2115–2126. [Google Scholar] [PubMed]
- Yu, D.-D.; Lv, M.-M.; Chen, W.-X.; Zhong, S.; Zhang, X.-H.; Chen, L.; Ma, T.-F.; Tang, J.-H.; Zhao, J.-H. Role of miR-155 in drug resistance of breast cancer. Tumour Biol. 2015, 36, 1395–1401. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Weinberg, R.A. Epithelial-Mesenchymal Plasticity: A Central Regulator of Cancer Progression. Trends Cell Biol. 2015, 25, 675–686. [Google Scholar] [CrossRef]
- Zhang, Y.; Weinberg, R.A. Epithelial-to-mesenchymal transition in cancer: Complexity and opportunities. Front. Med. 2018, 12, 361–373. [Google Scholar] [CrossRef] [PubMed]
- Peng, F.; Xiong, L.; Tang, H.; Peng, C.; Chen, J. Regulation of epithelial-mesenchymal transition through microRNAs: Clinical and biological significance of microRNAs in breast cancer. Tumour Biol. 2016, 37, 14463–14477. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Chen, C.J.; Guo, G.L. MiR-155 promotes the proliferation and migration of breast cancer cells via targeting SOCS1 and MMP16. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 7323–7332. [Google Scholar]
- Jiang, S.; Zhang, H.-W.; Lu, M.-H.; He, X.-H.; Li, Y.; Gu, H.; Liu, M.-F.; Wang, E.-D. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 2010, 70, 3119–3127. [Google Scholar] [CrossRef]
- Zhang, Y.; Wei, W.; Cheng, N.; Wang, K.; Li, B.; Jiang, X.; Sun, S. Hepatitis C virus-induced up-regulation of microRNA-155 promotes hepatocarcinogenesis by activating Wnt signaling. Hepatology 2012, 56, 1631–1640. [Google Scholar] [CrossRef] [PubMed]
- Prossomariti, A.; Piazzi, G.; D’Angelo, L.; Miccoli, S.; Turchetti, D.; Alquati, C.; Montagna, C.; Bazzoli, F.; Ricciardiello, L. miR-155 Is Downregulated in Familial Adenomatous Polyposis and Modulates WNT Signaling by Targeting AXIN1 and TCF4. Mol. Cancer Res. 2018, 16, 1965–1976. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Feng, Y.; Yan, H.; Chen, Y.; Wang, J.; Chua, B.; Stuart, C.; Yin, D. β-arrestin2/miR-155/GSK3β regulates transition of 5′-azacytizine-induced Sca-1-positive cells to cardiomyocytes. J. Cell. Mol. Med. 2014, 18, 1562–1570. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.-Y.; Xia, W.; Wang, J.C.; Kwong, K.Y.; Spohn, B.; Wen, Y.; Pestell, R.G.; Hung, M.-C. β-Catenin, a novel prognostic marker for breast cancer: Its roles in cyclin D1 expression and cancer progression. Proc. Natl. Acad. Sci. USA 2000, 97, 4262–4266. [Google Scholar] [CrossRef]
- Chen, J.; Jiang, Y.; Zhou, J.; Liu, S.; Gu, Y.; Jin, G.; Hu, Z.; Ma, H.; Shen, H.; Dai, J. Genetic Variants in the Promoter Region of. BioMed Res. Int. 2017, 2017, 2352874. [Google Scholar]
- Chen, K.; Rajewsky, N. Deep conservation of microRNA-target relationships and 3’UTR motifs in vertebrates, flies, and nematodes. Cold Spring Harb. Symp. Quant. Biol. 2006, 71, 149–156. [Google Scholar] [CrossRef]
- Fang, C.; Zeng, H.; Li, A.; Xu, X.; Long, X. Association of the pri-miR-124-1 rs531564 polymorphism with cancer risk: A meta-analysis. Mol. Clin. Oncol. 2015, 3, 892–896. [Google Scholar] [CrossRef][Green Version]
- Yu, H.; Xu, W.; Gong, F.; Chi, B.; Chen, J.; Zhou, L. MicroRNA-155 regulates the proliferation, cell cycle, apoptosis and migration of colon cancer cells and targets CBL. Exp. Ther. Med. 2017, 14, 4053–4060. [Google Scholar] [CrossRef]
- Shao, C.; Yang, F.; Qin, Z.; Jing, X.; Shu, Y.; Shen, H. The value of miR-155 as a biomarker for the diagnosis and prognosis of lung cancer: A systematic review with meta-analysis. BMC Cancer 2019, 19, 1103. [Google Scholar] [CrossRef]
- Iorio, M.V.; Croce, C.M. Causes and consequences of microRNA dysregulation. Cancer J. 2012, 18, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Volinia, S.; Calin, G.A.; Liu, C.G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef]
- Zeng, H.; Fang, C.; Nam, S.; Cai, Q.; Long, X. The clinicopathological significance of microRNA-155 in breast cancer: A meta-analysis. BioMed Res. Int. 2014, 2014, 724209. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.Y.; Dayan, S.; Wong, S.W.; Kaczmarek, A.; Hope, C.M.; Pederson, S.M.; Arnet, V.; Goodall, G.J.; Russell, D.; Sadlon, T.J.; et al. FOXP3 and miR-155 cooperate to control the invasive potential of human breast cancer cells by down regulating ZEB2 independently of ZEB1. Oncotarget 2018, 9, 27708–27727. [Google Scholar] [CrossRef]
- Chen, J.; Wang, B.C.; Tang, J.H. Clinical significance of microRNA-155 expression in human breast cancer. J. Surg. Oncol. 2012, 106, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Suresh, P.S.; Venkatesh, T.; Tsutsumi, R. In silico analysis of polymorphisms in microRNAs that target genes affecting aerobic glycolysis. Ann. Transl. Med. 2016, 4, 69. [Google Scholar] [PubMed]
- Kunej, T.; Godnic, I.; Horvat, S.; Zorc, M.; Calin, G.A. Cross talk between microRNA and coding cancer genes. Cancer J. 2012, 18, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, M.; Zuo, K.; Li, D.; Ye, M.; Ding, L.; Cai, H.; Fu, D.; Fan, Y.; Lv, Z. Upregulated miR-155 in papillary thyroid carcinoma promotes tumor growth by targeting APC and activating Wnt/β-catenin signaling. J. Clin. Endocrinol. Metab. 2013, 98, E1305–E1313. [Google Scholar] [CrossRef]
- Zhang, G.-J.; Xiao, H.-X.; Tian, H.-P.; Liu, Z.-L.; Xia, S.-S.; Zhou, T. Upregulation of microRNA-155 promotes the migration and invasion of colorectal cancer cells through the regulation of claudin-1 expression. Int. J. Mol. Med. 2013, 31, 1375–1380. [Google Scholar] [CrossRef]
- Lao, G.; Liu, P.; Wu, Q.; Zhang, W.; Liu, Y.; Yang, L.; Ma, C. Mir-155 promotes cervical cancer cell proliferation through suppression of its target gene LKB1. Tumour Biol. 2014, 35, 11933–11938. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- De Mayo, T.; Ziegler, A.; Morales, S.; Jara, L. Identification of a Rare Germline Heterozygous Deletion Involving the Polycistronic miR-17–92 Cluster in Two First-Degree Relatives from a BRCA 1/2 Negative Chilean Family with Familial Breast Cancer: Possible Functional Implications. Int. J. Mol. Sci. 2018, 19, 321. [Google Scholar] [CrossRef]
- Duan, R.; Pak, C.; Jin, P. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum. Mol. Genet. 2007, 16, 1124–1131. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Morales-Pison, S.; Jara, L.; Carrasco, V.; Gutiérrez-Vera, C.; Reyes, J.M.; Gonzalez-Hormazabal, P.; Carreño, L.J.; Tapia, J.C.; Contreras, H.R. Genetic Variation in MicroRNA-423 Promotes Proliferation, Migration, Invasion, and Chemoresistance in Breast Cancer Cells. Int. J. Mol. Sci. 2021, 23, 380. [Google Scholar] [CrossRef]

| Gene | Chromosomal Location | rsID | Nucleotide Change | Relative Position |
|---|---|---|---|---|
| hsa-miR-10b | Chr.2 | rs1867863 | NC_000002.12:g.176150242G>T | 61 nt 5′upstream from pre-miR-10b (ENST00000385011.1) |
| rs138423463 | NC_000002.12:g.176150514A>G | 102 nt 3′downstream from pre-miR-10b (ENST00000385011.1) | ||
| ND | ND | 189 nt 3′downstream from pre-miR-10b (ENST00000385011.1) | ||
| ND | ND | 199 nt 3′downstream from pre-miR-10b (ENST00000385011.1) | ||
| hsa-miR-21 | Chr.17 | rs570199250 | NC_000017.11:g.59841173_59841174insT | 99 nt 5′upstream from pre-miR-21 (ENST00000362134.1) |
| hsa-miR-125a | Chr.19 | rs12976445 | NC_000019.10:g.51693200T>C | 54 nt 5′upstream from pre-miR-125a (ENST00000385273.1) |
| rs372615282 | NC_000019.10:g.51693210T>C | 44 nt 5′upstream from pre-miR-125a (ENST00000385273.1) | ||
| rs78758318 | NC_000019.10:g.51693469G>A | 129 nt 3′downstream from pre-miR-125a (ENST00000385273.1) | ||
| rs11881781 | NC_000019.10:g.51693559G>C | 218 nt 3′downstream from pre-miR-125a (ENST00000385273.1) | ||
| hsa-miR-145 | Chr.5 | * | * | * |
| hsa-miR-155 | Chr.21 | rs190708267 | NC_000021.9:g.25574088C>T | 43 nt 3′downstream from pre-miR-155 (ENST00000385060.1) |
| hsa-miR-195 | Chr.17 | rs41283391 | NC_000017.11:g.7017569C>T | 45 nt 3′downstream from pre-miR-195 (ENST00000385194.1) |
| hsa-miR-221 | Chr. X | rs191213444 | NC_000023.11:g.45746349G>T | 83 nt 5′upstream from pre-miR-221 (ENST00000385135.1) |
| hsa-miR-222 | Chr. X | * | * | * |
| hsa-miR-335 | Chr.7 | rs201306521 | NC_000007.14:g.130495936_130495937insT | 174 nt 3′downstream from pre-miR-335 (ENST00000362173.1) |
| rs3807348 | NC_000007.14:g.130496266G>A | 62 nt 3′downstream from pre-miR-335 (ENST00000362173.1) | ||
| rs376491654 | NC_000007.14:g.130496217T>C | 13 nt 3′downstream from pre-miR-335 (ENST00000362173.1) | ||
| rs41272366 | NC_000007.14:g.130496224T>A | 19 nt 3′downstream from pre-miR-335 (ENST00000362173.1) | ||
| hsa-miR-520c | Chr.19 | * | * | * |
| hsa-miR-497 | Chr.17 | rs755634302 | NC_000017.11:g.7017940dup | Nucleotide 83 of mature miR-497 ENST00000385056.1 |
| SNP ID | Genotype or Allele | Controls (%) n = 1031 | BC Cases (%) n = 440 | OR [95% CI] | p Value a |
|---|---|---|---|---|---|
| rs376491654 (miR-335) | TT | 1031 (100) | 438 (99.5) | 1.0 (ref) | |
| TC | 0 | 2 (0.5) | - | ||
| CC | 0 | 0 | - | ||
| Allele T | 2062 (100) | 878 (99.7) | 1.0 (ref) | ||
| Allele C | 0 | 2 (0.3) | 11.73 [0.56–244.95] | 0.0890 | |
| rs755634302 (miR-497) | -/- | 500 (100) | 439 (99.7) | 1.0 (ref) | |
| InsA/- | 0 | 1 (0.3) | - | ||
| InsA/InsA | 0 | 0 | - | ||
| Allele - | 1000 (100) | 879 (99.8) | 1.0 (ref) | ||
| Allele InsA | 0 | 1 (0.2) | 3.41 [0.13–83.9] | 0.4680 | |
| rs190708267 (miR-155) | CC | 1031 (100) | 436 (99.1) | 1.0 (ref) | |
| CT | 0 | 4 (0.9) | - | ||
| TT | 0 | 0 | - | ||
| Allele C | 2062 (100) | 876 (99.5) | 1.0 (ref) | ||
| Allele T | 0 | 4 (0.5) | 21.17 [1.138–394.06] | 0.0079 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Landeros, N.; Gonzalez-Hormazabal, P.; Pérez-Moreno, P.; Tapia, J.C.; Jara, L. A Single Variant in Pri-miRNA-155 Associated with Susceptibility to Hereditary Breast Cancer Promotes Aggressiveness in Breast Cancer Cells. Int. J. Mol. Sci. 2022, 23, 15418. https://doi.org/10.3390/ijms232315418
Landeros N, Gonzalez-Hormazabal P, Pérez-Moreno P, Tapia JC, Jara L. A Single Variant in Pri-miRNA-155 Associated with Susceptibility to Hereditary Breast Cancer Promotes Aggressiveness in Breast Cancer Cells. International Journal of Molecular Sciences. 2022; 23(23):15418. https://doi.org/10.3390/ijms232315418
Chicago/Turabian StyleLanderos, Natalia, Patricio Gonzalez-Hormazabal, Pablo Pérez-Moreno, Julio C. Tapia, and Lilian Jara. 2022. "A Single Variant in Pri-miRNA-155 Associated with Susceptibility to Hereditary Breast Cancer Promotes Aggressiveness in Breast Cancer Cells" International Journal of Molecular Sciences 23, no. 23: 15418. https://doi.org/10.3390/ijms232315418
APA StyleLanderos, N., Gonzalez-Hormazabal, P., Pérez-Moreno, P., Tapia, J. C., & Jara, L. (2022). A Single Variant in Pri-miRNA-155 Associated with Susceptibility to Hereditary Breast Cancer Promotes Aggressiveness in Breast Cancer Cells. International Journal of Molecular Sciences, 23(23), 15418. https://doi.org/10.3390/ijms232315418






