Tau Isoforms: Gaining Insight into MAPT Alternative Splicing
Abstract
1. Introduction
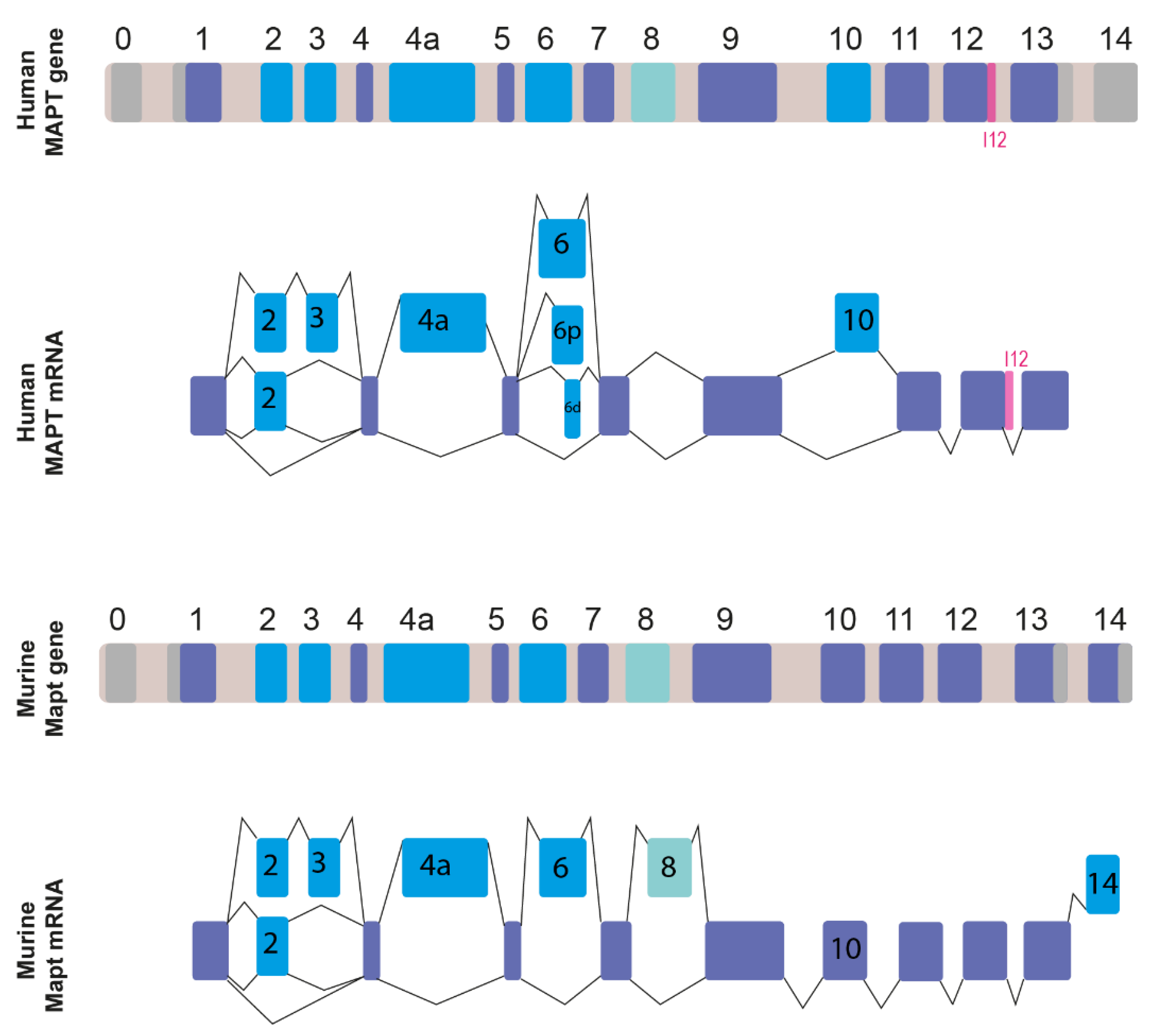
2. MAPT Gene Organization and Alternatively Spliced Transcripts
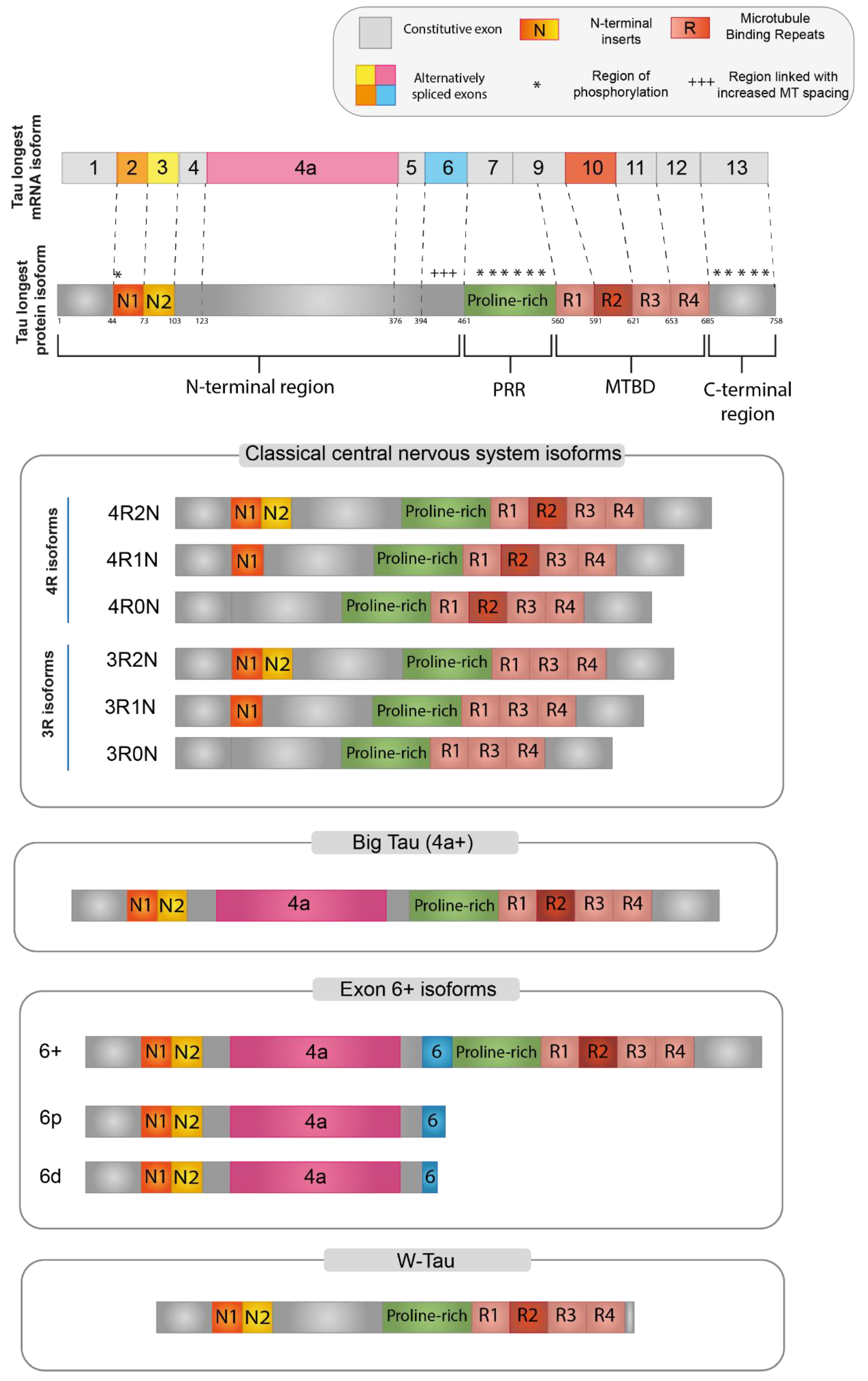
3. Tau Isoforms
3.1. Functional Domains Characterizing Tau Protein
3.2. Central Nervous System (CNS) Isoforms
3.3. Big-Tau
3.4. Isoforms including Exon 6
3.5. W-Tau
4. Tau and Cancer
5. Human MAPT Mutations Causing Aberrant Tau Splicing in Tauopathies
6. Alternative Splicing Regulation of Tau Exons
Tau Splicing and Ribonucleoproteins
| RBP | Tau Exon Regulation | Ref. |
|---|---|---|
| LUC7L3 | Exon 10 | [133] |
| THRAP3 | Exon 10 | [133] |
| SRSF1, SRSF2, SRSF3, SRSF4, SRSF6, SRSF7, SRSF9, SRSF11 | Exon 10 | [24,48,130,154] |
| EMG1 | Exon 10 | [133] |
| PRPF19 | Exon 10 | [133] |
| ARL6IP4 | Exon 10 | [133] |
| DHX15, 21 | Exon 10 | [133] |
| SLM1, SLM2 | Exon 6 | [151] |
| hnRNPU, D, A3, HI, C, R, A2B1, A1, G, E2 (PCBP2) | Exon 6, exon 10 | [133,151,155] |
| CLF4, CLF5, CLF6 | Exon 6, exon 10 | [39] |
| SC35 | Exon6, exon 10 | [151,156] |
| YT521B | Exon 6 | [151] |
| ASF | Exon 6 | [151] |
| Htra2β1 | Exon 6, exon 10 | [48,129,151] |
| 9G8 | Exon 6 | [151] |
| PTBP1 | Exon 6, exon 10 | [142,151] |
| PTBP2 | Exon 6 | [151] |
| SRSF3, SRSF5, SRSF6, SRSF4, SRSF9, | Exon6 | [151] |
| Swap | Exon 6 | [151] |
| Nova 1 | Exon 6 | [151] |
| U2AF | Exon 6 | [151] |
| SFPQ | Exon 10 | [141] |
| RBM4 | Exon 10 | [140] |
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Wang, Y.; Mandelkow, E. Tau in Physiology and Pathology. Nat. Rev. Neurosci. 2016, 17, 5–21. [Google Scholar] [CrossRef] [PubMed]
- Arendt, T. Tau and Tauopathies. Brain Res. Bull. 2016, 126, 238–292. [Google Scholar] [CrossRef] [PubMed]
- Chung, D.-E.C.; Roemer, S.; Petrucelli, L.; Dickson, D.W. Cellular and Pathological Heterogeneity of Primary Tauopathies. Mol. Neurodegener. 2021, 16, 57. [Google Scholar] [CrossRef]
- Strang, K.H.; Golde, T.E.; Giasson, B.I. MAPT Mutations, Tauopathy, and Mechanisms of Neurodegeneration. Lab. Investig. 2019, 99, 912–928. [Google Scholar] [CrossRef] [PubMed]
- Bonneau, C.; Gurard-Levin, Z.A.; Andre, F.; Pusztai, L.; Rouzier, R. Predictive and Prognostic Value of the TauProtein in Breast Cancer. Anticancer Res. 2015, 35, 5179–5184. [Google Scholar] [PubMed]
- Papin, S.; Paganetti, P. Emerging Evidences for an Implication of the Neurodegeneration-Associated Protein TAU in Cancer. Brain Sci. 2020, 10, 862. [Google Scholar] [CrossRef]
- Sekino, Y.; Han, X.; Babasaki, T.; Goto, K.; Inoue, S.; Hayashi, T.; Teishima, J.; Shiota, M.; Takeshima, Y.; Yasui, W.; et al. Microtubule-Associated Protein Tau (MAPT) Promotes Bicalutamide Resistance and Is Associated with Survival in Prostate Cancer. Urol. Oncol. 2020, 38, 795.e1–795.e8. [Google Scholar] [CrossRef]
- Smoter, M.; Bodnar, L.; Grala, B.; Stec, R.; Zieniuk, K.; Kozlowski, W.; Szczylik, C. Tau Protein as a Potential Predictive Marker in Epithelial Ovarian Cancer Patients Treated with Paclitaxel/Platinum First-Line Chemotherapy. J. Exp. Clin. Cancer Res. CR 2013, 32, 25. [Google Scholar] [CrossRef] [PubMed]
- Zarin, B.; Eshraghi, A.; Zarifi, F.; Javanmard, S.H.; Laher, I.; Amin, B.; Vaseghi, G. A Review on the Role of Tau and Stathmin in Gastric Cancer Metastasis. Eur. J. Pharmacol. 2021, 908, 174312. [Google Scholar] [CrossRef]
- Askanas, V.; Engel, W.K. Sporadic Inclusion-Body Myositis: Conformational Multifactorial Ageing-Related Degenerative Muscle Disease Associated with Proteasomal and Lysosomal Inhibition, Endoplasmic Reticulum Stress, and Accumulation of Amyloid-Β42 Oligomers and Phosphorylated Tau. Presse Med. 2011, 40, e219–e235. [Google Scholar] [CrossRef] [PubMed]
- Maurage, C.-A.; Bussière, T.; Sergeant, N.; Ghesteem, A.; Figarella-Branger, D.; Ruchoux, M.-M.; Pellissier, J.-F.; Delacourte, A. Tau Aggregates Are Abnormally Phosphorylated in Inclusion Body Myositis and Have an Immunoelectrophoretic Profile Distinct from Other Tauopathies. Neuropathol. Appl. Neurobiol. 2004, 30, 624–634. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.A.; Galvis-Escobar, S.; Abisambra, J.F. Tau-Mediated Dysregulation of RNA: Evidence for a Common Molecular Mechanism of Toxicity in Frontotemporal Dementia and Other Tauopathies. Neurobiol. Dis. 2020, 141, 104939. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.-Y.; Wang, Z.-T.; Tan, L.; Yu, J.-T. Tau Toxicity in Neurodegeneration. Mol. Neurobiol. 2022, 59, 3617–3634. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, G.G. Invited Review: Neuropathology of Tauopathies: Principles and Practice. Neuropathol. Appl. Neurobiol. 2015, 41, 3–23. [Google Scholar] [CrossRef]
- Spillantini, M.G.; Goedert, M. Tau Pathology and Neurodegeneration. Lancet Neurol. 2013, 12, 609–622. [Google Scholar] [CrossRef]
- Šimić, G.; Babić Leko, M.; Wray, S.; Harrington, C.; Delalle, I.; Jovanov-Milošević, N.; Bažadona, D.; Buée, L.; de Silva, R.; Di Giovanni, G.; et al. Tau Protein Hyperphosphorylation and Aggregation in Alzheimer’s Disease and Other Tauopathies, and Possible Neuroprotective Strategies. Biomolecules 2016, 6, 6. [Google Scholar] [CrossRef]
- Rösler, T.W.; Tayaranian Marvian, A.; Brendel, M.; Nykänen, N.-P.; Höllerhage, M.; Schwarz, S.C.; Hopfner, F.; Koeglsperger, T.; Respondek, G.; Schweyer, K.; et al. Four-Repeat Tauopathies. Prog. Neurobiol. 2019, 180, 101644. [Google Scholar] [CrossRef]
- Götz, J.; Halliday, G.; Nisbet, R.M. Molecular Pathogenesis of the Tauopathies. Annu. Rev. Pathol. 2019, 14, 239–261. [Google Scholar] [CrossRef]
- Andreadis, A. Misregulation of Tau Alternative Splicing in Neurodegeneration and Dementia. Prog. Mol. Subcell. Biol. 2006, 44, 89–107. [Google Scholar] [CrossRef]
- Ishunina, T.A. Alternative Splicing in Aging and Alzheimer’s Disease: Highlighting the Role of Tau and Estrogen Receptor α Isoforms in the Hypothalamus. Handb. Clin. Neurol. 2021, 182, 177–189. [Google Scholar] [CrossRef]
- Park, S.A.; Ahn, S.I.; Gallo, J.-M. Tau Mis-Splicing in the Pathogenesis of Neurodegenerative Disorders. BMB Rep. 2016, 49, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Gabarre, D.; Carnero-Espejo, A.; Ávila, J.; García-Escudero, V. What’s in a Gene? The Outstanding Diversity of MAPT. Cells 2022, 11, 840. [Google Scholar] [CrossRef] [PubMed]
- Sündermann, F.; Fernandez, M.-P.; Morgan, R.O. An Evolutionary Roadmap to the Microtubule-Associated Protein MAP Tau. BMC Genom. 2016, 17, 264. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gao, L.; Conrad, C.G.; Andreadis, A. Saitohin, Which Is Nested within the Tau Gene, Interacts with Tau and Abl and Its Human-Specific Allele Influences Abl Phosphorylation. J. Cell. Biochem. 2011, 112, 3482–3488. [Google Scholar] [CrossRef]
- Poorkaj, P.; Kas, A.; D’Souza, I.; Zhou, Y.; Pham, Q.; Stone, M.; Olson, M.V.; Schellenberg, G.D. A Genomic Sequence Analysis of the Mouse and Human Microtubule-Associated Protein Tau. Mamm. Genome Off. J. Int. Mamm. Genome Soc. 2001, 12, 700–712. [Google Scholar] [CrossRef]
- Andreadis, A. Tau Gene Alternative Splicing: Expression Patterns, Regulation and Modulation of Function in Normal Brain and Neurodegenerative Diseases. Biochim. Biophys. Acta BBA-Mol. Basis Dis. 2005, 1739, 91–103. [Google Scholar] [CrossRef]
- Bachmann, S.; Bell, M.; Klimek, J.; Zempel, H. Differential Effects of the Six Human TAU Isoforms: Somatic Retention of 2N-TAU and Increased Microtubule Number Induced by 4R-TAU. Front. Neurosci. 2021, 15, 643115. [Google Scholar] [CrossRef]
- Goedert, M. Tau Filaments in Neurodegenerative Diseases. FEBS Lett. 2018, 592, 2383–2391. [Google Scholar] [CrossRef] [PubMed]
- Andreadis, A.; Nisson, P.E.; Kosik, K.S.; Watkins, P.C. The Exon Trapping Assay Partly Discriminates against Alternatively Spliced Exons. Nucleic Acids Res. 1993, 21, 2217–2221. [Google Scholar] [CrossRef]
- Lee, G.; Cowan, N.; Kirschner, M. The Primary Structure and Heterogeneity of Tau Protein from Mouse Brain. Science 1988, 239, 285–288. [Google Scholar] [CrossRef]
- Caillet-Boudin, M.-L.; Buée, L.; Sergeant, N.; Lefebvre, B. Regulation of Human MAPT Gene Expression. Mol. Neurodegener. 2015, 10, 28. [Google Scholar] [CrossRef]
- Andreadis, A.; Brown, W.M.; Kosik, K.S. Structure and Novel Exons of the Human Tau Gene. Biochemistry 1992, 31, 10626–10633. [Google Scholar] [CrossRef] [PubMed]
- Couchie, D.; Shelanski, M.L.; Nunez, J. Primary Structure of High Molecular Weight Tau Present in the Peripheral Nervous System. Proc. Natl. Acad. Sci. USA 1992, 89, 4378–4381. [Google Scholar] [CrossRef] [PubMed]
- Goedert, M.; Spillantini, M.G.; Crowther, R.A. Cloning of a Big Tau Microtubule-Associated Protein Characteristic of the Peripheral Nervous System. Proc. Natl. Acad. Sci. USA 1992, 89, 1983–1987. [Google Scholar] [CrossRef] [PubMed]
- Andreadis, A.; Broderick, J.A.; Kosik, K.S. Relative Exon Affinities and Suboptimal Splice Site Signals Lead to Non-Equivalence of Two Cassette Exons. Nucleic Acids Res. 1995, 23, 3585–3593. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, K.; Arikan, M.C.; Andreadis, A. Modulation of the Membrane-Binding Domain of Tau Protein: Splicing Regulation of Exon 2. Brain Res. Mol. Brain Res. 2003, 116, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Arikan, M.C.; Memmott, J.; Broderick, J.A.; Lafyatis, R.; Screaton, G.; Stamm, S.; Andreadis, A. Modulation of the Membrane-Binding Projection Domain of Tau Protein: Splicing Regulation of Exon 3. Brain Res. Mol. Brain Res. 2002, 101, 109–121. [Google Scholar] [CrossRef]
- Georgieff, I.S.; Liem, R.K.; Couchie, D.; Mavilia, C.; Nunez, J.; Shelanski, M.L. Expression of High Molecular Weight Tau in the Central and Peripheral Nervous Systems. J. Cell Sci. 1993, 105, 729–737. [Google Scholar] [CrossRef]
- Leroy, O.; Wang, J.; Maurage, C.-A.; Parent, M.; Cooper, T.; Buée, L.; Sergeant, N.; Andreadis, A.; Caillet-Boudin, M.-L. Brain-Specific Change in Alternative Splicing of Tau Exon 6 in Myotonic Dystrophy Type 1. Biochim. Biophys. Acta BBA-Mol. Basis Dis. 2006, 1762, 460–467. [Google Scholar] [CrossRef][Green Version]
- Wei, M.-L.; Andreadis, A. Splicing of a Regulated Exon Reveals Additional Complexity in the Axonal Microtubule-Associated Protein Tau. J. Neurochem. 1998, 70, 11. [Google Scholar] [CrossRef]
- Chen, W.T.; Liu, W.K.; Yen, S.H. Expression of Tau Exon 8 in Different Species. Neurosci. Lett. 1994, 172, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Himmler, A. Structure of the Bovine Tau Gene: Alternatively Spliced Transcripts Generate a Protein Family. Mol. Cell. Biol. 1989, 9, 1389–1396. [Google Scholar] [CrossRef] [PubMed]
- Nelson, P.T.; Stefansson, K.; Gulcher, J.; Saper, C.B. Molecular Evolution of Tau Protein: Implications for Alzheimer’s Disease. J. Neurochem. 1996, 67, 1622–1632. [Google Scholar] [CrossRef] [PubMed]
- Holzer, M.; Craxton, M.; Jakes, R.; Arendt, T.; Goedert, M. Tau Gene (MAPT) Sequence Variation among Primates. Gene 2004, 341, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Ghetti, B.; Oblak, A.L.; Boeve, B.F.; Johnson, K.A.; Dickerson, B.C.; Goedert, M. Invited Review: Frontotemporal Dementia Caused by Microtubule-Associated Protein Tau Gene (MAPT) Mutations: A Chameleon for Neuropathology and Neuroimaging. Neuropathol. Appl. Neurobiol. 2015, 41, 24–46. [Google Scholar] [CrossRef] [PubMed]
- Lacovich, V.; Espindola, S.L.; Alloatti, M.; Pozo Devoto, V.; Cromberg, L.E.; Čarná, M.E.; Forte, G.; Gallo, J.-M.; Bruno, L.; Stokin, G.B.; et al. Tau Isoforms Imbalance Impairs the Axonal Transport of the Amyloid Precursor Protein in Human Neurons. J. Neurosci. Off. J. Soc. Neurosci. 2017, 37, 58–69. [Google Scholar] [CrossRef]
- Liu, F.; Gong, C.-X. Tau Exon 10 Alternative Splicing and Tauopathies. Mol. Neurodegener. 2008, 3, 8. [Google Scholar] [CrossRef]
- Gao, Q.S.; Memmott, J.; Lafyatis, R.; Stamm, S.; Screaton, G.; Andreadis, A. Complex Regulation of Tau Exon 10, Whose Missplicing Causes Frontotemporal Dementia. J. Neurochem. 2000, 74, 490–500. [Google Scholar] [CrossRef]
- Kosik, K.S.; Orecchio, L.D.; Bakalis, S.; Neve, R.L. Developmentally Regulated Expression of Specific Tau Sequences. Neuron 1989, 2, 1389–1397. [Google Scholar] [CrossRef]
- D’Souza, I.; Schellenberg, G.D. Determinants of 4-Repeat Tau Expression. Coordination between Enhancing and Inhibitory Splicing Sequences for Exon 10 Inclusion. J. Biol. Chem. 2000, 275, 17700–17709. [Google Scholar] [CrossRef]
- García-Escudero, V.; Ruiz-Gabarre, D.; Gargini, R.; Pérez, M.; García, E.; Cuadros, R.; Hernández, I.H.; Cabrera, J.R.; García-Escudero, R.; Lucas, J.J.; et al. A New Non-Aggregative Splicing Isoform of Human Tau Is Decreased in Alzheimer’s Disease. Acta Neuropathol. 2021, 142, 159–177. [Google Scholar] [CrossRef] [PubMed]
- Didonna, A. Tau at the Interface between Neurodegeneration and Neuroinflammation. Genes Immun. 2020, 21, 288–300. [Google Scholar] [CrossRef] [PubMed]
- Montalbano, M.; Jaworski, E.; Garcia, S.; Ellsworth, A.; McAllen, S.; Routh, A.; Kayed, R. Tau Modulates MRNA Transcription, Alternative Polyadenylation Profiles of HnRNPs, Chromatin Remodeling and Spliceosome Complexes. Front. Mol. Neurosci. 2021, 14, 742790. [Google Scholar] [CrossRef] [PubMed]
- Colnaghi, L.; Rondelli, D.; Muzi-Falconi, M.; Sertic, S. Tau and DNA Damage in Neurodegeneration. Brain Sci. 2020, 10, 946. [Google Scholar] [CrossRef]
- Sultan, A.; Nesslany, F.; Violet, M.; Bégard, S.; Loyens, A.; Talahari, S.; Mansuroglu, Z.; Marzin, D.; Sergeant, N.; Humez, S.; et al. Nuclear Tau, a Key Player in Neuronal DNA Protection. J. Biol. Chem. 2011, 286, 4566–4575. [Google Scholar] [CrossRef]
- Violet, M.; Delattre, L.; Tardivel, M.; Sultan, A.; Chauderlier, A.; Caillierez, R.; Talahari, S.; Nesslany, F.; Lefebvre, B.; Bonnefoy, E.; et al. A Major Role for Tau in Neuronal DNA and RNA Protection In Vivo under Physiological and Hyperthermic Conditions. Front. Cell. Neurosci. 2014, 8, 84. [Google Scholar] [CrossRef]
- Sierksma, A.; Lu, A.; Salta, E.; Vanden Eynden, E.; Callaerts-Vegh, Z.; D’Hooge, R.; Blum, D.; Buée, L.; Fiers, M.; De Strooper, B. Deregulation of Neuronal MiRNAs Induced by Amyloid-β or TAU Pathology. Mol. Neurodegener. 2018, 13, 54. [Google Scholar] [CrossRef]
- Gonçalves, R.A.; Wijesekara, N.; Fraser, P.E.; De Felice, F.G. Behavioral Abnormalities in Knockout and Humanized Tau Mice. Front. Endocrinol. 2020, 11, 124. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, K.-M.; Yang, L.; Dong, Q.; Yu, J.-T. Tauopathies: New Perspectives and Challenges. Mol. Neurodegener. 2022, 17, 28. [Google Scholar] [CrossRef]
- Betrie, A.H.; Ayton, S.; Bush, A.I.; Angus, J.A.; Lei, P.; Wright, C.E. Evidence of a Cardiovascular Function for Microtubule-Associated Protein Tau. J. Alzheimers Dis. JAD 2017, 56, 849–860. [Google Scholar] [CrossRef]
- Trabzuni, D.; Wray, S.; Vandrovcova, J.; Ramasamy, A.; Walker, R.; Smith, C.; Luk, C.; Gibbs, J.R.; Dillman, A.; Hernandez, D.G.; et al. MAPT Expression and Splicing Is Differentially Regulated by Brain Region: Relation to Genotype and Implication for Tauopathies. Hum. Mol. Genet. 2012, 21, 4094–4103. [Google Scholar] [CrossRef] [PubMed]
- Fischer, I.; Baas, P.W. Resurrecting the Mysteries of Big Tau. Trends Neurosci. 2020, 43, 493–504. [Google Scholar] [CrossRef]
- Gentile, A.; Mori, F.; Bernardini, S.; Centonze, D. Role of Amyloid-β CSF Levels in Cognitive Deficit in MS. Clin. Chim. Acta Int. J. Clin. Chem. 2015, 449, 23–30. [Google Scholar] [CrossRef]
- LoPresti, P.; Szuchet, S.; Papasozomenos, S.C.; Zinkowski, R.P.; Binder, L.I. Functional Implications for the Microtubule-Associated Protein Tau: Localization in Oligodendrocytes. Proc. Natl. Acad. Sci. USA 1995, 92, 10369–10373. [Google Scholar] [CrossRef] [PubMed]
- Brunello, C.A.; Merezhko, M.; Uronen, R.-L.; Huttunen, H.J. Mechanisms of Secretion and Spreading of Pathological Tau Protein. Cell. Mol. Life Sci. CMLS 2020, 77, 1721–1744. [Google Scholar] [CrossRef] [PubMed]
- Annadurai, N.; De Sanctis, J.B.; Hajdúch, M.; Das, V. Tau Secretion and Propagation: Perspectives for Potential Preventive Interventions in Alzheimer’s Disease and Other Tauopathies. Exp. Neurol. 2021, 343, 113756. [Google Scholar] [CrossRef]
- Miguel, L.; Rovelet-Lecrux, A.; Feyeux, M.; Frebourg, T.; Nassoy, P.; Campion, D.; Lecourtois, M. Detection of All Adult Tau Isoforms in a 3D Culture Model of IPSC-Derived Neurons. Stem Cell Res. 2019, 40, 101541. [Google Scholar] [CrossRef]
- Brandt, R.; Trushina, N.I.; Bakota, L. Much More Than a Cytoskeletal Protein: Physiological and Pathological Functions of the Non-Microtubule Binding Region of Tau. Front. Neurol. 2020, 11, 590059. [Google Scholar] [CrossRef]
- Kolarova, M.; García-Sierra, F.; Bartos, A.; Ricny, J.; Ripova, D. Structure and Pathology of Tau Protein in Alzheimer Disease. Int. J. Alzheimers Dis. 2012, 2012, 731526. [Google Scholar] [CrossRef]
- Gauthier-Kemper, A.; Suárez Alonso, M.; Sündermann, F.; Niewidok, B.; Fernandez, M.-P.; Bakota, L.; Heinisch, J.J.; Brandt, R. Annexins A2 and A6 Interact with the Extreme N Terminus of Tau and Thereby Contribute to Tau’s Axonal Localization. J. Biol. Chem. 2018, 293, 8065–8076. [Google Scholar] [CrossRef]
- da Costa, P.J.; Hamdane, M.; Buée, L.; Martin, F. Tau MRNA Metabolism in Neurodegenerative Diseases: A Tangle Journey. Biomedicines 2022, 10, 241. [Google Scholar] [CrossRef] [PubMed]
- Sibille, N.; Huvent, I.; Fauquant, C.; Verdegem, D.; Amniai, L.; Leroy, A.; Wieruszeski, J.-M.; Lippens, G.; Landrieu, I. Structural Characterization by Nuclear Magnetic Resonance of the Impact of Phosphorylation in the Proline-Rich Region of the Disordered Tau Protein. Proteins 2012, 80, 454–462. [Google Scholar] [CrossRef] [PubMed]
- He, H.J.; Wang, X.S.; Pan, R.; Wang, D.L.; Liu, M.N.; He, R.Q. The Proline-Rich Domain of Tau Plays a Role in Interactions with Actin. BMC Cell Biol. 2009, 10, 81. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Prokop, S.; Gorion, K.-M.M.; Kim, J.D.; Sorrentino, Z.A.; Bell, B.M.; Manaois, A.N.; Chakrabarty, P.; Davies, P.; Giasson, B.I. Tau Ser208 Phosphorylation Promotes Aggregation and Reveals Neuropathologic Diversity in Alzheimer’s Disease and Other Tauopathies. Acta Neuropathol. Commun. 2020, 8, 88. [Google Scholar] [CrossRef]
- Janning, D.; Igaev, M.; Sündermann, F.; Brühmann, J.; Beutel, O.; Heinisch, J.J.; Bakota, L.; Piehler, J.; Junge, W.; Brandt, R. Single-Molecule Tracking of Tau Reveals Fast Kiss-and-Hop Interaction with Microtubules in Living Neurons. Mol. Biol. Cell 2014, 25, 3541–3551. [Google Scholar] [CrossRef]
- Trushina, N.I.; Bakota, L.; Mulkidjanian, A.Y.; Brandt, R. The Evolution of Tau Phosphorylation and Interactions. Front. Aging Neurosci. 2019, 11, 256. [Google Scholar] [CrossRef]
- Gu, J.-L.; Liu, F. Tau in Alzheimer’s Disease: Pathological Alterations and an Attractive Therapeutic Target. Curr. Med. Sci. 2020, 40, 1009–1021. [Google Scholar] [CrossRef]
- Wegmann, S.; Biernat, J.; Mandelkow, E. A Current View on Tau Protein Phosphorylation in Alzheimer’s Disease. Curr. Opin. Neurobiol. 2021, 69, 131–138. [Google Scholar] [CrossRef]
- Drummond, E.; Wisniewski, T. Alzheimer’s Disease: Experimental Models and Reality. Acta Neuropathol. 2017, 133, 155–175. [Google Scholar] [CrossRef]
- Goedert, M.; Spillantini, M.G. Propagation of Tau Aggregates. Mol. Brain 2017, 10, 18. [Google Scholar] [CrossRef]
- Jeganathan, S.; von Bergen, M.; Brutlach, H.; Steinhoff, H.-J.; Mandelkow, E. Global Hairpin Folding of Tau in Solution. Biochemistry 2006, 45, 2283–2293. [Google Scholar] [CrossRef] [PubMed]
- Hefti, M.M.; Farrell, K.; Kim, S.; Bowles, K.R.; Fowkes, M.E.; Raj, T.; Crary, J.F. High-Resolution Temporal and Regional Mapping of MAPT Expression and Splicing in Human Brain Development. PLoS ONE 2018, 13, e0195771. [Google Scholar] [CrossRef] [PubMed]
- Qian, W.; Liu, F. Regulation of Alternative Splicing of Tau Exon 10. Neurosci. Bull. 2014, 30, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Oblinger, M.M.; Argasinski, A.; Wong, J.; Kosik, K.S. Tau Gene Expression in Rat Sensory Neurons during Development and Regeneration. J. Neurosci. Off. J. Soc. Neurosci. 1991, 11, 2453–2459. [Google Scholar] [CrossRef]
- LaPointe, N.E.; Horowitz, P.M.; Guillozet-Bongaarts, A.L.; Silva, A.; Andreadis, A.; Binder, L.I. Tau 6D and 6P Isoforms Inhibit Polymerization of Full-Length Tau In Vitro. Biochemistry 2009, 48, 12290–12297. [Google Scholar] [CrossRef][Green Version]
- Luo, M.; Leski, M.L.; Andreadis, A. Tau Isoforms Which Contain the Domain Encoded by Exon 6 and Their Role in Neurite Elongation. J. Cell. Biochem. 2004, 91, 880–895. [Google Scholar] [CrossRef]
- Wei, M.-L.; Memmott, J.; Screaton, G.; Andreadis, A. The Splicing Determinants of a Regulated Exon in the Axonal MAP Tau Reside within the Exon and in Its Upstream Intron. Mol. Brain Res. 2000, 80, 207–218. [Google Scholar] [CrossRef]
- Wilson, D.M.; Binder, L.I. Free Fatty Acids Stimulate the Polymerization of Tau and Amyloid Beta Peptides. In Vitro Evidence for a Common Effector of Pathogenesis in Alzheimer’s Disease. Am. J. Pathol. 1997, 150, 2181–2195. [Google Scholar]
- Darlix, A.; Hirtz, C.; Thezenas, S.; Maceski, A.; Gabelle, A.; Lopez-Crapez, E.; De Forges, H.; Firmin, N.; Guiu, S.; Jacot, W.; et al. The Prognostic Value of the Tau Protein Serum Level in Metastatic Breast Cancer Patients and Its Correlation with Brain Metastases. BMC Cancer 2019, 19, 110. [Google Scholar] [CrossRef]
- Gargini, R.; Segura-Collar, B.; Sánchez-Gómez, P. Novel Functions of the Neurodegenerative-Related Gene Tau in Cancer. Front. Aging Neurosci. 2019, 11, 231. [Google Scholar] [CrossRef]
- Han, X.; Sekino, Y.; Babasaki, T.; Goto, K.; Inoue, S.; Hayashi, T.; Teishima, J.; Sakamoto, N.; Sentani, K.; Oue, N.; et al. Microtubule-Associated Protein Tau (MAPT) Is a Promising Independent Prognostic Marker and Tumor Suppressive Protein in Clear Cell Renal Cell Carcinoma. Urol. Oncol. 2020, 38, 605.e9–605.e17. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Qu, M.-H.; Wang, X.-S.; Chen, L.; Wang, D.-L.; Liu, Y.; Hua, Q.; He, R.-Q. Binding to the Minor Groove of the Double-Strand, Tau Protein Prevents DNA from Damage by Peroxidation. PLoS ONE 2008, 3, e2600. [Google Scholar] [CrossRef] [PubMed]
- Benhelli-Mokrani, H.; Mansuroglu, Z.; Chauderlier, A.; Albaud, B.; Gentien, D.; Sommer, S.; Schirmer, C.; Laqueuvre, L.; Josse, T.; Buée, L.; et al. Genome-Wide Identification of Genic and Intergenic Neuronal DNA Regions Bound by Tau Protein under Physiological and Stress Conditions. Nucleic Acids Res. 2018, 46, 11405–11422. [Google Scholar] [CrossRef] [PubMed]
- Mansuroglu, Z.; Benhelli-Mokrani, H.; Marcato, V.; Sultan, A.; Violet, M.; Chauderlier, A.; Delattre, L.; Loyens, A.; Talahari, S.; Bégard, S.; et al. Loss of Tau Protein Affects the Structure, Transcription and Repair of Neuronal Pericentromeric Heterochromatin. Sci. Rep. 2016, 6, 33047. [Google Scholar] [CrossRef]
- Siano, G.; Varisco, M.; Caiazza, M.C.; Quercioli, V.; Mainardi, M.; Ippolito, C.; Cattaneo, A.; Di Primio, C. Tau Modulates VGluT1 Expression. J. Mol. Biol. 2019, 431, 873–884. [Google Scholar] [CrossRef]
- Meier, S.; Bell, M.; Lyons, D.N.; Rodriguez-Rivera, J.; Ingram, A.; Fontaine, S.N.; Mechas, E.; Chen, J.; Wolozin, B.; LeVine, H.; et al. Pathological Tau Promotes Neuronal Damage by Impairing Ribosomal Function and Decreasing Protein Synthesis. J. Neurosci. Off. J. Soc. Neurosci. 2016, 36, 1001–1007. [Google Scholar] [CrossRef]
- Vanderweyde, T.; Apicco, D.J.; Youmans-Kidder, K.; Ash, P.E.A.; Cook, C.; Lummertz da Rocha, E.; Jansen-West, K.; Frame, A.A.; Citro, A.; Leszyk, J.D.; et al. Interaction of Tau with the RNA-Binding Protein TIA1 Regulates Tau Pathophysiology and Toxicity. Cell Rep. 2016, 15, 1455–1466. [Google Scholar] [CrossRef]
- Chauderlier, A.; Gilles, M.; Spolcova, A.; Caillierez, R.; Chwastyniak, M.; Kress, M.; Drobecq, H.; Bonnefoy, E.; Pinet, F.; Weil, D.; et al. Tau/DDX6 Interaction Increases MicroRNA Activity. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 762–772. [Google Scholar] [CrossRef]
- Rossi, G.; Dalprà, L.; Crosti, F.; Lissoni, S.; Sciacca, F.L.; Catania, M.; Di Fede, G.; Mangieri, M.; Giaccone, G.; Croci, D.; et al. A New Function of Microtubule-Associated Protein Tau: Involvement in Chromosome Stability. Cell Cycle Georget. Tex 2008, 7, 1788–1794. [Google Scholar] [CrossRef]
- Frost, B.; Hemberg, M.; Lewis, J.; Feany, M.B. Tau Promotes Neurodegeneration through Global Chromatin Relaxation. Nat. Neurosci. 2014, 17, 357–366. [Google Scholar] [CrossRef]
- Caneus, J.; Granic, A.; Rademakers, R.; Dickson, D.W.; Coughlan, C.M.; Chial, H.J.; Potter, H. Mitotic Defects Lead to Neuronal Aneuploidy and Apoptosis in Frontotemporal Lobar Degeneration Caused by MAPT Mutations. Mol. Biol. Cell 2018, 29, 575–586. [Google Scholar] [CrossRef] [PubMed]
- Rossi, G.; Conconi, D.; Panzeri, E.; Redaelli, S.; Piccoli, E.; Paoletta, L.; Dalprà, L.; Tagliavini, F. Mutations in MAPT Gene Cause Chromosome Instability and Introduce Copy Number Variations Widely in the Genome. J. Alzheimers Dis. JAD 2013, 33, 969–982. [Google Scholar] [CrossRef] [PubMed]
- Cimini, S.; Giaccone, G.; Tagliavini, F.; Costantino, M.; Perego, P.; Rossi, G. P301L Tau Mutation Leads to Alterations of Cell Cycle, DNA Damage Response and Apoptosis: Evidence for a Role of Tau in Cancer. Biochem. Pharmacol. 2022, 200, 115043. [Google Scholar] [CrossRef]
- Rossi, G.; Redaelli, V.; Contiero, P.; Fabiano, S.; Tagliabue, G.; Perego, P.; Benussi, L.; Bruni, A.C.; Filippini, G.; Farinotti, M.; et al. Tau Mutations Serve as a Novel Risk Factor for Cancer. Cancer Res. 2018, 78, 3731–3739. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gong, X.; Zhang, Y. Network-Based Approach to Identify Prognosis-Related Genes in Tamoxifen-Treated Patients with Estrogen Receptor-Positive Breast Cancer. Biosci. Rep. 2021, 41, BSR20203020. [Google Scholar] [CrossRef]
- Pagano, A.; Breuzard, G.; Parat, F.; Tchoghandjian, A.; Figarella-Branger, D.; De Bessa, T.C.; Garrouste, F.; Douence, A.; Barbier, P.; Kovacic, H. Tau Regulates Glioblastoma Progression, 3D Cell Organization, Growth and Migration via the PI3K-AKT Axis. Cancers 2021, 13, 5818. [Google Scholar] [CrossRef]
- Pan, Y.; Pan, Y.; Cheng, Y.; Yang, F.; Yao, Z.; Wang, O. Knockdown of LncRNA MAPT-AS1 Inhibites Proliferation and Migration and Sensitizes Cancer Cells to Paclitaxel by Regulating MAPT Expression in ER-Negative Breast Cancers. Cell Biosci. 2018, 8, 7. [Google Scholar] [CrossRef]
- Yamauchi, A.; Kobayashi, A.; Oikiri, H.; Yokoyama, Y. Functional Role of the Tau Protein in Epithelial Ovarian Cancer Cells. Reprod. Med. Biol. 2017, 16, 143–151. [Google Scholar] [CrossRef]
- Martellucci, S.; Clementi, L.; Sabetta, S.; Muzi, P.; Mattei, V.; Bologna, M.; Angelucci, A. Tau Oligomers Accumulation Sensitizes Prostate Cancer Cells to Docetaxel Treatment. J. Cancer Res. Clin. Oncol. 2021, 147, 1957–1971. [Google Scholar] [CrossRef]
- van Swieten, J.C.; Bronner, I.F.; Azmani, A.; Severijnen, L.-A.; Kamphorst, W.; Ravid, R.; Rizzu, P.; Willemsen, R.; Heutink, P. The DeltaK280 Mutation in MAP Tau Favors Exon 10 Skipping in Vivo. J. Neuropathol. Exp. Neurol. 2007, 66, 17–25. [Google Scholar] [CrossRef]
- Yoshida, H.; Crowther, R.A.; Goedert, M. Functional Effects of Tau Gene Mutations DeltaN296 and N296H. J. Neurochem. 2002, 80, 548–551. [Google Scholar] [CrossRef] [PubMed]
- Hong, M.; Zhukareva, V.; Vogelsberg-Ragaglia, V.; Wszolek, Z.; Reed, L.; Miller, B.I.; Geschwind, D.H.; Bird, T.D.; McKeel, D.; Goate, A.; et al. Mutation-Specific Functional Impairments in Distinct Tau Isoforms of Hereditary FTDP-17. Science 1998, 282, 1914–1917. [Google Scholar] [CrossRef] [PubMed]
- Spillantini, M.G.; Goedert, M. Neurodegeneration and the Ordered Assembly of α-Synuclein. Cell Tissue Res. 2018, 373, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Bowles, K.R.; Silva, M.C.; Whitney, K.; Bertucci, T.; Berlind, J.E.; Lai, J.D.; Garza, J.C.; Boles, N.C.; Mahali, S.; Strang, K.H.; et al. ELAVL4, Splicing, and Glutamatergic Dysfunction Precede Neuron Loss in MAPT Mutation Cerebral Organoids. Cell 2021, 184, 4547–4563.e17. [Google Scholar] [CrossRef] [PubMed]
- Tomasiewicz, H.G.; Flaherty, D.B.; Soria, J.P.; Wood, J.G. Transgenic Zebrafish Model of Neurodegeneration. J. Neurosci. Res. 2002, 70, 734–745. [Google Scholar] [CrossRef]
- Paquet, D.; Bhat, R.; Sydow, A.; Mandelkow, E.-M.; Berg, S.; Hellberg, S.; Fälting, J.; Distel, M.; Köster, R.W.; Schmid, B.; et al. A Zebrafish Model of Tauopathy Allows In Vivo Imaging of Neuronal Cell Death and Drug Evaluation. J. Clin. Investig. 2009, 119, 1382–1395. [Google Scholar] [CrossRef]
- Lopez, A.; Lee, S.E.; Wojta, K.; Ramos, E.M.; Klein, E.; Chen, J.; Boxer, A.L.; Gorno-Tempini, M.L.; Geschwind, D.H.; Schlotawa, L.; et al. A152T Tau Allele Causes Neurodegeneration That Can Be Ameliorated in a Zebrafish Model by Autophagy Induction. Brain J. Neurol. 2017, 140, 1128–1146. [Google Scholar] [CrossRef]
- Barbereau, C.; Cubedo, N.; Maurice, T.; Rossel, M. Zebrafish Models to Study New Pathways in Tauopathies. Int. J. Mol. Sci. 2021, 22, 4626. [Google Scholar] [CrossRef]
- Giustiniani, J.; Chambraud, B.; Sardin, E.; Dounane, O.; Guillemeau, K.; Nakatani, H.; Paquet, D.; Kamah, A.; Landrieu, I.; Lippens, G.; et al. Immunophilin FKBP52 Induces Tau-P301L Filamentous Assembly In Vitro and Modulates Its Activity in a Model of Tauopathy. Proc. Natl. Acad. Sci. USA 2014, 111, 4584–4589. [Google Scholar] [CrossRef]
- Alavi Naini, S.M.; Yanicostas, C.; Hassan-Abdi, R.; Blondeel, S.; Bennis, M.; Weiss, R.J.; Tor, Y.; Esko, J.D.; Soussi-Yanicostas, N. Surfen and Oxalyl Surfen Decrease Tau Hyperphosphorylation and Mitigate Neuron Deficits in Vivo in a Zebrafish Model of Tauopathy. Transl. Neurodegener. 2018, 7, 6. [Google Scholar] [CrossRef]
- Sepulveda-Diaz, J.E.; Alavi Naini, S.M.; Huynh, M.B.; Ouidja, M.O.; Yanicostas, C.; Chantepie, S.; Villares, J.; Lamari, F.; Jospin, E.; van Kuppevelt, T.H.; et al. HS3ST2 Expression Is Critical for the Abnormal Phosphorylation of Tau in Alzheimer’s Disease-Related Tau Pathology. Brain J. Neurol. 2015, 138, 1339–1354. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Meng, Y.; Jin, Y. RNA Structures in Alternative Splicing and Back-Splicing. Wiley Interdiscip. Rev. RNA 2021, 12, e1626. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Shai, O.; Lee, L.J.; Frey, B.J.; Blencowe, B.J. Deep Surveying of Alternative Splicing Complexity in the Human Transcriptome by High-Throughput Sequencing. Nat. Genet. 2008, 40, 1413–1415. [Google Scholar] [CrossRef] [PubMed]
- Baralle, F.E.; Giudice, J. Alternative Splicing as a Regulator of Development and Tissue Identity. Nat. Rev. Mol. Cell Biol. 2017, 18, 437–451. [Google Scholar] [CrossRef]
- De Conti, L.; Baralle, M.; Buratti, E. Exon and Intron Definition in Pre-MRNA Splicing. Wiley Interdiscip. Rev. RNA 2013, 4, 49–60. [Google Scholar] [CrossRef]
- Long, R.M.; Urbinati, C.R. Monitoring the Temporal and Spatial Distribution of RNA in Living Yeast Cells. Methods Mol. Biol. 2008, 419, 187–196. [Google Scholar] [CrossRef]
- Geuens, T.; Bouhy, D.; Timmerman, V. The HnRNP Family: Insights into Their Role in Health and Disease. Hum. Genet. 2016, 135, 851–867. [Google Scholar] [CrossRef]
- Singh, G.; Cooper, T.A. Minigene Reporter for Identification and Analysis of Cis Elements and Trans Factors Affecting Pre-MRNA Splicing. BioTechniques 2006, 41, 177–181. [Google Scholar] [CrossRef]
- Jiang, Z.; Tang, H.; Havlioglu, N.; Zhang, X.; Stamm, S.; Yan, R.; Wu, J.Y. Mutations in Tau Gene Exon 10 Associated with FTDP-17 Alter the Activity of an Exonic Splicing Enhancer to Interact with Tra2 Beta. J. Biol. Chem. 2003, 278, 18997–19007. [Google Scholar] [CrossRef]
- Yu, Q.; Guo, J.; Zhou, J. A Minimal Length between Tau Exon 10 and 11 Is Required for Correct Splicing of Exon 10. J. Neurochem. 2004, 90, 164–172. [Google Scholar] [CrossRef]
- Donahue, C.P.; Muratore, C.; Wu, J.Y.; Kosik, K.S.; Wolfe, M.S. Stabilization of the Tau Exon 10 Stem Loop Alters Pre-MRNA Splicing. J. Biol. Chem. 2006, 281, 23302–23306. [Google Scholar] [CrossRef] [PubMed]
- Carpentier, C.; Ghanem, D.; Fernandez-Gomez, F.J.; Jumeau, F.; Philippe, J.V.; Freyermuth, F.; Labudeck, A.; Eddarkaoui, S.; Dhaenens, C.M.; Holt, I.; et al. Tau Exon 2 Responsive Elements Deregulated in Myotonic Dystrophy Type I Are Proximal to Exon 2 and Synergistically Regulated by MBNL1 and MBNL2. Biochim. Biophys. Acta 2014, 1842, 654–664. [Google Scholar] [CrossRef] [PubMed]
- Xing, S.; Wang, J.; Wu, R.; Hefti, M.M.; Crary, J.F.; Lu, Y. Identification of HnRNPC as a Novel Tau Exon 10 Splicing Factor Using RNA Antisense Purification Mass Spectrometry. RNA Biol. 2022, 19, 104–116. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Disney, M.D. Bottom-up Design of Small Molecules That Stimulate Exon 10 Skipping in Mutant MAPT Pre-MRNA. Chembiochem. Eur. J. Chem. Biol. 2014, 15, 2041–2044. [Google Scholar] [CrossRef]
- Chen, J.L.; Zhang, P.; Abe, M.; Aikawa, H.; Zhang, L.; Frank, A.J.; Zembryski, T.; Hubbs, C.; Park, H.; Withka, J.; et al. Design, Optimization, and Study of Small Molecules That Target Tau Pre-MRNA and Affect Splicing. J. Am. Chem. Soc. 2020, 142, 8706–8727. [Google Scholar] [CrossRef]
- Qian, W.; Liang, H.; Shi, J.; Jin, N.; Grundke-Iqbal, I.; Iqbal, K.; Gong, C.-X.; Liu, F. Regulation of the Alternative Splicing of Tau Exon 10 by SC35 and Dyrk1A. Nucleic Acids Res. 2011, 39, 6161–6171. [Google Scholar] [CrossRef]
- Izquierdo, J.M.; Valcárcel, J. Two Isoforms of the T-Cell Intracellular Antigen 1 (TIA-1) Splicing Factor Display Distinct Splicing Regulation Activities: Control of Tia-1 Isoform Ratio by Tia-1-Related Protein. J. Biol. Chem. 2007, 282, 19410–19417. [Google Scholar] [CrossRef]
- Apicco, D.J.; Ash, P.E.A.; Maziuk, B.; LeBlang, C.; Medalla, M.; Al Abdullatif, A.; Ferragud, A.; Botelho, E.; Ballance, H.I.; Dhawan, U.; et al. Reducing the RNA Binding Protein TIA1 Protects against Tau-Mediated Neurodegeneration In Vivo. Nat. Neurosci. 2018, 21, 72–80. [Google Scholar] [CrossRef]
- Apicco, D.J.; Zhang, C.; Maziuk, B.; Jiang, L.; Ballance, H.I.; Boudeau, S.; Ung, C.; Li, H.; Wolozin, B. Dysregulation of RNA Splicing in Tauopathies. Cell Rep. 2019, 29, 4377–4388.e4. [Google Scholar] [CrossRef]
- Kar, A.; Havlioglu, N.; Tarn, W.-Y.; Wu, J.Y. RBM4 Interacts with an Intronic Element and Stimulates Tau Exon 10 Inclusion. J. Biol. Chem. 2006, 281, 24479–24488. [Google Scholar] [CrossRef]
- Kar, A.; Fushimi, K.; Zhou, X.; Ray, P.; Shi, C.; Chen, X.; Liu, Z.; Chen, S.; Wu, J.Y. RNA Helicase P68 (DDX5) Regulates Tau Exon 10 Splicing by Modulating a Stem-Loop Structure at the 5’ Splice Site. Mol. Cell. Biol. 2011, 31, 1812–1821. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Gao, Q.-S.; Wang, Y.; Lafyatis, R.; Stamm, S.; Andreadis, A. Tau Exon 10, Whose Missplicing Causes Frontotemporal Dementia, Is Regulated by an Intricate Interplay of Cis Elements and Trans Factors. J. Neurochem. 2004, 88, 1078–1090. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, I.; Schellenberg, G.D. Arginine/Serine-Rich Protein Interaction Domain-Dependent Modulation of a Tau Exon 10 Splicing Enhancer: Altered Interactions and Mechanisms for Functionally Antagonistic FTDP-17 Mutations Delta280K AND N279K. J. Biol. Chem. 2006, 281, 2460–2469. [Google Scholar] [CrossRef] [PubMed]
- Ke, Y.D.; Ke, Y.; Dramiga, J.; Schütz, U.; Kril, J.J.; Ittner, L.M.; Schröder, H.; Götz, J. Tau-Mediated Nuclear Depletion and Cytoplasmic Accumulation of SFPQ in Alzheimer’s and Pick’s Disease. PLoS ONE 2012, 7, e35678. [Google Scholar] [CrossRef]
- Romanelli, M.G.; Diani, E.; Lievens, P.M.-J. New Insights into Functional Roles of the Polypyrimidine Tract-Binding Protein. Int. J. Mol. Sci. 2013, 14, 22906–22932. [Google Scholar] [CrossRef]
- Fochi, S.; Lorenzi, P.; Galasso, M.; Stefani, C.; Trabetti, E.; Zipeto, D.; Romanelli, M.G. The Emerging Role of the RBM20 and PTBP1 Ribonucleoproteins in Heart Development and Cardiovascular Diseases. Genes 2020, 11, 402. [Google Scholar] [CrossRef]
- Zhu, W.; Zhou, B.-L.; Rong, L.-J.; Ye, L.; Xu, H.-J.; Zhou, Y.; Yan, X.-J.; Liu, W.-D.; Zhu, B.; Wang, L.; et al. Roles of PTBP1 in Alternative Splicing, Glycolysis, and Oncogensis. J. Zhejiang Univ. Sci. B 2020, 21, 122–136. [Google Scholar] [CrossRef]
- Vilys, L.; Peciuliene, I.; Jakubauskiene, E.; Zinkeviciute, R.; Makino, Y.; Kanopka, A. U2AF-Hypoxia-Induced Fas Alternative Splicing Regulator. Exp. Cell Res. 2021, 399, 112444. [Google Scholar] [CrossRef]
- Filippello, A.; Lorenzi, P.; Bergamo, E.; Romanelli, M.G. Identification of Nuclear Retention Domains in the RBM20 Protein. FEBS Lett. 2013, 587, 2989–2995. [Google Scholar] [CrossRef]
- Lorenzi, P.; Sangalli, A.; Fochi, S.; Dal Molin, A.; Malerba, G.; Zipeto, D.; Romanelli, M.G. RNA-Binding Proteins RBM20 and PTBP1 Regulate the Alternative Splicing of FHOD3. Int. J. Biochem. Cell Biol. 2019, 106, 74–83. [Google Scholar] [CrossRef]
- Wang, J.; Tse, S.-W.; Andreadis, A. Tau Exon 6 Is Regulated by an Intricate Interplay of Trans Factors and Cis Elements, Including Multiple Branch Points: Splicing Regulation of a Tau Exon Affected in Myotonic Dystrophy 1. J. Neurochem. 2007, 100, 437–445. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.-L.; Yuan, R.-Y.; Hu, C.-J.; Hsu, C.Y. Amyloid-β Peptide Alteration of Tau Exon-10 Splicing via the GSK3β-SC35 Pathway. Neurobiol. Dis. 2010, 40, 378–385. [Google Scholar] [CrossRef] [PubMed]
- Sud, R.; Geller, E.T.; Schellenberg, G.D. Antisense-Mediated Exon Skipping Decreases Tau Protein Expression: A Potential Therapy For Tauopathies. Mol. Ther. Nucleic Acids 2014, 3, e180. [Google Scholar] [CrossRef]
- Kondo, S.; Yamamoto, N.; Murakami, T.; Okumura, M.; Mayeda, A.; Imaizumi, K. Tra2 Beta, SF2/ASF and SRp30c Modulate the Function of an Exonic Splicing Enhancer in Exon 10 of Tau Pre-MRNA. Genes Cells Devoted Mol. Cell. Mech. 2004, 9, 121–130. [Google Scholar] [CrossRef]
- Broderick, J.; Wang, J.; Andreadis, A. Heterogeneous Nuclear Ribonucleoprotein E2 Binds to Tau Exon 10 and Moderately Activates Its Splicing. Gene 2004, 331, 107–114. [Google Scholar] [CrossRef]
- Hernández, F.; Pérez, M.; Lucas, J.J.; Mata, A.M.; Bhat, R.; Avila, J. Glycogen Synthase Kinase-3 Plays a Crucial Role in Tau Exon 10 Splicing and Intranuclear Distribution of SC35. Implications for Alzheimer’s Disease. J. Biol. Chem. 2004, 279, 3801–3806. [Google Scholar] [CrossRef] [PubMed]
- Merezhko, M.; Uronen, R.-L.; Huttunen, H.J. The Cell Biology of Tau Secretion. Front. Mol. Neurosci. 2020, 13, 569818. [Google Scholar] [CrossRef] [PubMed]
| Minigene Name | Exon(s) | Properties | Application | Ref. |
|---|---|---|---|---|
| hTau | 1, 2, 4 | Exon 2 flanked by different length of its native introns, surrounded by E1 and E4 | Identification of cis and trans acting elements altering splicing of E2 | [132] |
| SV1/2L/4 | 2 | Exon 2 flanked by its native exons | In situ mutagenesis to identify cis acting elements in the exon and downstream intron | [36] |
| SP/2L | 2 | Exon 2 flanked by heterologous exons | In situ mutagenesis to identify cis acting elements in the exon and downstream intron | [36] |
| SVΔ2/3 | 3 | E3, 1.1 kbps of I2 and 1.4 kpbs of I3 | E3 default splicing pattern analysis, identification of cis-acting elements | [37] |
| pSVIRB SV6 | 6 | E6, 600 bps I5, 200 bps I6 | Exon-trapping assay | [40] |
| TauEx9-11d5 | 9-10-11 | E10 flanked by 269 and 246 bp of its native intron and exons 9–11. | Identification of cis-acting elements altering splicing of E10 | [129] |
| SI9/SI10 LI9/SI10 SI9/LI10 LI9/LI10 | 9-10-11 | E10 can be flanked by a long (L) or short (S) portion of the upstream and downstream intron | Identification of a required minimal distance for E10 splicing | [130] |
| Tau minigene | 9-10-11 | Identification of HnRNPC as a novel trans-acting factor on E10 | [133] | |
| RHCGlo Tau wt RHCGlo Tau Δ280K RHCGlo Tau N279K | 10 | Contains WT E10 or E10 with Δ280K, and N279K mutations | Identification of cis and trans-acting elements affecting E10 splicing | [128] |
| Luc M14 | 10 | Luciferase reporter, modified from TauEx9-11 d5 minigene | Identification of compound that reduced exon 10 inclusion; identify cis/trans elements | [130,131,134,135] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corsi, A.; Bombieri, C.; Valenti, M.T.; Romanelli, M.G. Tau Isoforms: Gaining Insight into MAPT Alternative Splicing. Int. J. Mol. Sci. 2022, 23, 15383. https://doi.org/10.3390/ijms232315383
Corsi A, Bombieri C, Valenti MT, Romanelli MG. Tau Isoforms: Gaining Insight into MAPT Alternative Splicing. International Journal of Molecular Sciences. 2022; 23(23):15383. https://doi.org/10.3390/ijms232315383
Chicago/Turabian StyleCorsi, Andrea, Cristina Bombieri, Maria Teresa Valenti, and Maria Grazia Romanelli. 2022. "Tau Isoforms: Gaining Insight into MAPT Alternative Splicing" International Journal of Molecular Sciences 23, no. 23: 15383. https://doi.org/10.3390/ijms232315383
APA StyleCorsi, A., Bombieri, C., Valenti, M. T., & Romanelli, M. G. (2022). Tau Isoforms: Gaining Insight into MAPT Alternative Splicing. International Journal of Molecular Sciences, 23(23), 15383. https://doi.org/10.3390/ijms232315383









