Role of Acrostyle Cuticular Proteins in the Retention of an Aphid Salivary Effector
Abstract
1. Introduction
2. Results
2.1. Stylin-03 Interacts with the Effector Mp10 in Yeast
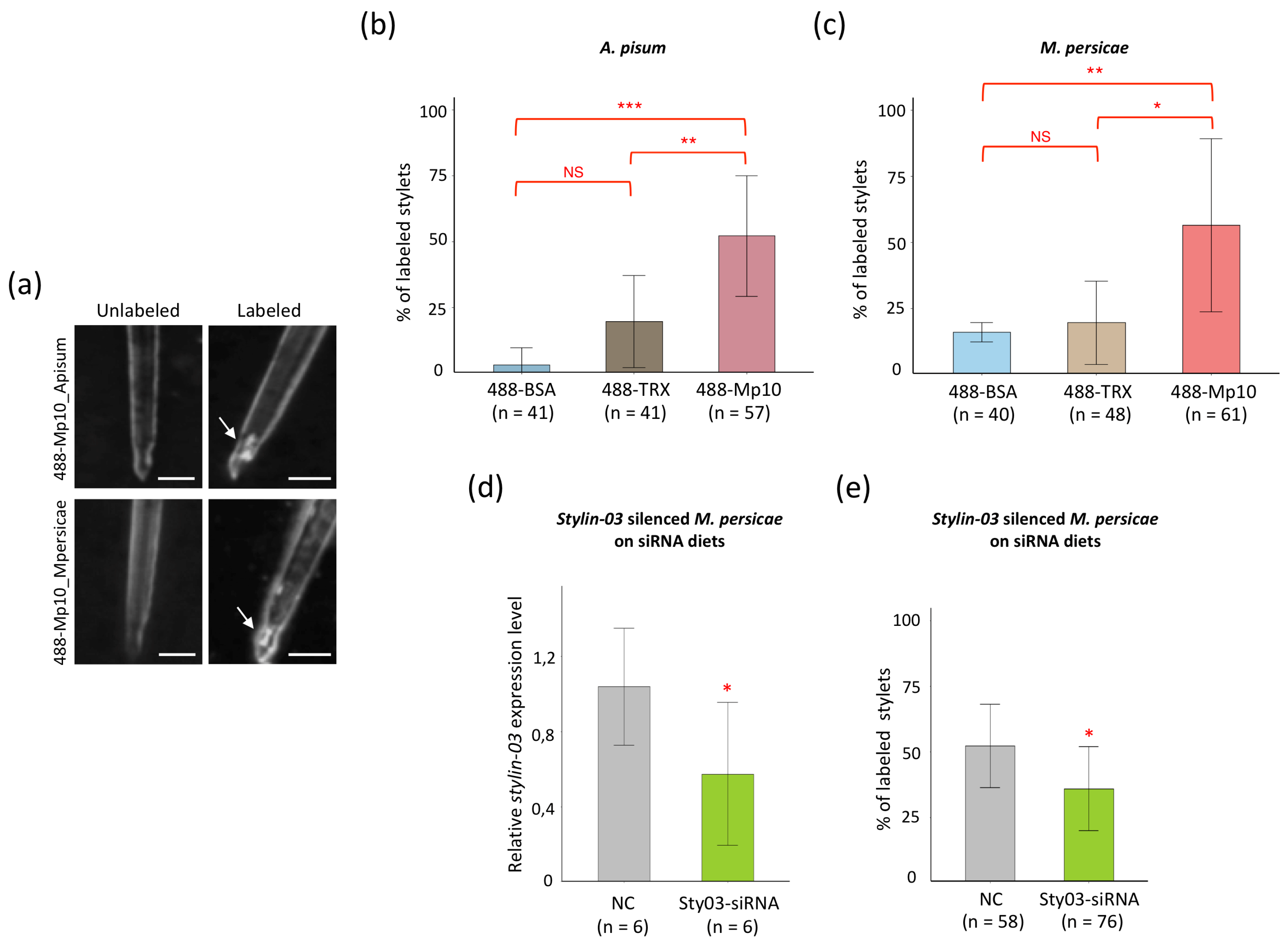
2.2. Mp10 Binds to the Acrostyle of A. Pisum and M. Persicae
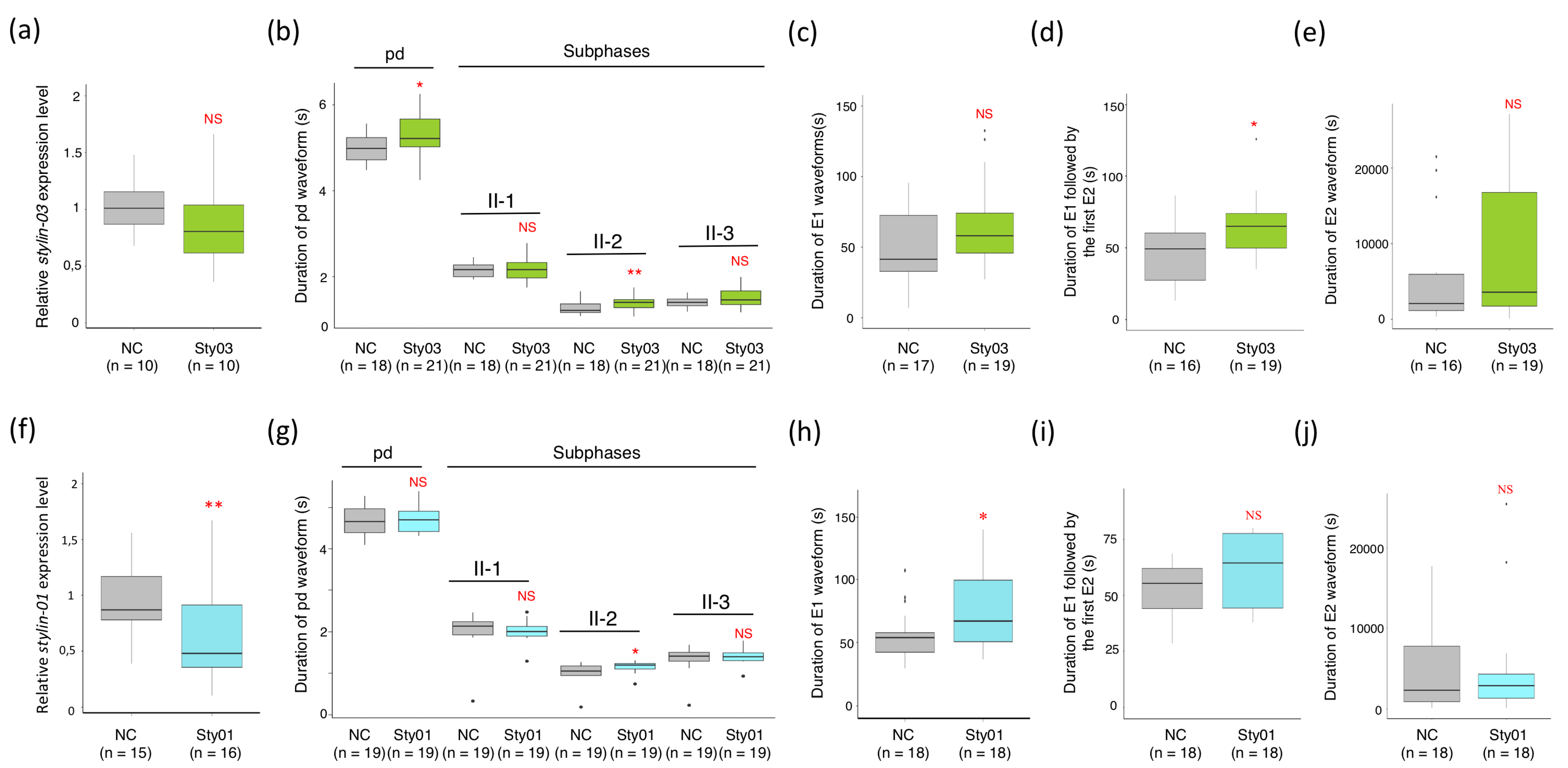
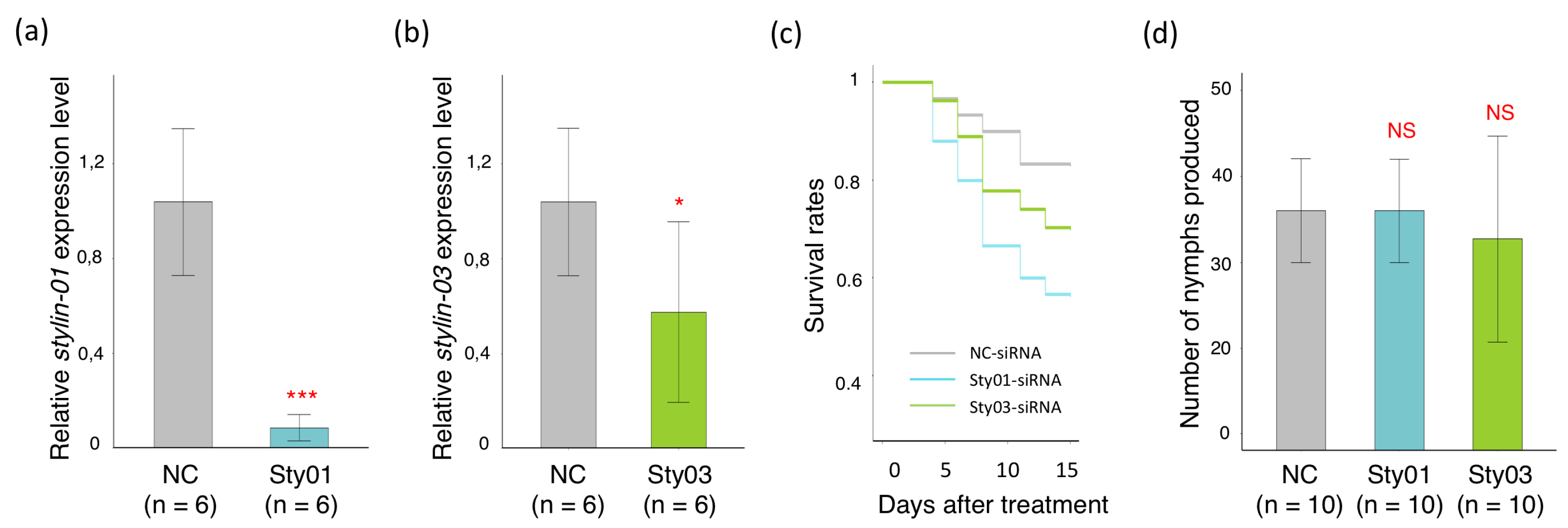
2.3. Impact of siRNA Targeting Stylin-03 or Stylin-01 Gene Expression on M. Persicae Feeding Behavior
2.4. Impact of siRNA Targeting Stylin-03 or Stylin-01 Gene Expression on M. Persicae Survival Rate and Fecundity
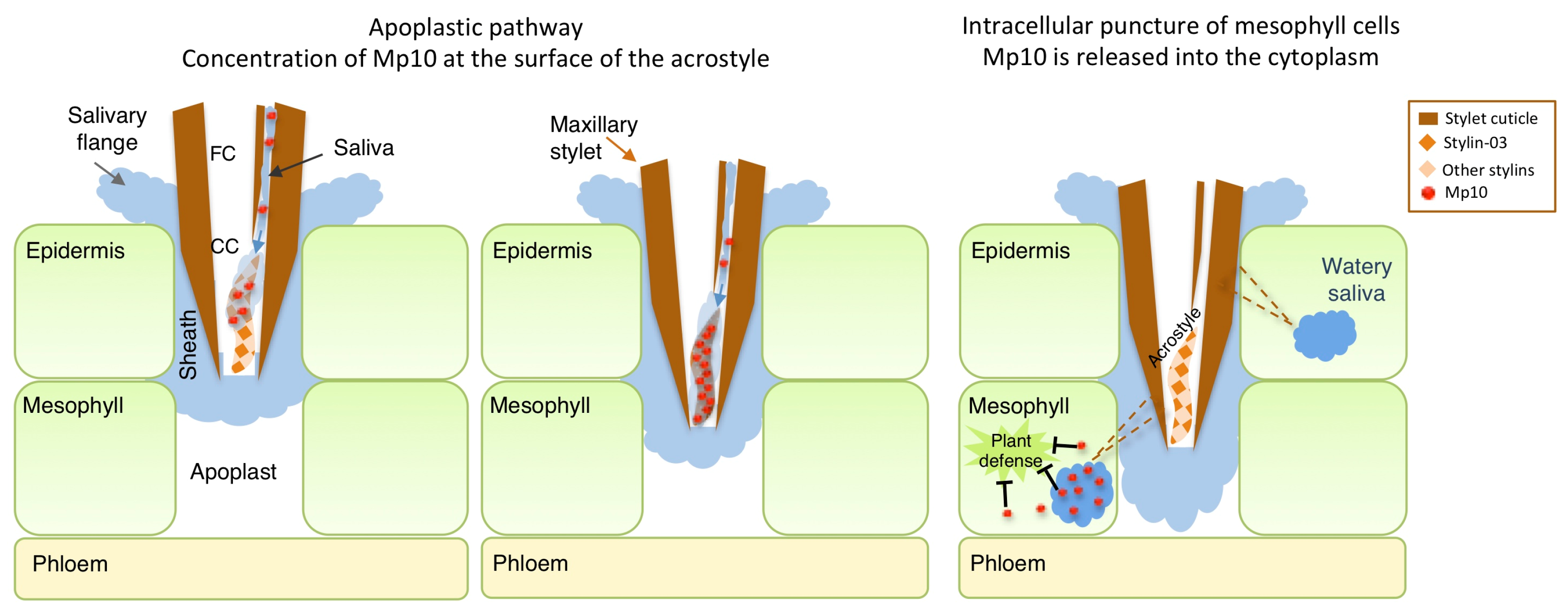
3. Discussion
4. Materials and Methods
4.1. Aphid Colonies
4.2. Yeast Two-Hybrid Constructs and Assays
4.3. Constructs, Production and Labeling of Mp10 for Detection on Insect Stylets
4.4. In Vitro Interactions between Mp10 and Dissected Aphid Stylets
4.5. RNA Interference
4.6. Quantitative PCR
4.7. Analysis of Aphid Feeding Behavior
4.8. Aphid Life Table Statistics
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dedryver, C.A.; Le Ralec, A.; Fabre, F. The conflicting relationships between aphids and men: A review of aphid damage and control strategies. Comptes Rendus Biol. 2010, 333, 539–553. [Google Scholar] [CrossRef]
- Tjallingii, W.F. Regulation of Phloem Sap Feeding by Aphids. In Regulatory Mechanisms in Insect Feeding; Chapman, R.F., De Boer, G., Eds.; Elsevier: Amsterdam, The Netherlands, 1995; pp. 190–209. [Google Scholar] [CrossRef]
- Nault, L.R. Arthropod transmission of plant viruses: A new synthesis. Ann. Entomol. Soc. Am. 1997, 90, 521–541. [Google Scholar] [CrossRef]
- Favret, C. Aphids species file. Version 5.0/5.0. 2022. Available online: http://aphid.speciesfile.org (accessed on 1 August 2022).
- Peccoud, J.; Ollivier, A.; Plantegenest, M.; Simon, J.-C. A continuum of genetic divergence from sympatric host races to species in the pea aphid complex. Proc. Natl. Acad. Sci. USA 2009, 106, 7495–7500. [Google Scholar] [CrossRef]
- Blackman, L.M.; Eastop, V.F. Aphids on the World’s Crops. An Identification and Information Guide, 2nd ed.; John Wiley & Sons: Chichester, UK, 2000; 476p. [Google Scholar]
- Nault, L.R.; Gyrisco, G.G. Relation of the feeding process of the pea aphid to the inoculation of Pea Enation Mosaic Virus 1. Ann. Entomol. Soc. Am. 1966, 59, 1185–1197. [Google Scholar] [CrossRef]
- Miles, P.W. Aphid saliva. Biol. Rev. 1999, 74, 41–85. [Google Scholar] [CrossRef]
- Jiménez, J.; Tjallingii, W.F.; Moreno, A.; Fereres, A. Newly distinguished cell punctures associated with transmission of the semipersistent phloem-limited Beet yellows virus. J. Virol. 2018, 92, e01076-18. [Google Scholar] [CrossRef]
- Tjallingii, W.F. Salivary secretions by aphids interacting with proteins of phloem wound responses. J. Exp. Bot. 2006, 57, 739–745. [Google Scholar] [CrossRef]
- Hogenhout, S.A.; Bos, J.I.B. Effector proteins that modulate plant–insect interactions. Curr. Opin. Plant. Biol. 2011, 14, 422–428. [Google Scholar] [CrossRef]
- Boulain, H.; Legeai, F.; Guy, E.; Morlière, S.; Douglas, N.E.; Oh, J.; Murugan, M.; Smith, M.; Jaquiéry, J.; Peccoud, J.; et al. Fast evolution and lineage-specific gene family expansions of aphid salivary effectors driven by interactions with host-plants. Genome Biol. Evol. 2018, 10, 1554–1572. [Google Scholar] [CrossRef]
- Ray, S.; Casteel, C.L. Effector-mediated plat-virus-vector interactions. Plant Cell. 2022, 34, 1514–1531. [Google Scholar] [CrossRef]
- Mutti, N.S.; Park, Y.; Reese, J.C.; Reeck, G.R. RNAi knockdown of a salivary transcript leading to lethality in the pea aphid, Acyrthosiphon pisum. J. Insect Sci. 2006, 6, 38. [Google Scholar] [CrossRef] [PubMed]
- Mutti, N.S.; Louis, J.; Pappan, L.K.; Pappan, K.; Begum, K.; Chen, M.S.; Park, Y.; Dittmer, N.; Marshall, J.; Reese, J.C.; et al. A protein from the salivary glands of the pea aphid, Acyrthosiphon pisum, is essential in feeding on a host plant. Proc. Natl. Acad. Sci. USA 2008, 105, 9965–9969. [Google Scholar] [CrossRef]
- Bos, J.I.B.; Prince, D.; Pitino, M.; Maffei, M.E.; Win, J.; Hogenhout, S.A. A functional genomics approach identifies candidate effectors from the aphid species Myzus persicae (green peach aphid). PLoS Genet. 2010, 6, e1001216. [Google Scholar] [CrossRef] [PubMed]
- Pitino, M.; Hogenhout, S.A. Aphid protein effectors promote aphid colonization in a plant species-specific manner. Mol. Plant-Microbe Interact. 2013, 26, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, P.A.; Stam, R.; Warbroek, T.; Bos, J.I.B. Mp10 and Mp42 from the aphid species Myzus persicae trigger plant defenses in Nicotiana benthamiana through different activities. Mol. Plant-Microbe Inter. 2014, 27, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Naessens, E.; Dubreuil, G.; Giordanengo, P.; Baron, O.L.; Minet-Kebdani, N.; Keller, H.; Coustau, C. A secreted MIF cytokine enables aphid feeding and represses plant immune responses. Curr. Biol. 2015, 25, 1898–1903. [Google Scholar] [CrossRef]
- Wang, W.; Luo, L.; Lu, H.; Chen, S.; Kang, L.; Cui, F. Angiotensin-converting enzymes modulate aphid-plant interactions. Sci. Rep. 2015, 5, 8885. [Google Scholar] [CrossRef]
- Wang, W.; Dai, H.; Zhang, Y.; Chandrasekar, R.; Luo, L.; Hiromasa, Y.; Sheng, C.; Peng, G.; Chen, S.; Tomich, J.M.; et al. Armet is an effector protein mediating aphid-plant interactions. Faseb J. 2015, 29, 2032–2045. [Google Scholar] [CrossRef]
- Guy, E.; Boulain, H.; Aigu, Y.; Le Pennec, C.; Chawki, K.; Morlière, S.; Schädel, K.; Kunert, G.; Simon, J.-C.; Sugio, A. Optimization of agroinfiltration in Pisum sativum provides a new tool for studying the salivary protein functions in the pea aphid complex. Front. Plant Sci. 2016, 7, 1171. [Google Scholar] [CrossRef]
- Mugford, S.T.; Barclay, E.; Drurey, C.; Findlay, K.C.; Hogenhout, S.A. An immuno-suppressive aphid saliva protein is delivered into the cytosol of plant mesophyll cells during feeding. Mol. Plant-Microbe Interact. 2016, 29, 854–861. [Google Scholar] [CrossRef]
- Uzest, M.; Gargani, D.; Dombrovsky, A.; Cazevieille, C.; Cot, D.; Blanc, S. The “acrostyle”: A newly described anatomical structure in aphid stylets. Arthropod Struct. Dev. 2010, 39, 221–229. [Google Scholar] [CrossRef]
- Tjallingii, W.F.; Esch, T.H. Fine structure of aphid stylet routes in plant tissues in correlation with EPG signals. Physiol. Entomol. 1993, 18, 317–328. [Google Scholar] [CrossRef]
- Webster, C.G.; Pichon, E.; van Munster, M.; Monsion, B.; Deshoux, M.; Gargani, D.; Calevro, F.; Jimenez, J.; Moreno, A.; Krenz, B.; et al. Identification of plant virus receptor candidates in the stylets of their aphid vectors. J. Virol. 2018, 92, e00432-18. [Google Scholar] [CrossRef] [PubMed]
- Deshoux, M.; Masson, V.; Arafah, K.; Voisin, S.; Guschinskaya, N.; van Munster, M.; Cayrol, B.; Webster, C.G.; Rahbé, Y.; Blanc, S.; et al. Cuticular structure proteomics in the pea aphid Acyrthosiphon pisum reveals new plant virus receptor candidates at the tip of maxillary stylets. J. Prot. Res. 2020, 19, 1319–1337. [Google Scholar] [CrossRef] [PubMed]
- Moreno, A.; Palacios, I.; Blanc, S.; Fereres, A. Intracellular salivation is the mechanism involved in the inoculation of Cauliflower mosaic virus by its major vectors, Brevicorynae brassicae and Myzus persicae. Ann. Entomol. Soc. Am. 2005, 98, 763–769. [Google Scholar] [CrossRef]
- Uzest, M.; Gargani, D.; Drucker, M.; Hebrard, E.; Garzo, E.; Candresse, T.; Fereres, A.; Blanc, S. A protein key to plant virus transmission at the tip of the insect vector stylet. Proc. Natl. Acad. Sci. USA 2007, 104, 17959–17964. [Google Scholar] [CrossRef]
- Tjallingii, W.F. Electrical recording of stylet penetration activities. In Aphids, Their Biology, Natural Enemies and Control; Minks, A.K., Harrewijn, P., Eds.; Elsevier: Amsterdam, The Netherlands, 1988; Volume 2B, pp. 95–108. [Google Scholar]
- Wróblewska-Kurdyk, A.; Gniłka, R.; Dancewicz, K.; Grudniewska, A.; Wawrzeńczyk, C.; Gabryś, B. β-Thujone and its derivatives modify the probing behavior of the peach potato aphid. Molecules 2019, 24, 1847. [Google Scholar] [CrossRef]
- Moreno, A.; Tjallingii, W.F.; Fernandez-Mata, G.; Fereres, A. Differences in the mechanism of inoculation between a semi-persistent and a non-persistent aphid-transmitted plant virus. J. Gen. Virol. 2012, 93, 662–667. [Google Scholar] [CrossRef]
- Palacios, I.; Drucker, M.; Blanc, S.; Leite, S.; Moreno, A.; Fereres, A. Cauliflower mosaic virus is preferentially acquired from the phloem by its aphid vectors. J. Gen. Virol. 2002, 83, 3163–3171. [Google Scholar] [CrossRef]
- Christiaens, O.; Swevers, L.; Smagghe, G. DsRNA degradation in the pea aphid (Acyrthosiphon pisum) associated with lack of response in RNAi feeding and injection assay. Peptides 2014, 53, 307–314. [Google Scholar] [CrossRef]
- Kanakala, S.; Ghanim, M. RNA interference in insect vectors for plant viruses. Viruses 2016, 8, 329. [Google Scholar] [CrossRef] [PubMed]
- Mulot, M.; Boissinot, S.; Monsion, B.; Rastegar, M.; Clavijo, G.; Halter, D.; Bochet, N.; Erdinger, M.; Brault, V. Comparative analysis of RNAi-based methods to down-regulate expression of two genes expressed at different levels in Myzus persicae. Viruses 2016, 8, 316. [Google Scholar] [CrossRef] [PubMed]
- Sapountzis, P.; Duport, G.; Balmand, S.; Gaget, K.; Jaubert-Possamai, S.; Febvay, G.; Charles, H.; Rahbe, Y.; Colella, S.; Calevro, F. New insight into the RNA interference response against cathepsin-L gene in the pea aphid, Acyrthosiphon pisum: Molting or gut phenotypes specifically induced by injection or feeding treatments. Insect Biochem. Mol. Biol. 2014, 51, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Guschinskaya, N.; Ressnikoff, D.; Arafah, K.; Voisin, S.; Bulet, P.; Uzest, M.; Rahbé, Y. Insect mouthpart transcriptome unveils extension of cuticular protein repertoire and complex organization. iScience 2020, 23, 100828. [Google Scholar] [CrossRef]
- Drurey, C.; Mathers, T.C.; Prince, D.C.; Wilson, C.; Caceres-Moreno, C.; Mugford, S.T.; Hogenhout, S.A. Chemosensory proteins in the CSP4 clade evolved as plant immunity suppressors before two suborders of plant-feeding hemipteran insects diverged. BioRxiv 2019, 173278. [Google Scholar] [CrossRef]
- Garzo, E.; Soria, C.; Gómez-Guillamón, M.L.; Fereres, A. Feeding behavior of Aphis gossypii on resistant accessions of different melon genotypes (Cucumis melo). Phytoparasitica 2002, 30, 129–140. [Google Scholar] [CrossRef]
- Sun, M.; Voorrips, R.E.; Steenhuis-Broers, G.; van’t Westende, W.; Vosman, B. Reduced phloem uptake of Myzus persicae on an aphid resistant pepper accession. BMC Plant Biol. 2018, 18, 138. [Google Scholar] [CrossRef]
- Will, T.; Tjallingii, W.F.; Thönnessen, A.; van Bel, A.J.E. Molecular sabotage of plant defense by aphid saliva. Proc. Natl. Acad. Sci. USA 2007, 104, 10536–10541. [Google Scholar] [CrossRef]
- Campanacci, V.; Lartigue, A.; Hällberg, B.M.; Jones, T.A.; Giudici-Orticoni, M.-T.; Tegoni, M.; Cambillau, C. Moth chemosensory protein exhibits drastic conformational changes and cooperativity on ligand binding. Proc. Nat. Acad. Sci. USA 2003, 100, 5069–5074. [Google Scholar] [CrossRef]
- Arnoldi, I.; Mancini, G.; Fumagalli, M.; Gastaldi, D.; D’Andrea, L.; Bandi, C.; Di Venere, M.; Iadarola, P.; Forneris, F.; Gabrieli, P. A salivary factor binds a cuticular protein and modulates biting by inducing morphological changes in the mosquito labrum. Curr. Biol. 2022, 32, 3493–3504. [Google Scholar] [CrossRef]
- Buezo Montero, S.; Gabrieli, P.; Montarsi, F.; Borean, A.; Capelli, S.; De Silvestro, G.; Forneris, F.; Pombi, M.; Breda, A.; Capelli, G.; et al. IgG antibody responses to the Aedes albopictus 34k2 salivary protein as novel candidate marker of human exposure to the tiger mosquito. Front. Cell. Infect. Microbiol. 2020, 10, 377. [Google Scholar] [CrossRef] [PubMed]
- Mulot, M.; Monsion, B.; Boissinot, S.; Rastegar, M.; Meyer, S.; Bochet, N.; Brault, V. Transmission of Turnip yellows virus by Myzus persiace is reduced by feeding aphids on double-stranded RNA targeting the Ephrin receptor protein. Front Microbiol. 2018, 9, 457. [Google Scholar] [CrossRef] [PubMed]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.H.; Karlen, Y.; Bakker, O.; van den Hoff, M.J.B.; and Moorman, A.F.M. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemeans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for publication of Quantitaive real-time PCR experiments. Clin Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using Real-Time Quantitative PCR and the 2−ΔΔCT method. Methods. 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Sarria, E.; Cid, M.; Garzo, E.; Fereres, A. Excel workbook for automatic parameter clculation of EPG data. Comput. Electron. Agric. 2009, 67, 35–42. [Google Scholar] [CrossRef]
- Backus, E.A.; Cline, A.R.; Ellerseick, M.R.; Serrano, M.S. Lygus hesperus (Hemiptera: Miridae) feeding on cotton: New methods and parameters for analysis of nonsequential electrical penetration graph data. Ann. Entomol. Soc. Am. 2007, 100, 296–310. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deshoux, M.; Monsion, B.; Pichon, E.; Jiménez, J.; Moreno, A.; Cayrol, B.; Thébaud, G.; Mugford, S.T.; Hogenhout, S.A.; Blanc, S.; et al. Role of Acrostyle Cuticular Proteins in the Retention of an Aphid Salivary Effector. Int. J. Mol. Sci. 2022, 23, 15337. https://doi.org/10.3390/ijms232315337
Deshoux M, Monsion B, Pichon E, Jiménez J, Moreno A, Cayrol B, Thébaud G, Mugford ST, Hogenhout SA, Blanc S, et al. Role of Acrostyle Cuticular Proteins in the Retention of an Aphid Salivary Effector. International Journal of Molecular Sciences. 2022; 23(23):15337. https://doi.org/10.3390/ijms232315337
Chicago/Turabian StyleDeshoux, Maëlle, Baptiste Monsion, Elodie Pichon, Jaime Jiménez, Aránzazu Moreno, Bastien Cayrol, Gaël Thébaud, Sam T. Mugford, Saskia A. Hogenhout, Stéphane Blanc, and et al. 2022. "Role of Acrostyle Cuticular Proteins in the Retention of an Aphid Salivary Effector" International Journal of Molecular Sciences 23, no. 23: 15337. https://doi.org/10.3390/ijms232315337
APA StyleDeshoux, M., Monsion, B., Pichon, E., Jiménez, J., Moreno, A., Cayrol, B., Thébaud, G., Mugford, S. T., Hogenhout, S. A., Blanc, S., Fereres, A., & Uzest, M. (2022). Role of Acrostyle Cuticular Proteins in the Retention of an Aphid Salivary Effector. International Journal of Molecular Sciences, 23(23), 15337. https://doi.org/10.3390/ijms232315337






