The Phosphoserine Phosphatase Alters the Free Amino Acid Compositions and Fecundity in Cyrtorhinus lividipennis Reuter
Abstract
:1. Introduction
2. Results
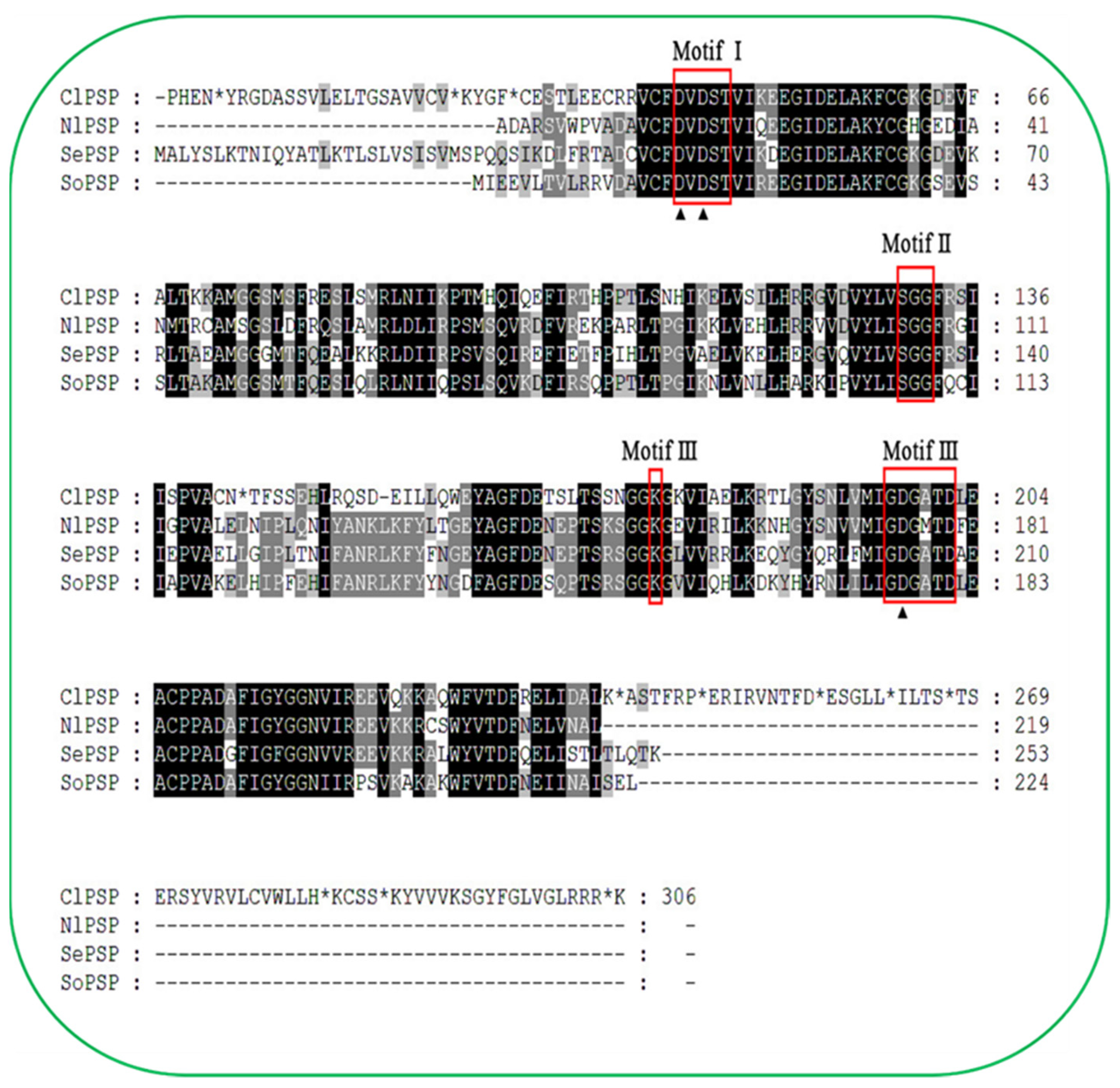
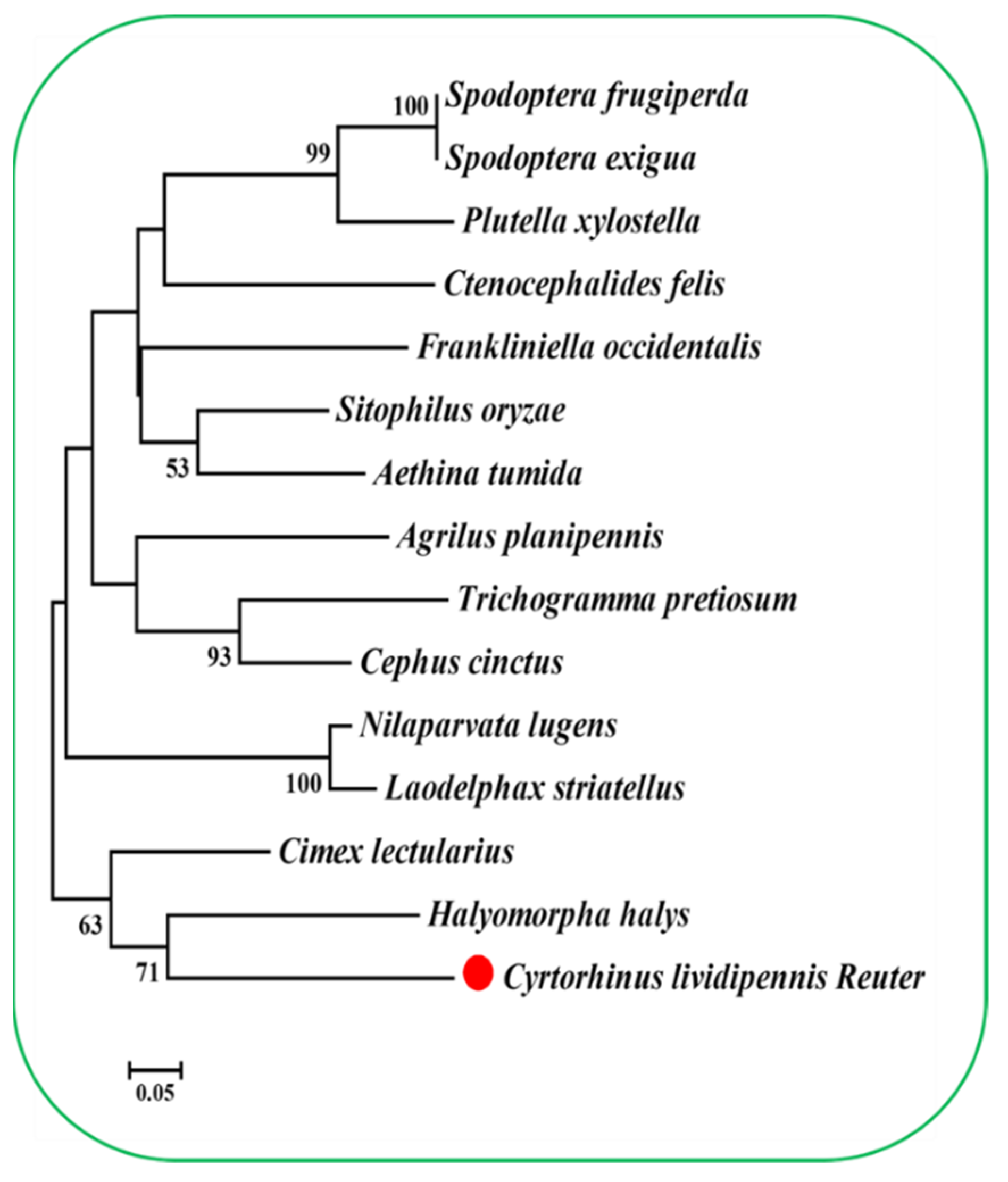
2.1. Conserved Domain, Cloning, and Sequencing Analysis of PSP in C. lividipennis
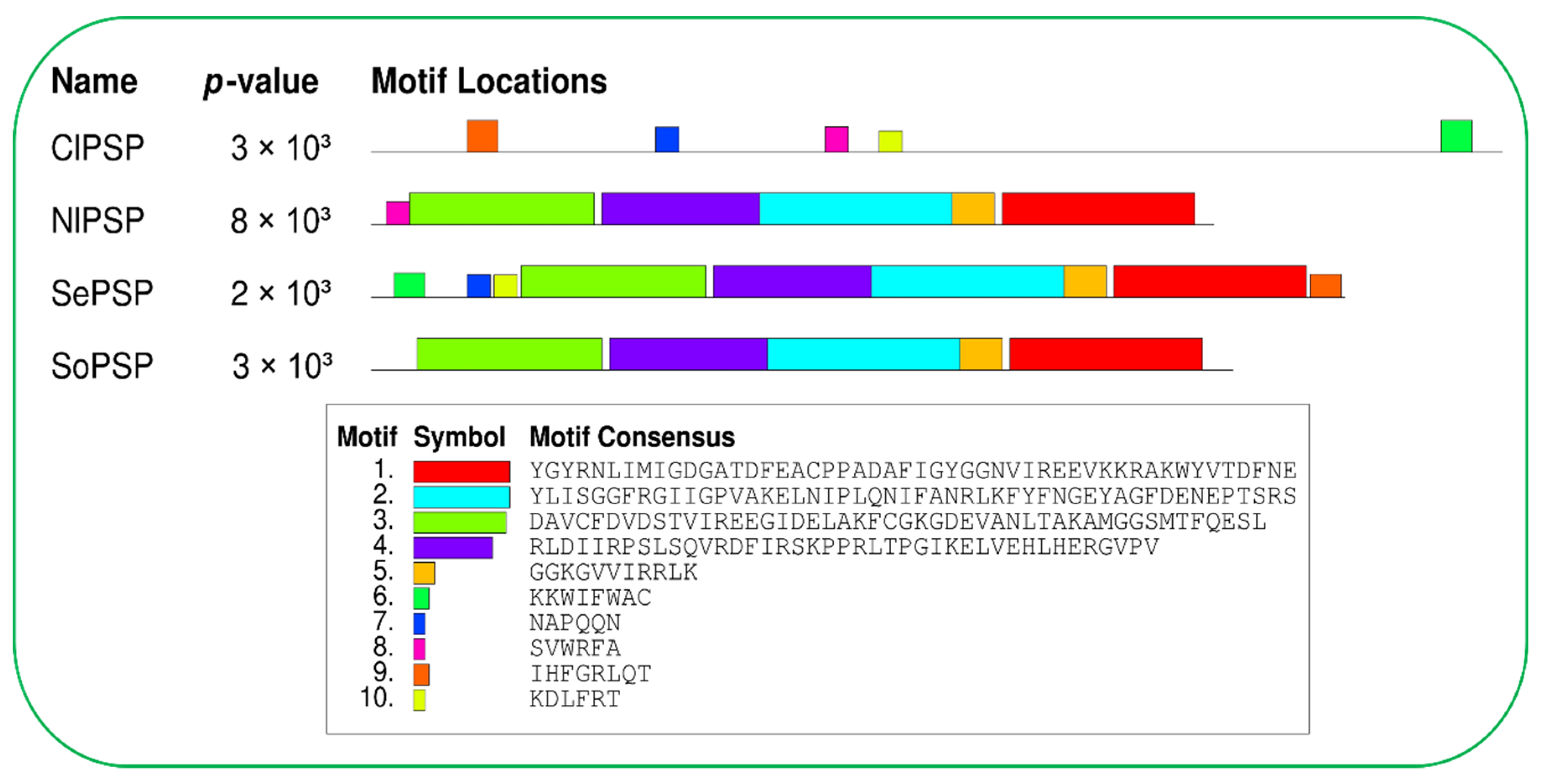
2.2. Motifs and Interactive Protein Analysis
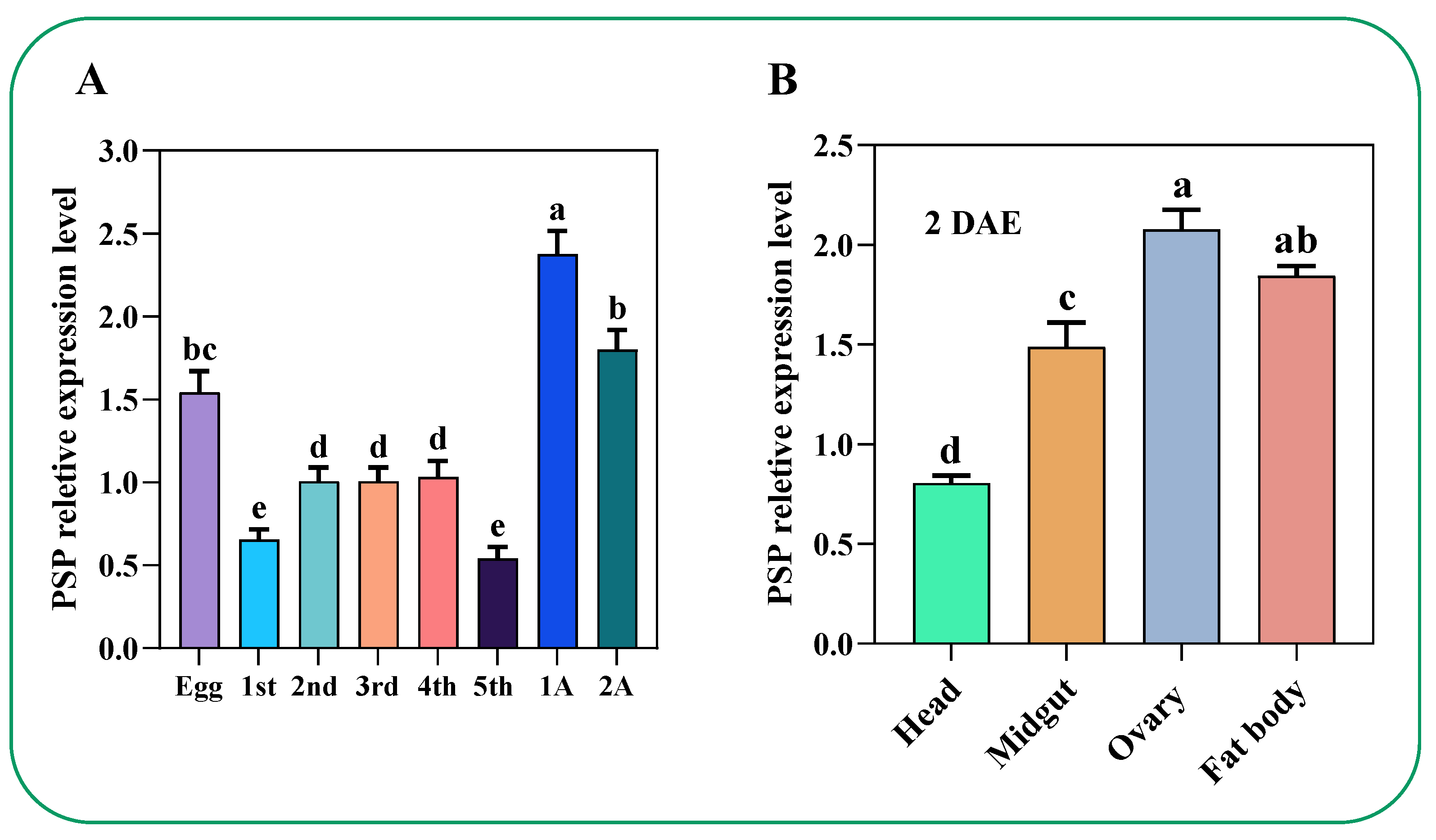
2.3. Temporal and Spatial Expression Profile of PSP Gene in C. lividipennis
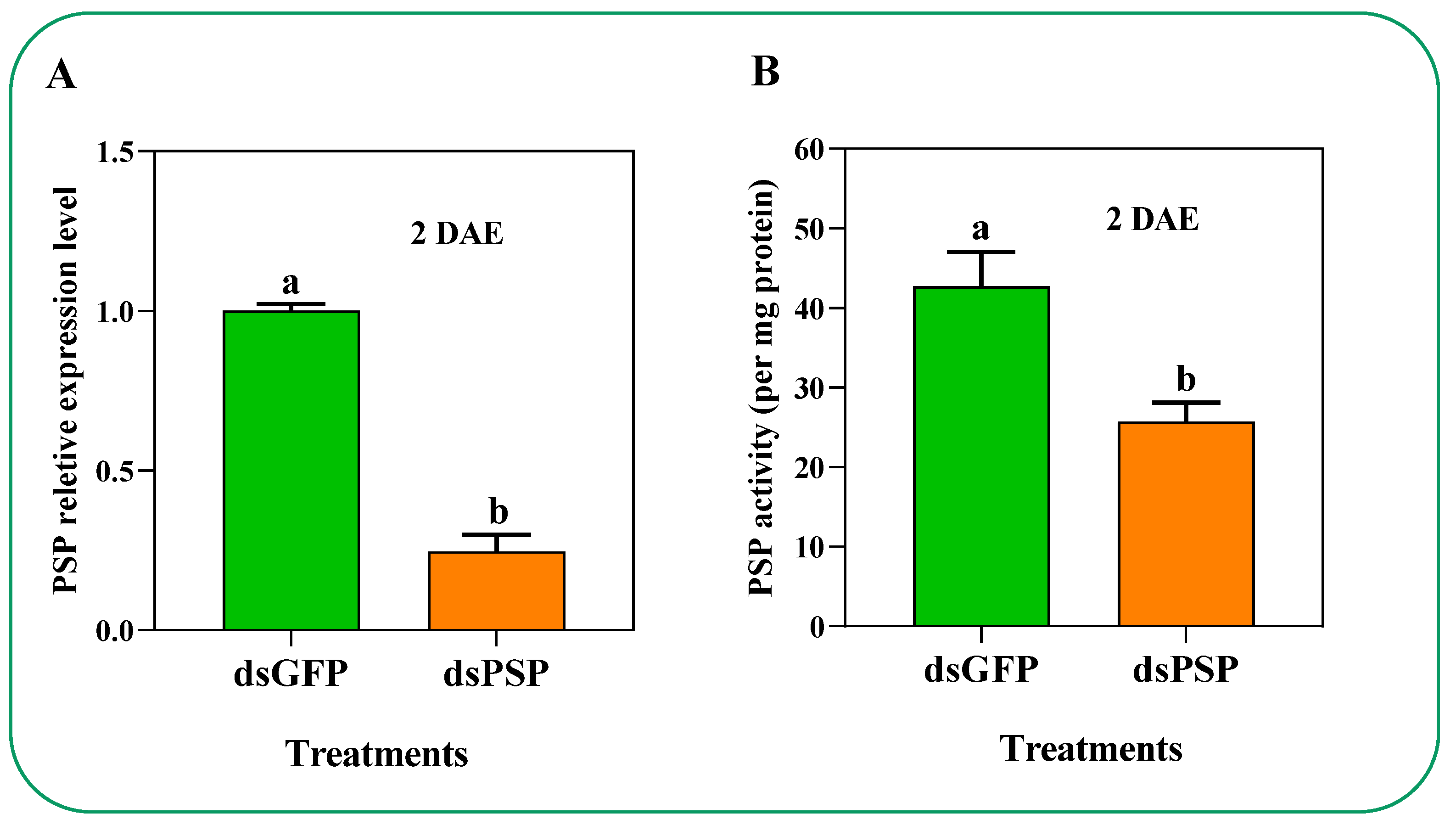
2.4. Effect of dsPSP Treatment on the PSP Expression, Activity, and Free Amino Acid Compositions in C. lividipennis Female Adult
2.5. Silencing of PSP Gene Affects Total Protein, Vg Expression, Vg Protein Synthesis, and Body Weight in C. lividipennis Female Adult
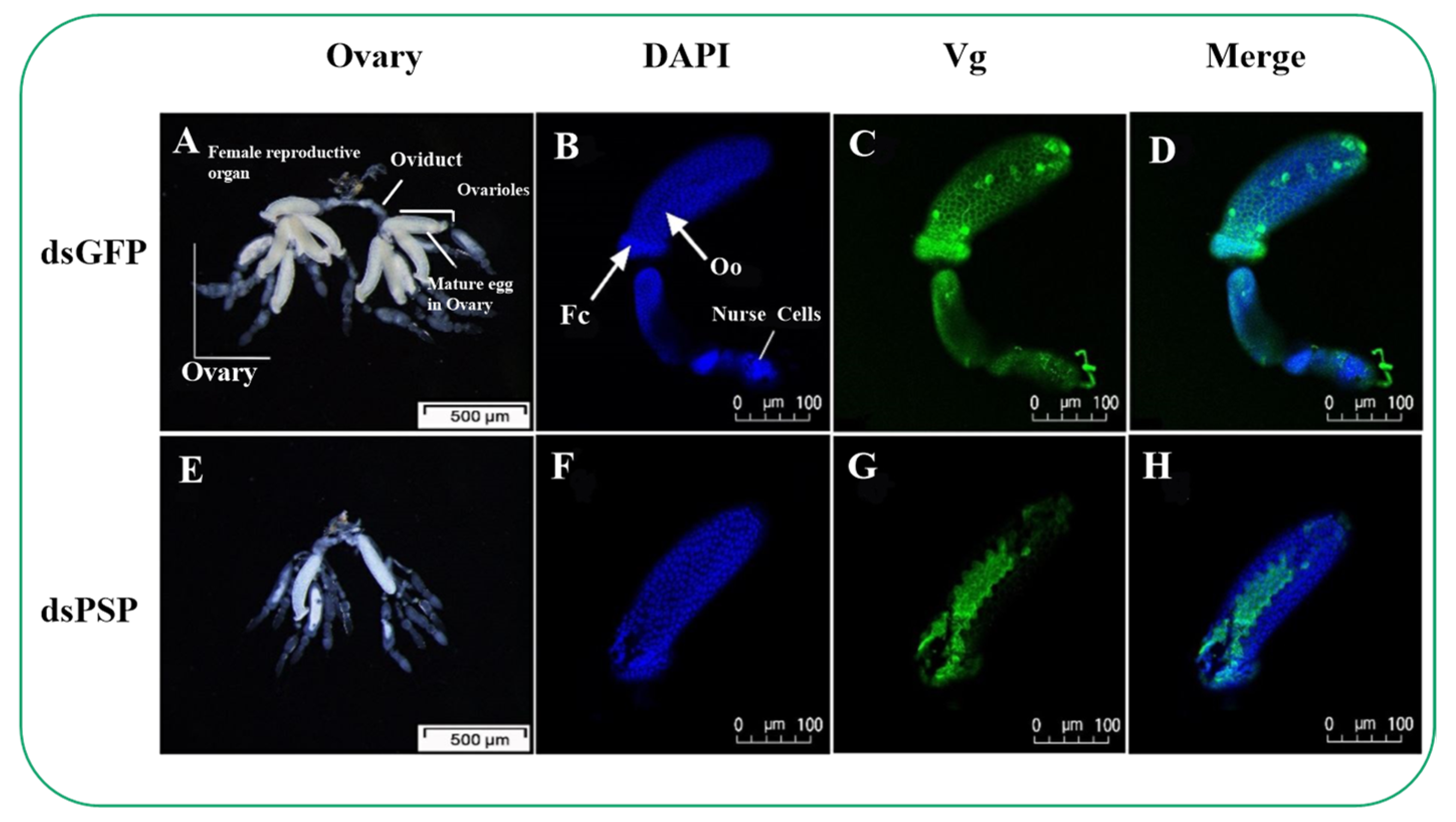
2.6. Silencing of PSP Gene Blocks Ovarian Development and Ovariole Vg Uptake
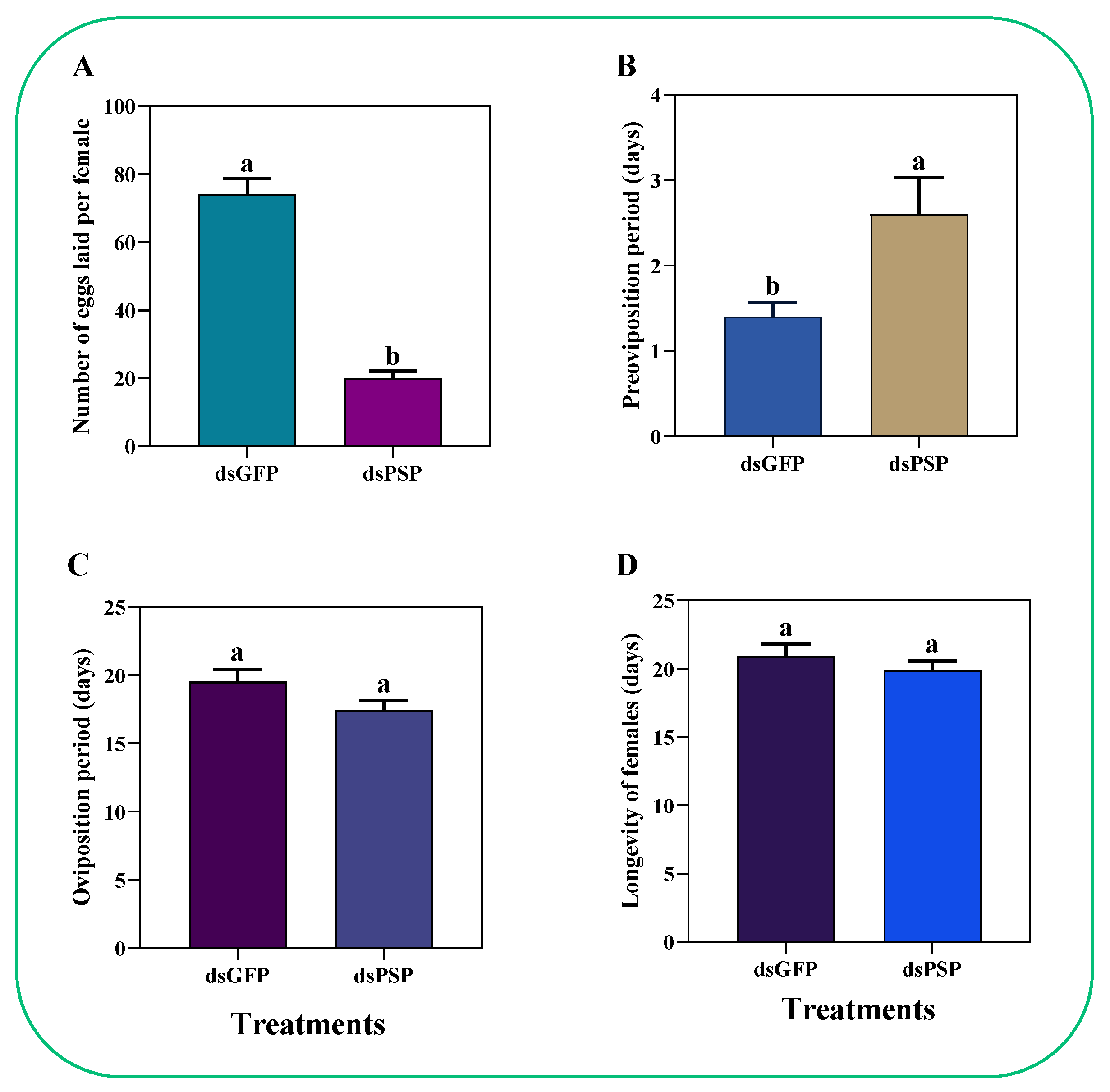
2.7. Silencing of the PSP Gene Led to Reduced Fecundity
3. Discussion
3.1. PSP Regulates the Physiological Development
3.2. Potential Role of ClPSP in Physiological Parameters of C. lividipennis
3.3. ClPSP Regulates the Reproductive Biology of C. lividipennis
4. Materials and Methods
4.1. Rice Variety and Insects
4.2. Cyrtorhinus lividipennis Samples Preparation
4.3. Total RNA Isolation, Complementary DNA (cDNA) Synthesis, and Real-Time Polymerase Chain Reaction (qRT-PCR)
4.4. ClPSP Cloning, Multiple Sequence Alignment, and Evolutionary Analysis
4.5. Conserved Motifs, Interactive Protein, and Gene Ontology
4.6. PSP Enzyme Activity Assays
4.7. dsPSP Synthesis and PSP Gene Silencing
4.8. Free Amino Acid Analysis
4.9. Female Body Mass and Isolation of Ovaries
4.10. Western Blot Analysis
4.11. Immunofluorescence Microscopy
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Burroughs, A.M.; Allen, K.N.; Dunaway-Mariano, D.; Aravind, L. Evolutionary Genomics of the HAD Superfamily: Understanding the Structural Adaptations and Catalytic Diversity in a Superfamily of Phosphoesterases and Allied Enzymes. J. Mol. Biol. 2006, 361, 1003–1034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, T.-Y.; Kim, Y.-S.; Oh, B.-H.; Woo, E. Identification of a novel ligand binding site in phosphoserine phosphatase from the hyperthermophilic archaeon Thermococcus onnurineus. Proteins: Struct. Funct. Bioinform. 2013, 81, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Dunaway-Mariano, D.; Allen, K.N. HAD Superfamily Phosphotransferase Substrate Diversification: Structure and Function Analysis of HAD Subclass IIB Sugar Phosphatase BT4131. Biochemistry 2005, 44, 8684–8696. [Google Scholar] [CrossRef] [PubMed]
- Jaeken, J.; Detheux, M.; Fryns, J.P.; Collet, J.F.; Alliet, P.; Van Schaftingen, E. Phosphoserine phosphatase deficiency in a patient with Williams syndrome. J. Med Genet. 1997, 34, 594–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tabatabaie, L.; Klomp, L.; Berger, R.; De Koning, T.J. L-serine synthesis in the central nervous system: A review on serine deficiency disorders. Mol. Genet. Metab. 2010, 99, 256–262. [Google Scholar] [CrossRef]
- Collet, J.F.; Stroobant, V.; Van Schaftingen, E.V. Mechanistic studies of phosphoserine phosphatase, an enzyme related to P-type ATPases. J. Biol. Chem. 1999, 274, 33985–33990. [Google Scholar] [CrossRef] [Green Version]
- Bachelor, M.A.; Lu, Y.; Owens, D.M. L-3-Phosphoserine phosphatase (PSPH) regulates cutaneous squamous cell carcinoma proliferation independent of L-serine biosynthesis. J. Dermatol. Sci. 2011, 63, 164–172. [Google Scholar] [CrossRef] [Green Version]
- Lowry, M.; Hall, D.E.; Hall, M.S.; Brosnan, J.T. Renal metabolism of amino acids in vivo: Studies on serine and glycine fluxes. Am. J. Physiol. Physiol. 1987, 252, F304–F309. [Google Scholar] [CrossRef]
- Snell, K.; Fell, D.A. Metabolic control analysis of mammalian serine metabolism. Adv. Enzym. Regul. 1990, 30, 13–32. [Google Scholar] [CrossRef]
- Fell, D.A.; Snell, K. Control analysis of mammalian serine biosynthesis. Feedback inhibition on the final step. Biochem. J. 1988, 256, 97–101. [Google Scholar] [CrossRef]
- Wang, W.; Cho, H.S.; Kim, R.; Jancarik, J.; Yokota, H.; Nguyen, H.H.; Grigoriev, I.; Wemmer, D.E.; Kim, S.-H. Structural Characterization of the Reaction Pathway in Phosphoserine Phosphatase: Crystallographic “snapshots” of Intermediate States. J. Mol. Biol. 2002, 319, 421–431. [Google Scholar] [CrossRef] [PubMed]
- Bainbridge, B.; Verma, R.K.; Eastman, C.; Yehia, B.; Rivera, M.; Moffatt, C.; Bhattacharyya, I.; Lamont, R.J.; Kesavalu, L. Role of Porphyromonas gingivalis phosphoserine phosphatase enzyme SerB in inflammation, immune response, and induction of alveolar bone resorption in rats. Infect. Immun. 2010, 78, 4560–4569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hawkinson, J.E.; Acosta-Burruel, M.; Ta, N.D.; Wood, P.L. Novel phosphoserine phosphatase inhibitors. Eur. J. Pharmacol. 1997, 337, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.-K.; Ko, Y.; Yu, K.R.; Kim, J.H.; Lee, J.-Y.; Chae, C.H.; Ji, S.; Kim, C.H.; Lee, H.K.; Choi, E.B.; et al. Identification of 3-acyl-2-phenylamino-1,4-dihydroquinolin-4-one derivatives as inhibitors of the phosphatase SerB653 in Porphyromonas gingivalis, implicated in periodontitis. Bioorganic Med. Chem. Lett. 2012, 22, 2084–2088. [Google Scholar] [CrossRef]
- Suzuki, C.; Tanigawa, M.; Soma, H.; Nagata, Y.J.J.o.i.b. D-Serine metabolism and role in the silkworm, Bombyx mori. J. Insect Biotechnol. Sericol. 2012, 81, 1_021–1_027. [Google Scholar]
- Suzuki, C.; Tanigawa, M.; Tanaka, H.; Horiike, K.; Kanekatsu, R.; Tojo, M.; Nagata, Y. Effect of d-serine on spermatogenesis and extracellular signal-regulated protein kinase (ERK) phosphorylation in the testis of the silkworm, Bombyx mori. J. Insect Physiol. 2014, 67, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Tanigawa, M.; Suzuki, C.; Niwano, K.; Kanekatsu, R.; Tanaka, H.; Horiike, K.; Hamase, K.; Nagata, Y. Participation of d -serine in the development and reproduction of the silkworm Bombyx mori. J. Insect Physiol. 2016, 87, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Lou, Y.G.; Zhang, G.R.; Zhang, W.Q.; Hu, Y.; Zhang, J. Biological control of rice insect pests in China. Biol. Control. 2013, 67, 8–20. [Google Scholar] [CrossRef]
- Sigsgaard, L. Early season natural control of the brown planthopper, Nilaparvata lugens: The contribution and interaction of two spider species and a predatory bug. Bull. Entomol. Res. 2007, 97, 533. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Jiang, L.; Zheng, S.; Chen, Y.; Zhang, J.Y.; Stanley, D.; Miao, H.; Ge, L.Q. Silencing of a putative alanine aminotransferase (ALT) gene influences free amino acid composition in hemolymph and fecundity of the predatory bug, Cyrtorhinus lividipennis Reuter. Arch. Insect Biochem. Physiol. 2021, 108, e21836. [Google Scholar] [CrossRef]
- Zhu, H.; Zheng, S.; Xu, J.; Wu, Q.; Song, Q.; Ge, L. The amino acid-mediated TOR pathway regulates reproductive potential and population growth in Cyrtorhinus lividipennis reuter (Hemiptera: Miridae). Front. Physiol. 2020, 11, 617237. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Heong, K.; Hu, C. Effects of various non-rice hosts on the growth, reproduction and predation of mirid bug, Cyrtorhinus lividipennis. Chin. J. Rice Sci. 1996, 10, 18–22. [Google Scholar]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, w202–w208. [Google Scholar] [PubMed]
- Yu, C.S.; Cheng, C.W.; Su, W.C.; Chang, K.C.; Huang, S.W.; Hwang, J.K.; Lu, C.H. CELLO2GO: A web server for protein subCELlular LOcalization prediction with functional gene ontology annotation. Plos One 2014, 9, e99368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, K.; Mohri, S.; Furuya, S. D-3-phosphoglycerate dehydrogenase from the silkworm Bombyx mori: Identification, functional characterization, and expression. Arch. Insect Biochem. Physiol. 2021, 106, e21751. [Google Scholar]
- Hu, C.-A.A.; Khalil, S.; Zhaorigetu, S.; Liu, Z.; Tyler, M.; Wan, G.; Valle, D. Human Δ1-pyrroline-5-carboxylate synthase: Function and regulation. Amino Acids 2008, 35, 665–672. [Google Scholar] [CrossRef] [Green Version]
- Nakano, I.; Dougherty, J.D.; Kim, K.; Klement, I.; Geschwind, D.H.; Kornblum, H.I. Phosphoserine Phosphatase Is Expressed in the Neural Stem Cell Niche and Regulates Neural Stem and Progenitor Cell Proliferation. Stem Cells 2007, 25, 1975–1984. [Google Scholar] [CrossRef]
- Chiba, Y.; Oshima, K.; Arai, H.; Ishii, M.; Igarashi, Y. Discovery and Analysis of Cofactor-dependent Phosphoglycerate Mutase Homologs as Novel Phosphoserine Phosphatases in Hydrogenobacter thermophilus. J. Biol. Chem. 2012, 287, 11934–11941. [Google Scholar] [CrossRef] [Green Version]
- Kumari, P.; Babuta, M.; Bhattacharya, A.; Gourinath, S. Structural and functional characterisation of phosphoserine phosphatase, that plays critical role in the oxidative stress response in the parasite Entamoeba histolytica. J. Struct. Biol. 2019, 206, 254–266. [Google Scholar] [CrossRef]
- Grant, G.A. Regulatory Mechanism of Mycobacterium tuberculosis Phosphoserine Phosphatase SerB2. Biochemistry 2017, 56, 6481–6490. [Google Scholar] [CrossRef] [PubMed]
- Haque, M.R.; Hirowatari, A.; Saruta, F.; Furuya, S.; Yamamoto, K. Molecular survey of the phosphoserine phosphatase involved in L-serine synthesis by silkworms (Bombyx mori). Insect Mol. Biol. 2019, 29, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Veeranna, V.; Shetty, T. Phosphoserine phosphatase of human brain: Partial purification, characterization, regional distribution, and effect of certain modulators including psychoactive drugs. Neurochem. Res. 1990, 15, 1203–12010. [Google Scholar] [CrossRef] [PubMed]
- Kuang, W.; Yan, C.; Zhan, Z.; Guan, L.; Wang, J.; Chen, J.; Li, J.; Ma, G.; Zhou, X.; Jin, L.J.V.J. Transcriptional responses of Daphnis nerii larval midgut to oral infection by Daphnis nerii cypovirus-23. Virol. J. 2021, 18, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Sonn, J.Y.; Lee, J.; Sung, M.K.; Ri, H.; Choi, J.K.; Lim, C.; Choe, J.J.P.o.t.N.A.o.S. Serine metabolism in the brain regulates starvation-induced sleep suppression in Drosophila melanogaster. Biol. Sci. 2018, 115, 7129–7134. [Google Scholar] [CrossRef] [Green Version]
- Chen, P. Amino acid and protein metabolism in insect development. In Advances in Insect Physiology; Elsevier: Amsterdam, The Netherlands, 1966; Volume 3, pp. 13–53. [Google Scholar]
- Lu, W.; Xu, Q.; Zhu, J.; Liu, C.; Ge, L.Q.; Yang, G.; Liu, F. Inductions of reproduction and population growth in the generalist predator Cyrtorhinus lividipennis (Hemiptera: Miridae) exposed to sub-lethal concentrations of insecticides. Pest Manag. Sci. 2017, 73, 1709–1718. [Google Scholar] [CrossRef] [PubMed]
- North, M.J.; Lockwood, B.C. 5-amino acid and protein metabolism. Biochem. Mol. Biol. Parasites 1995, 67–88. [Google Scholar] [CrossRef]
- Haque Lehmann, M. Diverse roles of phosphatidate phosphatases in insect development and metabolism. Insect Biochem. Mol. Biology 2021, 133, 103469. [Google Scholar] [CrossRef]
- Lou, Y.G.; Cheng, J.A. Role of rice volatiles in the foraging behaviour of the predator Cyrtorhinus lividipennis for the rice brown planthopper Nilaparvata Lugens. Biol. Control. 2003, 48, 73–86. [Google Scholar] [CrossRef]
- Bentur, J.S.; Kalode, M.B. Technique for rearing the predatory mirid bug Cyrtorhinus lividipennis (Reut) on Corcyra eggs. Curr. Sci. 1985, 54, 513–514. [Google Scholar]
- Pomari-Fernandes, A.; Bueno, A.; Queiroz, A.; De Bortoli, S. Biological parameters and parasitism capacity of Telenomus remus Nixon (Hymenoptera: Platygastridae) reared on natural and factitious hosts for successive generations. Afr. J. Agric. Res. 2015, 10, 3225–3233. [Google Scholar]
- Heong, K.L.; Bleih, S.; Amor, A.L. Predation of Cyrtorhinus lividipennis Reuter on eggs of the green leafhopper and brown planthopper in rice. Popul. Ecol. 1990, 32, 2–255. [Google Scholar] [CrossRef]
- Cortesero, M.A.; Stapel, J.O.; Lewis, W.J. Understanding and manipulating plant attributes to enhance biological control. Biol. Control. 2000, 17, 35–49. [Google Scholar] [CrossRef] [Green Version]
- Walde, S.J.J.E. How quality of host plant affects a predator-prey interaction in biological control. Ecology 1995, 76, 1206–1219. [Google Scholar] [CrossRef]
- Shah, F.A.; Wang, C.S.; Butt, T.M. Nutrition influences growth and virulence of the insect-pathogenic fungus Metarhizium anisopliae. FEMS Microbiol. Lett. 2005, 251, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Grandison, R.C.; Piper, M.D.W.; Partridge, L. Amino-acid imbalance explains extension of lifespan by dietary restriction in Drosophila. Nature 2009, 462, 1061–1064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, T.; Zhang, G.; Zeng, F.; Mao, J.; Liang, H.; Liu, F. Molecular Cloning of the Vitellogenin Gene and the Effects of Vitellogenin Protein Expression on the Physiology of Harmonia axyridis (Coleoptera: Coccinellidae). Sci. Rep. 2017, 7, 13926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Wang, Y.; Liu, Y.; Zhao, M.; Jin, J.; Zhou, Z.; Guo, J. Identification and Expression Patterns of Three Vitellogenin Genes and Their Roles in Reproduction of the Alligatorweed Flea Beetle Agasicles hygrophila (Coleoptera: Chrysomelidae). Front. Physiol. 2019, 10, 368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasegawa, Y.; Ogawa, T.; Nakanishi, M.; Tojo, M.; Okumura, M.; Nakagaki, M.; Nagata, Y. Effects of O-phospho-L-serine on the larval growth and development in the silkworm, Bombyx mori. J. Insect Biotechnol. Sericology 2009, 78, 2_75–72_79. [Google Scholar]
- Ahmad, S.; Zhu, H.; Chen, Y.; Xi, C.; Shah, A.Z.; Ge, L. Comprehensive Bioinformatics and Expression Analysis of the TLP Gene Family Revealed Its Role in Regulating the Response of Oryza sativa to Nilaparvata lugens, Laodelphax striatellus, and Jinggangmycin. Agronomy 2022, 12, 1297. [Google Scholar]
- Ahmad, S.; Chen, Y.; Shah, A.Z.; Wang, H.; Xi, C.; Zhu, H.; Ge, L. The Homeodomain-Leucine Zipper Genes Family Regulates the Jinggangmycin Mediated Immune Response of Oryza sativa to Nilaparvata lugens, and Laodelphax striatellus. Bioengineering 2022, 9, 398. [Google Scholar] [CrossRef] [PubMed]
- Mujtaba, M.; Wang, D.; Carvalho, L.B.; Oliveira, J.L.; Pereira, A.D.E.S.; Sharif, R.; Jogaiah, S.; Paidi, M.K.; Wang, L.; Ali, Q.; et al. Nanocarrier-Mediated Delivery of miRNA, RNAi, and CRISPR-Cas for Plant Protection: Current Trends and Future Directions. ACS Agric. Sci. Technol. 2021, 1, 417–435. [Google Scholar] [CrossRef]
- Ahmad, S.; Chen, Y.; Zhang, J.; Stanley, D.; Song, Q.; Ge, L.Q. Octopamine signaling is involved in the female postmating state in Nilaparvata lugens Stål (Hemiptera: Delphacidae). Arch. Insect Biochem. Physiol. 2021, 107, e21825. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jiang, L.; Ahamd, S.; Chen, Y.; Zhang, J.; Stanley, D.; Miao, H.; Ge, L. The octopamine receptor, OA2B2, modulates stress resistance and reproduction in Nilaparvata lugens Stål (Hemiptera: Delphacidae). Insect Mol. Biol. 2022, 31, 33–48. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, D.; Yao, Q.; Zhang, J.; Dong, X.; Tian, H.; Chen, J.; Zhang, W. Feeding-based RNA interference of a trehalose phosphate synthase gene in the brown planthopper, Nilaparvata lugens. Insect Mol. Biol. 2010, 19, 777–786. [Google Scholar] [CrossRef]
- Xu, H.-J.; Xue, J.; Lu, B.; Zhang, X.-C.; Zhuo, J.-C.; He, S.-F.; Ma, X.-F.; Jiang, Y.-Q.; Fan, H.-W.; Xu, J.-Y.; et al. Two insulin receptors determine alternative wing morphs in planthoppers. Nature 2015, 519, 464–467. [Google Scholar] [CrossRef] [PubMed]
- Wan, P.-J.; Fu, K.-Y.; Lü, F.-G.; Guo, W.-C.; Li, G.Q. Knockdown of a putative alanine aminotransferase gene affects amino acid content and flight capacity in the Colorado potato beetle Leptinotarsa decemlineata. Amino Acids 2015, 47, 1445–1454. [Google Scholar] [CrossRef] [PubMed]
- Ge, L.; Zhou, Z.; Sun, K.; Huang, B.; Stanley, D.; Song, Q.S. The antibiotic jinggangmycin increases brown planthopper (BPH) fecundity by enhancing rice plant sugar concentrations and BPH insulin-like signaling. Chemosphere 2020, 249, 126463. [Google Scholar] [CrossRef] [PubMed]
- Ge, L.Q.; Jiang, L.; Zheng, S.; Zhou, Y.; Wu, Q.; Liu, F. Frizzled 2 functions in the regulation of TOR-mediated embryonic development and fecundity in Cyrtorhinus lividipennis Reuter. Front. Physiol. 2020, 11, 1159. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Chen, X.; Liu, W.-T.; Zhou, Q. TOR pathway-mediated juvenile hormone synthesis regulates nutrient-dependent female reproduction in Nilaparvata lugens (Stål). Int. J. Mol. Sci. 2016, 17, 438. [Google Scholar] [PubMed]
- Tang, Q.y.; Zhang, C.X. Data Processing System (DPS) software with experimental design, statistical analysis and data mining developed for use in entomological research. Insect Sci. 2012, 20, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Qi, Y.; Yang, Z.; Cheng, L.; Sharif, R.; Raza, A.; Chen, P.; Hou, D.; Li, Y. Exploring the Agrobacterium-mediated transformation with CRISPR/Cas9 in cucumber (Cucumis sativus L.). Mol. Biol. Rep. 2022, 49, 11481–11490. [Google Scholar] [CrossRef] [PubMed]
- Champion de Crespigny, F.E.; Wedell, N. The impact of anaesthetic technique on survival and fertility in Drosophila. Physiol. Entomol. 2008, 33, 310–315. [Google Scholar] [CrossRef]


| Amino Acids | Concentration (μmol/mL) | ||
|---|---|---|---|
| PBS | dsGFP | dsPSP | |
| Asparagine (Asp) | 15.05 ± 1.52 a | 15.77 ± 1.66 a | 14.32 ± 1.09 a |
| Glutamicacid (Glu) | 18.41 ± 1.36 a | 18.62 ± 1.69 a | 17.81 ± 1.75 b |
| Serine (Ser) * | 14.65 ± 1.42 a | 14.47 ± 0.94 a | 8.14 ± 0.95 b |
| Arginine (Arg) * | 18.87 ± 1.65 a | 18.47 ± 1.46 a | 13.46 ± 1.26 b |
| Glycine (Gly) | 10.09 ± 0.78 a | 9.86 ± 0.74 a | 9.76 ± 0.79 a |
| Proline (Pro) | 11.60 ± 1.02 a | 10.14 ± 0.95 a | 12.71 ± 0.93 a |
| Alanine (Ala) | 20.98 ± 1.89 a | 21.74 ± 2.05 a | 19.39 ± 1.84 a |
| Threonine (Thr) * | 8.68 ± 0.97 a | 8.72 ± 1.02 a | 11.23 ± 0.92 b |
| Valine (Val) | 0.46 ± 0.036 b | 0.43 ± 0.024 b | 1.57 ± 0.13 a |
| Methionine (Met) | 11.34 ± 1.12 a | 11.98 ± 1.07 a | 9.85 ± 0.92 a |
| Cystine (Cys) * | 56.53 ± 3.12 b | 56.67 ± 3.56 b | 71.27 ± 4.39 a |
| Isoleucine (Ile) | 10.41 ± 1.16 a | 10.76 ± 0.96 a | 9.73 ± 0.82 a |
| Leucine (Leu) | 15.05 ± 1.29 b | 16.00 ± 1.03 b | 16.24 ± 1.34 a |
| Phenylalanine (Phe) | 10.83 ± 0.95 a | 10.51 ± 1.12 a | 9.48 ± 0.87 a |
| Histidine (His) | 5.83 ± 0.36 a | 4.73 ± 0.23 a | 4.94 ± 0.47 a |
| Lysine (Lys) | 10.16 ± 0.93 a | 10.03 ± 0.85 a | 8.82 ± 0.85 a |
| Tyrosine (Tyr) * | 8.68 ± 0.78 b | 8.39 ± 0.88 b | 12.00 ± 1.09 a |
| Total | 246.4 ± 13.43 a | 247.8 ± 11.57 a | 249.5 ± 12.53 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmad, S.; Zhang, J.; Wang, H.; Zhu, H.; Dong, Q.; Zong, S.; Wang, T.; Chen, Y.; Ge, L. The Phosphoserine Phosphatase Alters the Free Amino Acid Compositions and Fecundity in Cyrtorhinus lividipennis Reuter. Int. J. Mol. Sci. 2022, 23, 15283. https://doi.org/10.3390/ijms232315283
Ahmad S, Zhang J, Wang H, Zhu H, Dong Q, Zong S, Wang T, Chen Y, Ge L. The Phosphoserine Phosphatase Alters the Free Amino Acid Compositions and Fecundity in Cyrtorhinus lividipennis Reuter. International Journal of Molecular Sciences. 2022; 23(23):15283. https://doi.org/10.3390/ijms232315283
Chicago/Turabian StyleAhmad, Sheraz, Jieyu Zhang, Huaiqi Wang, Haowen Zhu, Qiaoqiao Dong, Suman Zong, Tingting Wang, Yu Chen, and Linquan Ge. 2022. "The Phosphoserine Phosphatase Alters the Free Amino Acid Compositions and Fecundity in Cyrtorhinus lividipennis Reuter" International Journal of Molecular Sciences 23, no. 23: 15283. https://doi.org/10.3390/ijms232315283
APA StyleAhmad, S., Zhang, J., Wang, H., Zhu, H., Dong, Q., Zong, S., Wang, T., Chen, Y., & Ge, L. (2022). The Phosphoserine Phosphatase Alters the Free Amino Acid Compositions and Fecundity in Cyrtorhinus lividipennis Reuter. International Journal of Molecular Sciences, 23(23), 15283. https://doi.org/10.3390/ijms232315283







